Background: Disordered miR-26b expression has been linked to the development of many diseases induced by hepatitis B virus (HBV).
Results: miR-26b inhibits HBV replication, and HBV infection suppresses miR-26b expression.
Conclusion: miR-26b acts as a host restriction factor to attenuate HBV transcription and replication.
Significance: The functional interplay between HBV infection and miR-26b may influence the outcome of viral proliferation and virus-induced diseases.
Keywords: Hepatitis Virus, Host-Pathogen Interaction, Infectious Disease, MicroRNA (miRNA), Viral Transcription, CHORDC1, Hepatitis B Virus, Host Factor, MicroRNA-26b, Viral Infection
Abstract
Hepatitis B virus (HBV) causes acute and chronic hepatitis in humans, and HBV infection is a major threat to global health. HBV replication is regulated by a series of host factors, including microRNAs (miRNAs), which are highly conserved small noncoding RNAs that participate in a variety of physiological and pathological processes. Here, we report that a chemically synthesized mimic of miR-26b inhibited HBV antigen expression, transcription, and replication, whereas antisense knockdown of endogenous miR-26b enhanced HBV replication in HepG2 cells. Overexpression and knockdown experiments showed that miR-26b significantly decreased HBV enhancer/promoter activities. We identified the cysteine- and histidine-rich domain containing 1 (CHORDC1) as a novel host factor target of miR-26b. CHORDC1 protein but not mRNA was markedly decreased by miR-26b overexpression via base-pairing with complementary sequences in the 3′UTR of its mRNA. Overexpression and knockdown studies showed that CHORDC1 increased viral antigen expression, transcription, and replication by elevating HBV enhancer/promoter activities. Conversely, HBV infection suppressed miR-26b expression and increased CHORDC1 protein levels in human liver cells. Another mature miRNA of the hsa-miR-26 family, miR-26a, had a similar function as miR-26b in targeting CHORDC1 and affecting HBV production. These results suggest that suppression of miR-26b expression up-regulates its target gene CHORDC1, which increases HBV enhancer/promoter activities and promotes viral transcription, gene expression, and replication. Our study could provide new insights into miRNA expression and the persistence of HBV infection.
Introduction
Hepatitis B virus (HBV)2 chronically infects more than 350 million people worldwide, and HBV infection is a common cause of hepatic disease and liver cancer (1). HBV is the prototype virus of the Hepadnaviridae family, and it is classified into eight genotypes (A to H) based on sequence (2). This virus contains a capsid-associated and relaxed circular partially double-stranded DNA genome. During hepatocyte infection, HBV genomic DNA is transported into the nucleus and repaired to form a covalently closed circular DNA molecule, which serves as the template for viral RNA transcription (3). In the HBV life cycle, viral DNA synthesis occurs via reverse transcription of viral pregenomic RNA (pgRNA). HBV pgRNA is packaged together with viral polymerase by viral core protein and is subsequently converted into relaxed circular DNA by the polymerase (3, 4). Therefore, it is obvious that the level of HBV transcription from the promoters is an important determinant of viral biosynthesis. There are four overlapping open reading frames (ORFs) coding for seven viral proteins in the HBV genome, and viral transcription is controlled by four promoter elements (preS1, preS2, core, and X) (5). The two enhancer elements En1 and En2 are located in the upstream regulatory region of the X promoter and core promoter, respectively (designated as En1/Xp and En2/Cp). They can up-regulate the activities of all four viral promoters (6–8).
MicroRNAs (miRNAs) are small single-stranded RNAs (∼21 nucleotides) that typically mediate messenger RNA (mRNA) cleavage or translational repression through imperfect complementary pairing to the 3′-untranslated regions (UTRs) of the target mRNAs. They control many developmental and cellular processes in eukaryotic organisms (9, 10). Recent data indicate that host miRNAs can be involved in host-virus interactions and significantly affect the viral life cycle (11). HBV replication is enhanced after knocking down the key components of miRNA machinery, suggesting possible negative regulation of miRNAs on HBV replication (12). HBV can regulate cellular miRNA expression to create a favorable environment for its replication and survival. The highly expressed liver-specific miRNA-122 plays an important role in HBV replication; miR-122 is down-regulated by HBV infection, and miR-122 can repress HBV replication by directly binding to the viral target sequence (13). Loss of miR-122 induced by HBV infection can stimulate HBV replication through cyclin G1 (CCNG1)-modulated p53 activity (14). Several other miRNAs affect HBV progression by targeting multiple host factors or directly binding to HBV sequences (15). Despite inhibition of HBV infection by miRNA machinery, some miRNAs appear to have positive effects on HBV replication. The miR-372/373 promotes HBV expression by targeting the nuclear factor I/B (16), and miR-1 enhances HBV replication by targeting the host gene histone deacetylase 4 (17). Considerable progress has been made in the understanding of miRNAs and HBV replication, but the complex regulatory networks involving miRNAs have not been evaluated comprehensively in HBV replication.
Extensive studies are elucidating miR-26b function. Disordered miR-26b expression is correlated with many diseases, and this alteration may play a role in disease development. Significantly elevated miR-26b levels in the defined pathological areas of human postmortem brains can contribute to the neuronal pathology of Alzheimer disease (18). Respiratory syncytial virus infection of A549 cells induces miR-26b expression, and it may affect the antiviral host response (19). The miR-26b expression is frequently abnormal in tumors, indicating that miR-26b plays a significant role in cancer development (20). The miR-26b level is significantly up-regulated in bladder cancer (21), whereas it is down-regulated in liver cancer (22), colorectal cancer (23), and human breast cancer (24, 25). miR-26b could serve as a tumor suppressor in colorectal cancer and in glioma (23, 26). Chronic HBV infection is one of the main causes of hepatocellular carcinoma (HCC), and many miRNAs have essential roles in liver cancer progression by directly targeting a large number of crucial genes in HCC cells (27). Down-regulation of the miR-26b level in liver cancer (22) suggests that miR-26b might play an important role in HBV infection and pathogenesis. However, the functional interaction between miR-26b and HBV infection has not been investigated.
In this study, we investigated the regulation of HBV replication by miR-26b. We found that miR-26b inhibited HBV replication by attenuating viral enhancer/promoter activities. We identified cysteine- and histidine-rich domain containing 1 (CHORDC1) as a novel miR-26b target that could be involved in regulating HBV transcription and replication. HBV infection down-regulated miR-26b levels in human liver cells. Our results indicate a functional correlation between miR-26b and HBV replication. This new evidence supports a role for miRNAs in HBV infection and could provide a novel anti-HBV therapeutic strategy.
EXPERIMENTAL PROCEDURES
Plasmids, miRNAs, and Antibodies
The pHBV1.3 (serotype adw, genotype B) plasmid was generated from the HBV genome (GenBankTM accession number JN406371; www.ncbi.nlm.nih.gov), digested with EcoRI/SalI, and inserted into pBluescript II. HBV En1/Xp (958–1353 nucleotides) and En2/Cp (1401–1801 nucleotides) elements were amplified from the pHBV1.3 plasmid by PCR, digested with KpnI/HindIII, and inserted into the pGL3-Basic vector immediately preceding the firefly luciferase gene. Coding regions for GSK3B, CEBPG, STK39, CCND2, CHORDC1, and CCNG1 were generated by PCR amplification using HepG2 cDNA as template. The PCR products were digested with EcoRI/XhoI and cloned directly into the pCMV-Tag2B expression vector. CHORDC1 3′UTR-luc, CCND2-3′UTR-luc, and STK39-3′UTR-luc reporter plasmids were constructed by insertion of the 3′UTRs containing the predicted miR-26b target sequences downstream of the firefly luciferase ORF as described previously (28). The following primer pairs for the cloning of luciferase reporters are used: CHORDC1, 3′UTR-luc, 5′-TTAAAGCTTGAAGGAAGGCTATTACATTA-3′ (sense) and 5′-TAACTCGAGTAAAGTACAATATATAGTCA-3′ (antisense); CCND2, 3′UTR-luc, 5′-TTAAAGCTTTGACATTCCCATCACAACAT-3′ (sense) and 5′-TAACTCGAGACTGCTTCTGTATTCCTCAT-3′ (antisense); and STK39, 3′UTR-luc, 5′-CCCAAGCTTTACTTATAAAATTAAG-3′ (sense) and 5′-CCGCTCGAGTCTTGAACCTTAACAGC-3′ (antisense). Target sequence mutations were generated by site-directed mutagenesis using specific primers. The following primer pairs for the mutagenesis of luciferase reporters are used: CHORDC1, 3′UTR Mut1, 5′-TGTGTCCTATTGAACAAGAGGCTGGAAAGTAGCCC-3′ (sense) and 5′-CCAGCCTCTTGTTCAATAGGACACAACTATGGTTC-3′ (antisense); CHORDC1, 3′UTR Mut2, 5′-TTCTTACAGTTGAACAAAATATTTAAGGAAGAGAT-3′ (sense) and 5′-TAAATATTTTGTTCAACTGTAAGAACACAAATCCA-3′ (antisense); CHORDC1, 3′UTR Mut3, 5′-TGTTTACAACTTGAACAATTTTTAAATTATGTCAA-3′ (sense) and 5′-TAAAAATTGTTCAAGTTGTAAACAAATTCTAATTTG-3′ (antisense); CCND2, 3′UTR Mut, 5′-GTATTCAGCGTTGAACAATTTTTCTTCCTCTCCACTT-3′ (sense) and 5′-AGAAAAATTGTTCAACGCTGAATACAACTTTGCAA-3′ (antisense); STK39, 3′UTR Mut1, 5′-TATTAGCCAAAGAACAATTCTAGTTTTAAAACTGAC-3′ (sense) and 5′-AAACTAGAATTGTTCTTTGGCTAATAAATCTTAATT-3′ (antisense); and STK39, 3′UTR Mut2, 5′-TCTGAGTTTTTGAACAAATTTTGCAGAATACCCAGG-3′ (sense) and 5′-GCAAAATTTGTTCAAAAACTCAGATTCTGATATTT-3′ (antisense). All constructs were confirmed by DNA sequencing.
Chemically synthesized miRNA mimics (miR-26b, miR-26b Mut, miR-26a, miR-1, miR-122, and miR-Ctrl) and miRNA inhibitors (miR-26b inhibitor and miR-inhibitor-Ctrl) were purchased from RiboBio (Guangzhou, China). The short hairpin RNA (shRNA)-based RNAi expression vectors (shCHORDC1 and shCtrl) were generated by Genepharma (Shanghai, China). The specific target sequences in endogenous CHORDC1 mRNA by shRNAs are 5′-GGGAAACACATGTGGACTAAA-3′ (shRNA#1), 5′-GAGAAGGAATTTGATCAAAAT-3′ (shRNA#2), and 5′-GGACCCACATGGAGTACATAA-3′ (shRNA#3). shCtrl was used to express nontargeting control shRNA.
Antibody against CHORDC1 was purchased from Novus Biologicals; antibody against CCNG1 and ATF2 were purchased from ProteinTech Group (Wuhan, China); antibody against β-actin was purchased from CWBIO (Beijing, China), and antibody against lamin A/C was purchased from Santa Cruz Biotechnology.
Cell Culture and Transfection
Human hepatoma HepG2 cells and HepG2.2.15 cells (ATCC) were grown separately in DMEM. Hepatic L02 cells (ATCC) were grown in RPMI 1640 medium. The culture medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate. Cells were grown at 37 °C in 5% CO2. Cells were diluted at a ratio of 1:2 every 2 days to maintain the exponential growth phase. Cells were plated in 6-well plates (4 × 105 cells/well) or 24-well plates (1 × 105 cells/well) and grown to ∼80% confluence at the time of transfection. The miRNA mimics, inhibitors, and plasmids were transfected into cells at indicated concentrations using Lipofectamine 2000 reagent (Invitrogen).
Quantitative Real Time PCR Analysis
Quantitative real time PCR analysis was performed to determine mature miRNA and mRNA levels. Total RNA was isolated with TRIzol (Invitrogen) and treated with DNase I (Takara). For quantitative mature miR-26a/b detection, total RNA (2 μg) was reverse-transcribed with Bulge-Loop miRNA-specific reverse transcription primers (RiboBio, Guangzhou, China) and Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). Quantitative PCRs were performed with FastStart Universal SYBR Green Master (Roche Applied Science) and Bulge-Loop primers (RiboBio) on the StepOne real time PCR system (Applied Biosystems) with small nuclear RNA U6 as the normalization control. To detect cellular mRNAs and HBV pgRNA, RNA samples were reverse-transcribed using 6-nucleotide random primers and Moloney murine leukemia virus reverse transcriptase (Promega). Quantitative PCRs were performed with FastStart Universal SYBR Green Master Mix (Roche Applied Science) and specific primers on the StepOne real time PCR system (Applied Biosystems). The mRNAs levels were normalized to the GAPDH expression level.
PCR was performed at 95 °C for 3 min followed by 40 cycles at 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 20 s. Each set of PCRs was performed in triplicate, and the CT values of each PCR were obtained. The ΔΔCT method was used to calculate the ratios of gene expression relative to the control set in the experiments.
The following primer pairs were used: HBV pgRNA, 5′-TGGtk;1ATTCGCACTCCTCCAGCTT-3′ (sense) and 5′-GGGACCTGCCTCGTCGTCTA-3′ (antisense); CHORDC1, 5′-TGCCTCCCTAAAACAAGCACT-3′ (sense) and 5′-TTTCTTCTACAACAGCTCCAGT-3′ (antisense); CCND2, 5′-GCGGAGAAGCTGTGCATTTA-3′ (sense) and 5′-GATCATCGACGGTGGGTACA-3′ (antisense); STK39, 5′-GGGGGTGATGTTACCCGAAA-3′ (sense) and 5′-ATAAGGCGCTGCTCCTGTTG-3′ (antisense); CCNG1, 5′-TCCTTCAAGAGAACTTGCCACT-3′ (sense) and 5′-GCTCTTGCCAGAAGGTCAGA-3′ (antisense); CTDSP1, 5′-GCGAGCTCTTTGAATGTGTG-3′ (sense) and 5′-GGCTCAGGTCCTTCACGTAG-3′ (antisense); GAPDH, 5′-AAGGCTGTGGGCAAGG-3′ (sense) and 5′-TGGAGGAGTGGGTGTCG-3′ (antisense).
HBV Replication Analysis
HepG2 cells were grown in 6-well plates, transfected with pHBV1.3 as indicated, and cell culture medium was collected 48 h post-transfection. The HBsAg and HBeAg viral protein levels were determined in the supernatant using ELISA kits (Shanghai KeHua Biotech, Shanghai, China).
Cells were collected at 72 h post-transfection, and RNA was extracted and treated with DNase I (Takara) to remove DNA. HBV pgRNA level was determined by real time PCR. GAPDH was amplified as an internal control.
HBV DNA from intracellular core particles was extracted at 72 h post-transfection. Cells were lysed in 1 ml of lysis buffer (50 mm Tris, pH 7.5, 0.5% Nonidet P-40, 1 mm EDTA, and 100 mm NaCl) at 4 °C for 1 h and subsequently incubated with 10 μl of 1 m MgCl2 and 10 μl of DNase I (10 mg/ml, Takara) at 37 °C for 2 h. DNA that was not protected by HBV core protein was digested with DNase I. Viral cores were then precipitated by adding 35 μl of 0.5 m EDTA and 225 μl of 35% polyethylene glycol, followed by centrifugation at 1200 × g for 15 min. The pellet was resuspended in buffer A (10 mm Tris, 100 mm NaCl, 1 mm EDTA, 1% SDS, and 2.5 mg/ml proteinase K) and incubated for 16 h. Capsid-associated viral DNA released from the lysed cores was extracted with phenol and chloroform, precipitated with isopropyl alcohol, and quantified using a set of TaqMan real time PCR primers (Invitrogen) for HBV DNA detection as follows: 5′-AGAAACAACACATAGCGCCTCAT-3′ (sense), 5′-TGCCCCATGCTGTAGATCTTG-3′ (antisense), and the HBV probe 5′-TGTGGGTCACCATATTCTTGGG-3′. The PCR was performed as follows: 1 cycle at 95 °C for 10 min and 40 cycles at 95 °C for 15 s and 60 °C for 30 s. Plasmid pHBV1.3 was diluted properly and used as a standard, and all samples were analyzed in triplicate. Real time PCR results were analyzed and expressed as relative HBV DNA levels after converting the viral copy numbers to fold changes.
Luciferase Reporter Gene Assays
The firefly luciferase reporter plasmids were transfected into cells using Lipofectamine 2000 reagent. A Renilla luciferase reporter vector pRL-TK was cotransfected as an internal control. The Dual-Luciferase reporter assay system (Promega, Madison, WI) was used to measure the luciferase activity of each sample 48 h post-transfection, and Renilla luciferase activity was determined as internal control for transfection efficiency. The results were normalized to the vector or mock-control samples.
Western Blot Analysis
Whole-cell lysates were prepared by lysing the cells in PBS, pH 7.4, that contained 0.01% Triton X-100, 0.01% EDTA, and 10% protease inhibitor mixture (Roche Applied Science). The protein concentration of each sample was determined using a Bradford assay kit (Bio-Rad). A 100-g aliquot of each sample was subjected to 12% SDS-PAGE and subsequently transferred to a nitrocellulose membrane (Amersham Biosciences).Western blot analysis was performed using antibodies as indicated in the figures. Antibodies were detected using SuperSignal chemiluminescent reagent (Pierce) and analyzed with an LAS-4000 instrument (FujiFilm, Tokyo, Japan).
Nuclear Extraction
To separate and collect the cytosolic and nuclear protein fractions, whole cells were washed with ice-cold PBS and were collected by centrifugation. The resulting pellets were resuspended in hypotonic buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.5 mm dithiothreitol (DTT), 10% protease mixture inhibitor) for 15 min on ice and were vortexed for 10 s. Nuclei were pelleted by centrifugation at 13,000 × g for 1 min, and the pellets and cytosolic protein-containing supernatants were collected. Cytosolic and nuclear protein fractions were then examined by Western blot analysis.
MTS Cell Viability Assay
HepG2 cells were seeded into 96-well plates at 5000 cells/well. After cell adhesion, the plasmids were then transfected into HepG2 cells using Lipofectamine 2000 reagent (Invitrogen). After an incubation of 48 h, the MTS assay was performed using the MTS cell viability kit (Promega) according to the manufacturer's instructions. Cell viability was then calculated as percentage of the control group.
Statistical Analysis
All experiments were reproducible and were performed in triplicate. Each set of experiments was repeated three times with similar results; a representative experiment was chosen for presentation. The data presented are expressed as mean ± S.D. For analysis of statistical difference between the two groups, a Student's two-tailed unpaired t test was applied. For analysis of statistical difference between three or more groups, a one-way analysis of variance with Bonferroni's multiple comparison test was applied. p values are indicated by asterisks (**, p < 0.01; *, p < 0.05).
RESULTS
MicroRNA-26b Inhibits HBV Gene Expression and Replication in Hepatoma Cells
We first investigated the effect of miR-26b on HBV gene expression and replication in hepatoma cells. The miR-26b mimic was used for transfection, and miR-Ctrl mimic was used as the negative control. Quantitative real time PCR analysis showed that transfection of HepG2 cells with miR-26b mimic up-regulated the miR-26b level compared with that of miR-Ctrl (Fig. 1A). Meanwhile, the protein level of ATF2, a known miR-26b target gene (29), could be efficiently diminished by miR-26b mimic transfection in HepG2 cells (Fig. 1A). A replication-competent clone of pHBV1.3 (serotype adw, genotype B) and miR-26b mimic was transfected into HepG2 cells to investigate the role of miR-26b in HBV replication. The levels of HBeAg and HBsAg viral proteins in the culture supernatant decreased after treatment with miR-26b mimic compared with those of the miR-Ctrl mimic (Fig. 1B). The levels of HBV pgRNA and capsid-associated DNA were also reduced by expression of exogenous miR-26b (Fig. 1C).
FIGURE 1.
miR-26b inhibits HBV gene expression and replication in hepatoma cells. A, HepG2 cells were transfected with 50 nm miR-26b mimic (miR-26b) or the nonspecific control miR-Ctrl. After 48 h, miR-26b level was analyzed by real time PCR and ATF2 protein level was analyzed by Western blot. B and C, HepG2 cells were cotransfected with pHBV1.3 and 50 nm of miR-26b mimic. The secretion of HBeAg and HBsAg was measured after 48 h by ELISA (B), and the amount of HBV pgRNA and capsid-associated DNA was determined after 72 h by real time PCR (C). D, HepG2 cells were transfected with 100 nm miR-26b inhibitor (miR-26b-Inh) and the nonspecific miRNA inhibitor control (miR-Inh-Ctrl). After 48 h, miR-26b level was analyzed by real time PCR and ATF2 protein level was analyzed by Western blot. E and F, HepG2 cells were cotransfected with pHBV1.3 and 100 nm miR-26b inhibitor. The level of HBeAg and HBsAg in the supernatant was detected by ELISA (E), and the amount of HBV pgRNA and capsid-associated DNA was detected by real time PCR (F). *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
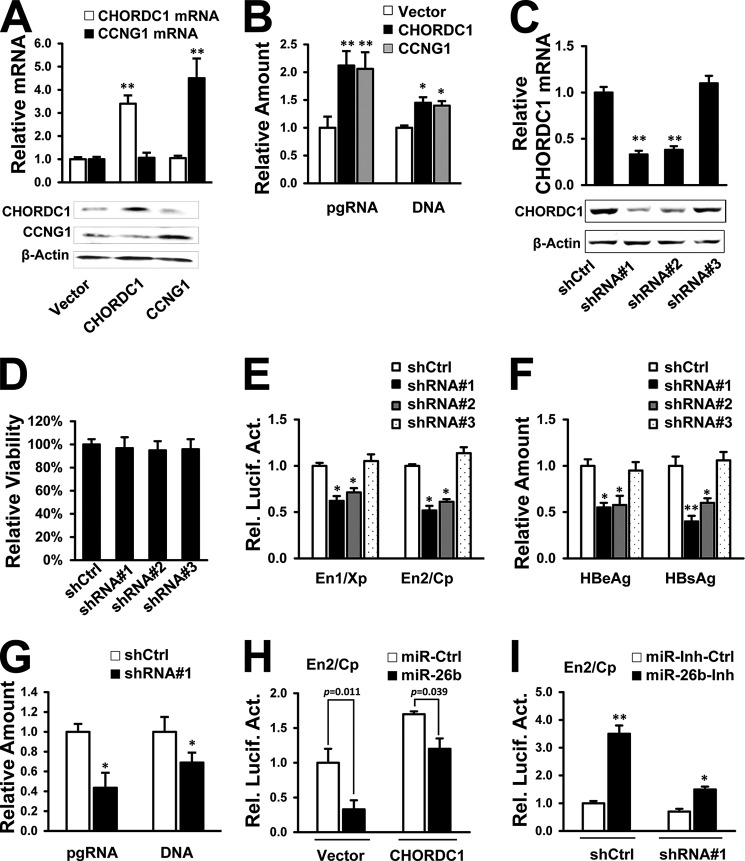
The miR-26b inhibitor (miR-26b-Inh) was synthesized for antisense inhibition of endogenous miR-26b, and miRNA inhibitor control (miR-Inh-Ctrl) was used as the negative control. The level of endogenous miR-26b declined after transfection of miR-26b inhibitor in HepG2 cells, and ATF2 protein level could be up-regulated by miR-26b inhibitor (Fig. 1D). The pHBV1.3 and miR-26b inhibitors were cotransfected into HepG2 cells, and treatment with miR-26b inhibitor resulted in higher levels of HBeAg and HBsAg viral proteins in culture supernatant compared with those of the negative control (Fig. 1E). Depletion of miR-26b also increased intracellular levels of HBV pgRNA and capsid-associated DNA in HepG2 cells (Fig. 1F). In addition, reduced secretion of HBeAg and HBsAg proteins was observed in miR-26b mimic-transfected HepG2.2.15, an HBV-positive cell line (serotype ayw, genotype D) (data not shown). Taken together, these results indicate that miR-26b inhibits HBV protein expression and viral DNA replication in human hepatoma cell lines. The miR-26b-mediated inhibition of HBV appears to be genotype-independent.
MicroRNA-26b Attenuates HBV Enhancer/Promoter Activities
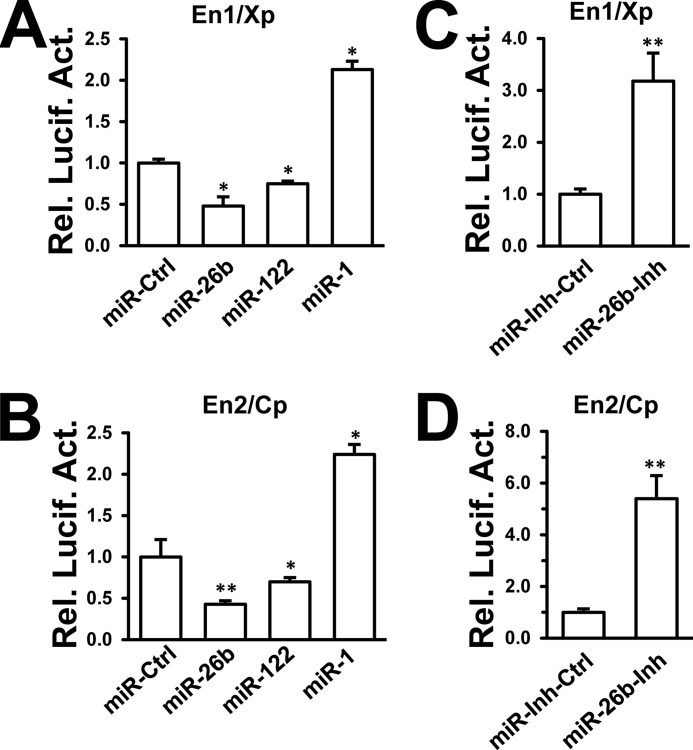
Two HBV enhancers are believed to be the central elements regulating viral transcription and replication (6–8). Several miRNAs, including miRNA-122 and miRNA-1, were reported to modulate HBV enhancer/promoter activities, thereby regulating HBV replication (14, 17, 30). We chose miR-122 and miR-1 as positive controls to assess whether miR-26b affects HBV replication at the transcriptional level. HepG2 cells were cotransfected with each of the miRNA mimics (miR-26b, miR-122, and miR-1) and each of the HBV enhancer/promoter reporters (En1/Xp and En2/Cp). The results showed that the miR-122 mimic reduced HBV En1/Xp and En2/Cp activities (14), and the miR-1 mimic stimulated viral enhancer/promoter activities (17). The miR-26b mimic obviously reduced both HBV En1/Xp and En2/Cp activities in HepG2 cells (Fig. 2, A and B). By contrast, the miR-26b inhibitor significantly stimulated En1/Xp and En2/Cp activities in HepG2 cells (Fig. 2, C and D). Considering the crucial role of enhancer/promoter activities for HBV infection, our results implied that miR-26b may inhibit HBV gene expression and replication by interfering with viral transcription.
FIGURE 2.
miR-26b decreases HBV enhancer/promoter activities. A and B, HepG2 cells were cotransfected with different miRNA mimics and HBV En1/Xp (A) or En2/Cp (B) reporter. pRL-TK was cotransfected as an internal control in the luciferase assay. En1/Xp or En2/Cp luciferase activities were measured after 48 h using a dual-luciferase assay kit. En1/Xp-luc and En2/Cp-luc were normalized to Renilla luciferase. C and D, HepG2 cells were cotransfected with miRNA inhibitors and En1/Xp (C) or En2/Cp (D) reporter. Luciferase activities were measured after 48 h using a dual-luciferase assay kit. *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
Screening for Potential miR-26b Target Genes Involved in HBV Enhancer/Promoter Activities and Protein Expression
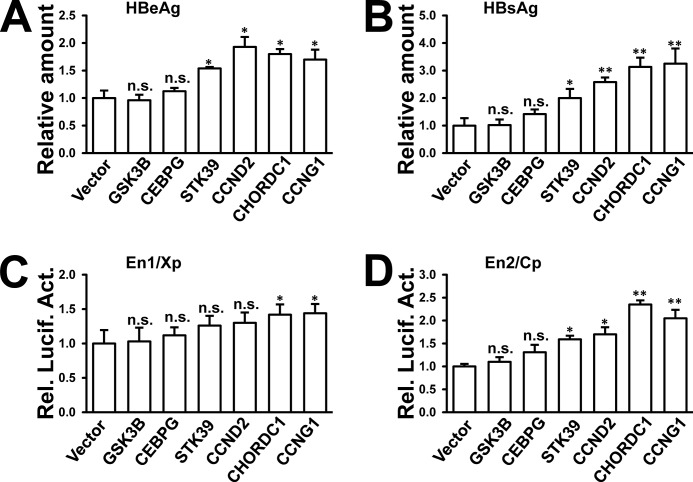
We analyzed the miR-26b target genes that might influence HBV replication. Two on-line databases for miRNA target prediction (TargetScanHuman and miRDB) were used to predict miR-26b targets. Hundreds of common potential targets were found by those two databases. However, none were reported to participate in regulation of HBV replication. To search for potential candidates that might control HBV replication, several open reading frames (ORFs) of predicted targets with high scores were chosen and cloned into a protein expression vector. The predicted targets included glycogen synthase kinase 3β (GSK3B), CCAAT/enhancer binding protein γ (CEBPG), serine/threonine kinase 39 (STK39), cyclin D2 (CCND2), and CHORDC1. The ORF of CCNG1, which was reported to enhance HBV transcription and replication (14), was cloned into the same vector and used as a positive control. The constructs were confirmed by DNA sequencing. The miR-26b inhibits HBV transcription and protein expression; therefore, overexpression of potential miR-26b targets was considered to up-regulate viral enhancer/promoter activities and viral protein expression. Each enhancer/promoter reporter or pHBV1.3 was cotransfected with the expression constructs into HepG2 cells. The results showed that STK39, CCND2, CHORDC1, and CCNG1 increased the levels of HBeAg and HBsAg in the culture supernatant (Fig. 3, A and B). In the viral enhancer/promoter reporter assays, CHORDC1 and CCNG1 could enhance HBV En1/Xp and En2/Cp activities (Fig. 3, C and D). However, GSK3B and CEBPG had no apparent effect on HBV promoter/enhancer activities or viral protein expression (Fig. 3, A–D). These results suggest that miR-26b may target more than one host gene to attenuate HBV infection.
FIGURE 3.
Screening for potential miR-26b targets that increase HBV protein expression and enhancer/promoter activities. A–D, pHBV1.3 or each enhancer/promoter reporter was cotransfected into HepG2 cells with expression vectors for GSK3B, CEBPG, CCND2, STK39, CHORDC1, or CCNG1. Empty vector was included as a negative control. Secretion of HBeAg (A) and HBsAg (B) was measured after 48 h by ELISA. En1/Xp (C) and En2/Cp (D) luciferase activities were measured using a dual-luciferase assay kit. n.s., not significant; *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
Identification of Novel Targets of miR-26b
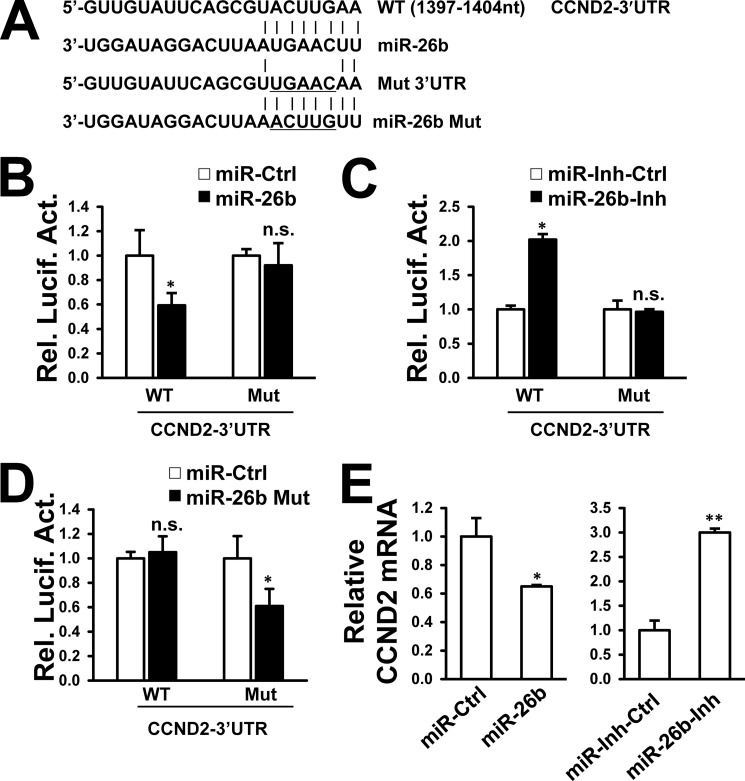
We focused primarily on the most effective potential target CHORDC1 in the regulation of HBV replication. Three putative miR-26b complementary regions were identified in the 3′UTR of the CHORDC1 sequence (at positions 242–249, 422–429, and 569–576) (Fig. 4A). To determine whether miR-26b regulates CHORDC1 expression by post-transcriptional targeting of its 3′UTR, the CHORDC1 3′UTR was cloned into a firefly luciferase reporter plasmid. Three distinct mutations were generated in the CHORDC1 3′UTR at predicted seed-matching sites to test the interaction between miR-26b and CHORDC1 3′UTR (Fig. 4A). Individual wild-type (WT) and CHORDC1 3′UTR mutants were transfected into HepG2 cells along with miR-26b mimic or miR-26b inhibitor. The miR-26b mimic effectively reduced luciferase activity controlled by the CHORDC1 3′UTR compared with that of control. By contrast, miR-26b-mediated attenuation of luciferase activity was disrupted by mutation of the CHORDC1 3′UTR-binding sites (Fig. 4B). The miR-26b inhibitor significantly stimulated luciferase activity controlled by the CHORDC1 3′UTR (Fig. 4C). Mutation of the miR-26b-binding sites in CHORDC1 3′UTR disrupted miR-26b inhibitor-mediated increase in luciferase activity (Fig. 4C). To confirm the sequence-specific effect of miR-26b on the 3′UTR-binding sites, miR-26b Mut mimic was synthesized in our study to target the mutated binding sites of miR-26b in the 3′UTR (Fig. 4A). As predicted, miR-26b Mut mimic inhibited the luciferase activity controlled by the mutant CHORDC1 3′UTR but not the WT CHORDC1 3′UTR (Fig. 4D). These results indicate that miR-26b could suppress CHORDC1 expression by sequence-specific binding to its 3′UTR.
FIGURE 4.
CHORDC1 is a target of miR-26b. A, predicted miR-26b-binding sites in the 3′UTR of CHORDC1 mRNA. Perfect matches in seed regions are indicated by a line. Mutations (underlined) were generated in the binding sites of 3′UTR and miR-26b seed region for the reporter gene assay. B–D, miR-26b mimic (B), miR-26b inhibitor (C), or miR-26b mutant (Mut) mimic (D) was cotransfected with a firefly luciferase reporter plasmid carrying either the wild-type or mutant 3′UTR of CHORDC1 into HepG2 cells. Luciferase activities were measured after 48 h using a dual-luciferase assay kit, and normalized to Renilla luciferase. E, HepG2 cells were transfected with miR-26b mimic or miR-26b inhibitor. CHORDC1 mRNA and protein levels were analyzed 48 h post-transfection by real time PCR and Western blot, respectively. n.s., not significant; *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
We investigated whether the endogenous levels of CHORDC1 mRNA and protein were affected by miR-26b mimic and inhibitor. Transfection of miR-26b mimic markedly reduced CHORDC1 protein expression but not the mRNA level in HepG2 cells (Fig. 4E, left). Conversely, miR-26b inhibitor-mediated knockdown of endogenous miR-26b increased CHORDC1 protein expression but did not significantly influence mRNA level (Fig. 4E, right). These results indicate that miR-26b could suppress CHORDC1 expression via translational inhibition but not mRNA degradation.
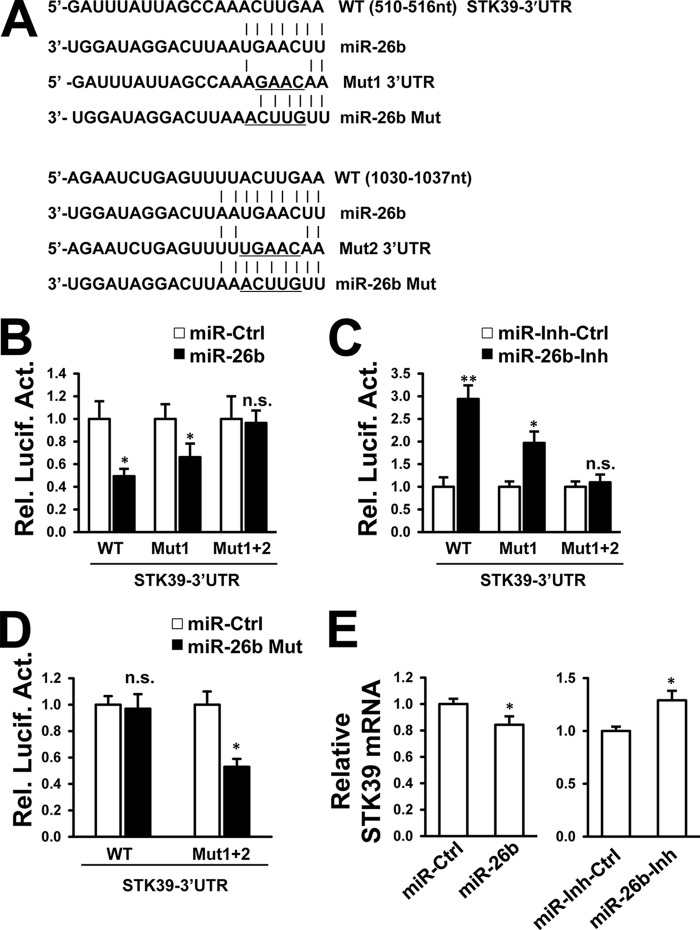
We also examined whether miR-26b can target 3′UTRs of CCND2 and STK39 mRNA. Putative miR-26b complementary regions and mutation sites in the CCND2 (Fig. 5A) and STK39 (Fig. 6A) sequences are shown. Transfection of the miR-26b mimic reduced the luciferase activity controlled by the 3′UTR of CCND2 mRNA (Fig. 5B) and STK39 mRNA (Fig. 6B) in HepG2 cells. Loss of endogenous miR-26b by transfection of miR-26b inhibitor resulted in the opposite effects (Figs. 5C and 6C). When the predicted seed-matching sites were generated to diminish the interaction between miR-26b and the 3′UTR sequences, miR-26b lost its ability to regulate luciferase activity controlled by the mutant 3′UTRs of CCND2 mRNA (Fig. 5, B and C) and STK39 mRNA (Fig. 6, B and C). On the contrary, miR-26b Mut mimic could only inhibit luciferase activity controlled by mutant 3′UTRs of CCND2 (Fig. 5D) and STK39 (Fig. 6D). The endogenous CCND2 mRNA level was efficiently reduced by miR-26b (Fig. 5E), and the endogenous STK39 mRNA level was slightly reduced by miR-26b (Fig. 6E). These results suggest that miR-26b could target CCND2 and STK39 by mRNA degradation. Because a single miRNA has the ability to recognize multiple target mRNAs with seed-complementary sequences, these data suggest that miR-26b can regulate multiple gene targets involved in HBV transcription and thus inhibit viral replication.
FIGURE 5.
CCND2 is a target of miR-26b. A, predicted miR-26b-binding site is located in the 3′UTR of CCND2 mRNA. Perfect matches in seed regions are indicated by a line. Mutations (underlined) were generated in the binding sites of 3′UTR and miR-26b seed region for the reporter gene assay. B–D, miR-26b mimic (B), miR-26b inhibitor (C), or miR-26b mutant (Mut) mimic (D) was cotransfected with a firefly luciferase reporter plasmid carrying either the wild-type or mutant 3′UTR of CCND2 into HepG2 cells. Luciferase activities were measured after 48 h using a dual-luciferase assay kit, and normalized to Renilla luciferase. E, HepG2 cells were transfected with miR-26b mimic or inhibitor and controls. CCND2 mRNA level was detected by real time PCR at 48 h post-transfection. n.s., not significant; *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
FIGURE 6.
STK39 is a target of miR-26b. A, predicted miR-26b-binding sites located in the 3′UTR of STK39 mRNA. Perfect matches in seed regions are indicated by a line. Mutations (underlined) were generated in the binding sites of 3′UTR and miR-26b seed region for the reporter gene assay. B–D, miR-26b mimic (B), miR-26b inhibitor (C), or miR-26b mutant (Mut) mimic (D) was cotransfected with a firefly luciferase reporter plasmid carrying either the wild-type or mutant 3′UTR of STK39 into HepG2 cells. Luciferase activities were measured after 48 h using a dual-luciferase assay kit, and normalized to Renilla luciferase. D and E, HepG2 cells were transfected with miR-26b mimic (D) or inhibitor (E) and controls. STK39 mRNA level was detected by real time PCR at 48 h post-transfection. n.s., not significant; *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
CHORDC1 Stimulates HBV Replication
Experiments were performed to confirm the role of CHORDC1 in HBV replication. Transcription and translation of CHORDC1 and CCNG1 expression vectors were confirmed by real time PCR and Western blot analysis (Fig. 7A). The initial screening showed that overexpression of CHORDC1 and the positive control CCNG1 apparently enhanced HBV enhancer/promoter activities and protein expression (Fig. 3, A–D). These additional experiments showed that HBV pgRNA transcription and capsid-associated DNA synthesis were elevated by CHORDC1 and CCNG1 expression in pHBV1.3-transfected HepG2 cells (Fig. 7B).
FIGURE 7.
CHORDC1 stimulates HBV replication. A, HepG2 cells were transfected with expression constructs for CHORDC1, CCNG1, or empty vector. After 48 h, the mRNA levels were analyzed by real time PCR, and the protein levels were measured by Western blot analysis. B, HepG2 cells were cotransfected with pHBV1.3 and CHORDC1 or CCNG1. The amount of HBV pgRNA and capsid-associated DNA was measured after 72 h by real time PCR. C, HepG2 cells were plated in 6-well plates and transfected with different CHORDC1 shRNA vectors (shRNA#1, shRNA#2, and shRNA#3) or shCtrl as mock. After 48 h, CHORDC1 mRNA level was measured by real time PCR, and protein level was measured by Western blot analysis. D, HepG2 cells were transfected with different CHORDC1 shRNA vectors. After an incubation of 48 h, the MTS assay was performed using the MTS cell viability kit (Promega) according to the manufacturer's instructions. Cell viability was then calculated as percentage of the control group. E and F, different CHORDC1 shRNA vectors were cotransfected with viral enhancer/promoter reporters (E) or pHBV1.3 (F) into HepG2 cells. After 48 h, luciferase activities were measured using a dual-luciferase assay kit (E) and the secretion of HBsAg and HBeAg was measured by ELISA (F). G, CHORDC1 shRNA#1 vector was cotransfected with pHBV1.3 into HepG2 cells, and the amount of HBV pgRNA and capsid-associated DNA was measured by real time PCR after 72 h post-transfection. H, HBV En2/Cp reporter and miR-26b mimic were cotransfected into HepG2 cells, along with CHORDC1 expression vector or control vector. Luciferase activities were measured after 48 h using a dual-luciferase assay kit. I, HBV En2/Cp reporter and miR-26b inhibitor were cotransfected into HepG2 cells, along with CHORDC1 shRNA#1 or shCtrl. Luciferase activities were measured after 48 h using a dual-luciferase assay kit. *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
Expression vectors containing shRNA are an effective tool for RNA interference in mammalian cells (31). The shRNAs targeting CHORDC1 were synthesized and transfected into HepG2 cells. Both shRNA#1 and shRNA#2 efficiently depleted CHORDC1 mRNA and protein expression compared with the control shRNA (Fig. 7C). CHORDC1 shRNA#3 was unable to knock down the endogenous gene expression (Fig. 7C) and used as a negative control in the subsequent experiments. The transfection of all CHORDC1 shRNAs had little influence on cell viability of HepG2 cells (Fig. 7D). The HBV enhancer/promoter activities (Fig. 7E) and antigen secretion (Fig. 7F) were reduced when CHORDC1 protein levels declined in HepG2 cells. Knockdown of endogenous CHORDC1 by shRNA#1 also decreased the intracellular levels of HBV pgRNA and viral capsid-associated DNA (Fig. 7G). Furthermore, the inhibitory effect of miR-26b mimic on viral En2/Cp activity could be partially reversed by transfection of CHORDC1 expression vector (Fig. 7H). Cotransfection of shCHORDC1 with miR-26b inhibitor could attenuate the increased activity of En2/Cp reporter (Fig. 7I). These experiments suggest that CHORDC1 protein is required to stimulate HBV gene expression, pgRNA transcription, and DNA replication.
HBV Suppresses miR-26b Expression in Human Liver Cells
Disrupted miR-26b expression is often correlated with disease development. To investigate the possible regulation of miR-26b by HBV, pHBV1.3 (serotype adw, genotype B) was transfected into human normal liver L02 cells and hepatoma HepG2 cells. HBV infection down-regulated the miR-26b level in both L02 cells (Fig. 8A) and HepG2 cells (Fig. 8B) in a dose-dependent manner. The miR-26b resides in an intron of CTDSP1 mRNA, and it is transcribed as part of its host transcription unit (32). In HepG2 cells, CTDSP1 mRNA declined along with miR-26b in a dose-dependent manner after pHBV1.3 transfection (Fig. 8C). Protein expression of endogenous CHORDC1, the target of miR-26b, was up-regulated in a dose-dependent manner by HBV (Fig. 8D), and HBV induced up-regulation of CHORDC1 could be diminished by miR-26b in HepG2 cells (Fig. 8E). CHORDC1 protein expression induced by HBV only existed in cytoplasm of HepG2 cells (Fig. 8F). These results suggest that HBV infection can reduce the miR-26b level in human liver cells. The down-regulation of miR-26b by HBV infection indicates that HBV could increase viral production by up-regulating its target gene in liver cells.
FIGURE 8.
HBV suppresses miR-26b expression in human liver cells. A, L02 cells were plated in 6-well plates, grown to 80% confluence, and transfected with different amounts of pHBV1.3. The miR-26b level was analyzed after 48 h by real time PCR. B and C, HepG2 cells were transfected with different amounts of pHBV1.3 as indicated. Total RNA was extracted after 48 h to determine the miR-26b level (B) and the CTDSP1 mRNA level (C) by real time PCR. U6 and GAPDH were amplified as internal control, respectively. D, HepG2 cells were transfected with different amount of pHBV1.3 as indicated. Whole-cell lysates were analyzed after 96 h to determine CHORDC1 protein levels by Western blot analysis. E, pHBV1.3 was cotransfected into HepG2 cells with miR-26b mimic or miRNA mimic control. After 96 h, CHORDC1 protein levels were detected by Western blot analysis. F, HepG2 cells were transfected with pHBV1.3 or control vector, and cytosolic and nuclear extracts were prepared 96 h after transfection and were subjected to Western blot analysis. β-Actin and lamin A/C were used as markers for the cytosolic and nuclear fractions, respectively. Levels of CHORDC1 protein were detected by Western blotting. *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
MicroRNA-26a Targets CHORDC1 and Affects HBV Replication
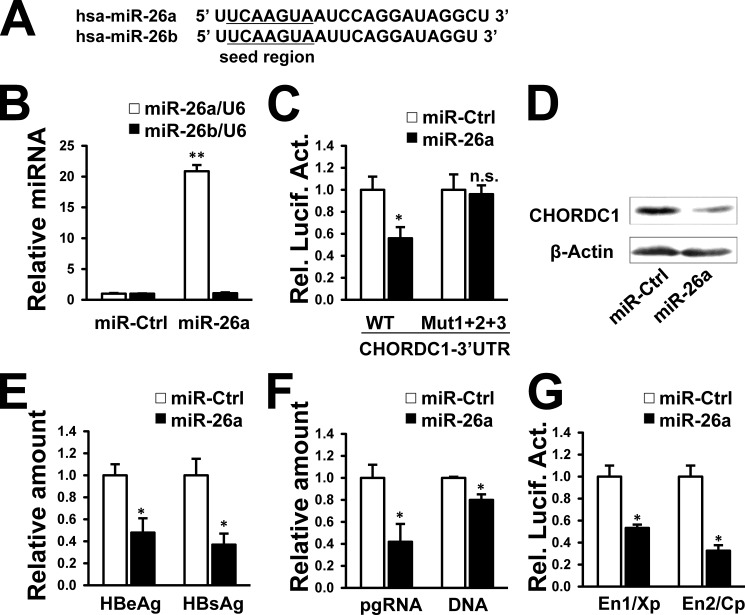
The human miR-26 family is composed of miR-26a-1, miR-26a-2, and miR-26b (20). Hsa-miR-26a is the mature form of miR-26a-1 and miR-26a-2. It has high similarity with miR-26b except for two nucleotides that are different (Fig. 9A). Both miR-26a and miR-26b are predicted to target the 3′UTR of CHORDC1. We assessed whether miR-26a could bind to the CHORDC1 3′UTR, inhibit CHORDC1 expression, and suppress HBV gene expression and replication. Transfection of the miR-26a mimic in HepG2 cells resulted in a higher level of mature miR-26a but not miR-26b as determined by real time PCR (Fig. 9B). The miR-26a mimic also reduced the luciferase activity controlled by the CHORDC1 3′UTR, but it did not reduce the reporter activity controlled by the mutated CHORDC1 3′UTR (Fig. 9C). Endogenous CHORDC1 protein was depleted after transfection of miR-26a mimic compared with that of the miRNA control (Fig. 9D). The suppression of CHORDC1 by miR-26a indicated that miR-26a might affect HBV transcription and replication. Indeed, miR-26a attenuated the secretion of HBeAg and HBsAg (Fig. 9E) and reduced the levels of HBV pgRNA and capsid-associated DNA in pHBV1.3-transfected HepG2 cells (Fig. 9F). HBV En1/Xp and En2/Cp activities were also markedly reduced by miR-26a in HepG2 cells (Fig. 9G). Thus, the mature forms of the miR-26 family (miR-26a and miR-26b) act as suppressors of HBV transcription and replication.
FIGURE 9.
miR-26a targets CHORDC1 and affects HBV replication. A, mature miR-26a and miR-26b sequences. B, HepG2 cells were transfected with miR-26a mimic and miRNA mimic control. Expression of miR-26a and miR-26b was measured by real time PCR. C, HepG2 cells were cotransfected with wild-type or mutated 3′UTR of CHORDC1 luciferase reporters and miR-26a mimic. Luciferase activities were measured after 48 h using a dual-luciferase assay kit. D, HepG2 cells were transfected with miR-26a mimic or miRNA mimic control. CHORDC1 protein levels were detected by Western blot analysis. E and F, HepG2 cells were cotransfected with pHBV1.3 and miR-26a mimic. After 48 h, secretion of HBsAg and HBeAg was measured by ELISA (E), and the amount of HBV pgRNA and capsid-associated DNA was measured by real time PCR (F). G, HepG2 cells were cotransfected with miR-26a mimic and En1/Xp or En2/Cp reporter. Luciferase activities were measured after 48 h using a dual-luciferase assay kit. n.s., not significant; *, p < 0.05; **, p < 0.01 on the basis of experiments performed in triplicate.
DISCUSSION
This study mainly investigated the interaction between cellular miR-26b and HBV. The results indicated that miR-26b inhibited HBV replication by targeting multiple host genes, including CHORDC1 in hepatoma cells. Conversely, HBV infection suppressed miR-26b expression in human liver cells.
There is evidence that miR-26b has a role in the development of normal tissues and many diseases (20). We determined that miR-26b acts as a host restriction factor to attenuate HBV replication in hepatoma cells. These results provide new evidence for the role of miRNAs in HBV replication. There is a dynamic interaction between miR-26b and HBV. miR-26b inhibits HBV transcription and disrupts viral replication, and HBV infection reduces the level of miR-26b. HBV-induced loss of miR-26b can facilitate viral replication, and the interaction between miR-26b and HBV may determine the persistence of HBV infection in host cells.
Patients with chronic HBV infection are at high risk to ultimately develop HCC (1, 2). Disruption of miR-26b expression often occurs during disease development, especially during cancer development (18–26). These reports inspired us to investigate whether HBV affects the miR-26b level. Transfection of a replication-competent HBV clone consistently down-regulated the miR-26b level in both normal L02 cells derived from liver tissue and liver carcinoma HepG2 cells. The exact mechanism involved in HBV-mediated miR-26b down-regulation is under investigation. A possible explanation is that miR-26b repression can partly, if not entirely, result from CTDSP1 mRNA transcription, which is also suppressed by HBV infection. However, the fold-reduction of the miR-26b level (Fig. 8B) was greater than that of the CTDSP1 mRNA level (Fig. 8C). Therefore, other pathways regulating the level of miR-26b in HBV-infected cells could not be excluded.
In cancer development, chromosomal deletion often indicates the absence of a functional tumor-suppressor gene in the lost region (33). The miR-26b/CTDSP1 is located at chromosome 2q35, a region that is frequently deleted in HCC (34). During HBV infection, the down-regulated miR-26b level may play an important role in virus-induced development of HCC.
MicroRNAs are post-transcriptional regulators of gene expression by affecting the translation and stability of mRNAs (35). We identified three novel miR-26b targets, including CHORDC1, CCND2, and STK39. Delivery of miR-26b efficiently decreased protein expression controlled by the 3′UTRs of these three mRNAs. The miR-26b degrades CCND2 mRNA, but it does not affect CHORDC1 mRNA stability. This suggests that miR-26b down-regulates CHORDC1 protein expression by inhibiting mRNA translation at some stage after the translation initiation step, without affecting mRNA abundance.
Proteins containing CHORDs are a novel class of eukaryotic zinc-binding proteins, which are required for disease resistance signaling in barley and development in Caenorhabditis elegans (36). CHORDC1, also called CHP1 or morgana, is one of only two genes encoding this class of proteins in vertebrates (37, 38). Mammalian CHORDC1 is a novel heat-shock protein 90 (HSP90) co-chaperone, and this interaction may be involved in the response to altered energy balance (39–41). The down-regulation of CHORDC1 leads to centrosome amplification and promotes tumorigenesis in mice (38). The function of human CHORDC1 has not been extensively investigated. We found that CHORDC1 simulated HBV gene expression and DNA replication by activating HBV enhancer/promoter elements. CHORDC1 was involved in miR-26-mediated HBV regulation. CHORDC1 protein level was increased by HBV in HepG2 cells and the up-regulation of CHORDC1 protein could be diminished by the transfection of miR-26b. However, CHORDC1 is supposed to regulate HBV transcription indirectly, because CHORDC1 protein only exists in cytoplasm in HepG2 cells before and after HBV infection. Further studies should be performed to investigate how CHORDC1 enhances HBV replication and the potential biological role of CHORDC1 in HBV-mediated cancer development.
STK39, also called SPAK, is a serine/threonine kinase that specifically activates the p38 pathway and may act as a novel mediator of stress-activated signals (42). This study showed that STK39 appears to play a positive role in HBV replication. The up-regulation of viral synthesis by STK39 might be mediated by activating the MAPK p38 pathway, which was previously reported to mediate enhanced HBV replication in response to inhibitors of nucleotide biosynthesis (43). CCND2 is the cyclin that determines the G1/S transition in the cell cycle. Overexpression of CCND2 is associated with tumor progression in human squamous cell carcinoma lines (44). This study showed that CCND2 positively regulated HBV production in hepatoma cells. In fact, CCND2 is not the only cyclin that can regulate HBV replication. The cyclin CCNG1 is a target gene of miR-122, and it can enhance HBV replication by modulating p53-mediated viral transcription (14).
Because of its small genome size and relatively low coding capacity, HBV requires extensive networks of host factors to facilitate successful infection and pathogenesis. This study identified three novel host factors that enhanced HBV transcriptional activity, which may also participate in the development of viral-induced diseases. These genes might be promising drug targets for the development of novel HBV therapies. Genome-wide screening to identify all miR-26b target genes involved in HBV replication is still needed to elucidate the overall function of miR-26b in HBV replication.
A problem remains unsolved in our study. Efficient transcription of HBV requires a number of ubiquitous cellular factors, liver-enriched transcription factors and nuclear receptors (5). We have tried to find out which transcription factor or nuclear receptor is directly involved in the suppression of HBV gene expression by miR-26b and CHORDC1. Unfortunately, none of the examined factors, including hepatocyte nuclear factor 4α and farnesoid X receptor α, was responsible for the regulation, because both miR-26b and CHORDC1 still efficiently regulated the enhancer/promoter activities when the binding sites of hepatocyte nuclear factor 4α or farnesoid X receptor α in En1/Xp and En2/Cp were all mutated (data not shown). CHORDC1 is a HSP90 cochaperone (39–41), and whether this interaction is involved in HBV replication was also investigated. H186A mutation of CHORDC1 was shown to have a weaker nucleotide-dependent interaction with HSP90 compared with wild-type CHORDC1 (45). However, neither H186A mutation of CHORDC1 nor knockdown of HSP90 expression by siRNA (46) could efficiently attenuate the enhancement of HBV production by CHORDC1 (data not shown). Future studies are needed to answer this question.
In conclusion, our data suggest that miR-26b down-regulates HBV expression, transcription, and replication by targeting cellular factors including CHORDC1 in hepatoma cells. Conversely, HBV infection reduces miR-26b levels in human liver cells. Human miR-26a plays a similar role as miR-26b in HBV replication. Our results suggest a new role for miR-26a/b in HBV infection and a possible novel strategy for the treatment of chronic hepatitis B infection.
This work was supported by Major State Basic Research Development Program of China Grant 2013CB911102, National Natural Science Foundation of China Grants 81461130019 and 81271821, and National Mega Project on Major Infectious Diseases Prevention Grant 2012ZX10004503-004.
- HBV
- hepatitis B virus
- miRNA
- microRNA
- CHORDC1
- cysteine- and histidine-rich domain containing 1
- HCC
- hepatocellular carcinoma
- pgRNA
- pregenomic RNA
- En1/Xp
- enhancer 1/X promoter
- En2/Cp
- enhancer 2/core promoter
- HBeAg
- hepatitis B e antigen
- HBsAg
- hepatitis B surface antigen
- GSK3B
- glycogen synthase kinase 3β
- CEBPG
- CCAAT/enhancer-binding protein γ
- STK39
- serine/threonine kinase 39
- CCND2
- cyclin D2
- CCNG1
- cyclin G1
- MTS
- methanethiosulfonate.
REFERENCES
- 1. Liaw Y. F., Chu C. M. (2009) Hepatitis B virus infection. Lancet 373, 582–592 [DOI] [PubMed] [Google Scholar]
- 2. Liang T. J. (2009) Hepatitis B: the virus and disease. Hepatology 49, S13–S21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seeger C., Mason W. S. (2000) Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64, 51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Will H., Reiser W., Weimer T., Pfaff E., Büscher M., Sprengel R., Cattaneo R., Schaller H. (1987) Replication strategy of human hepatitis B virus. J. Virol. 61, 904–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quasdorff M., Protzer U. (2010) Control of hepatitis B virus at the level of transcription. J. Viral Hepat. 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 6. Doitsh G., Shaul Y. (2004) Enhancer I predominance in hepatitis B virus gene expression. Mol. Cell. Biol. 24, 1799–1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su H., Yee J. K. (1992) Regulation of hepatitis B virus gene expression by its two enhancers. Proc. Natl. Acad. Sci. U.S.A. 89, 2708–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antonucci T. K., Rutter W. J. (1989) Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J. Virol. 63, 579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krol J., Loedige I., Filipowicz W. (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 [DOI] [PubMed] [Google Scholar]
- 10. Jackson R. J., Standart N. (2007) How do microRNAs regulate gene expression? Sci. STKE 2007, e1. [DOI] [PubMed] [Google Scholar]
- 11. Skalsky R. L., Cullen B. R. (2010) Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 64, 123–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Jiang L., Ji X., Yang B., Zhang Y., Fu X. D. (2013) Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J. Biol. Chem. 288, 18484–18493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y., Shen A., Rider P. J., Yu Y., Wu K., Mu Y., Hao Q., Liu Y., Gong H., Zhu Y., Liu F., Wu J. (2011) A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 25, 4511–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang S., Qiu L., Yan X., Jin W., Wang Y., Chen L., Wu E., Ye X., Gao G. F., Wang F., Chen Y., Duan Z., Meng S. (2012) Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1)-modulated P53 activity. Hepatology 55, 730–741 [DOI] [PubMed] [Google Scholar]
- 15. Thirion M., Ochiya T. (2013) Roles of microRNAs in the hepatitis B virus infection and related diseases. Viruses 5, 2690–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo H., Liu H., Mitchelson K., Rao H., Luo M., Xie L., Sun Y., Zhang L., Lu Y., Liu R., Ren A., Liu S., Zhou S., Zhu J., Zhou Y., Huang A., Wei L., Guo Y., Cheng J. (2011) MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology 54, 808–819 [DOI] [PubMed] [Google Scholar]
- 17. Zhang X., Zhang E., Ma Z., Pei R., Jiang M., Schlaak J. F., Roggendorf M., Lu M. (2011) Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology 53, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 18. Absalon S., Kochanek D. M., Raghavan V., Krichevsky A. M. (2013) MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 33, 14645–14659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bakre A., Mitchell P., Coleman J. K., Jones L. P., Saavedra G., Teng M., Tompkins S. M., Tripp R. A. (2012) Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 93, 2346–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao J., Liu Q. G. (2011) The role of miR-26 in tumors and normal tissues (Review). Oncol. Lett. 2, 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gottardo F., Liu C. G., Ferracin M., Calin G. A., Fassan M., Bassi P., Sevignani C., Byrne D., Negrini M., Pagano F., Gomella L. G., Croce C. M., Baffa R. (2007) Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 25, 387–392 [DOI] [PubMed] [Google Scholar]
- 22. Ji J., Shi J., Budhu A., Yu Z., Forgues M., Roessler S., Ambs S., Chen Y., Meltzer P. S., Croce C. M., Qin L. X., Man K., Lo C. M., Lee J., Ng I. O., Fan J., Tang Z. Y., Sun H. C., Wang X. W. (2009) MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 361, 1437–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang C., Tong J., Huang G. (2013) Nicotinamide phosphoribosyltransferase (Nampt) is a target of microRNA-26b in colorectal cancer cells. PLoS One 8, e69963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X. X., Li X. J., Zhang B., Liang Y. J., Zhou C. X., Cao D. X., He M., Chen G. Q., He J. R., Zhao Q. (2011) MicroRNA-26b is underexpressed in human breast cancer and induces cell apoptosis by targeting SLC7A11. FEBS Lett. 585, 1363–1367 [DOI] [PubMed] [Google Scholar]
- 25. Verghese E. T., Drury R., Green C. A., Holliday D. L., Lu X., Nash C., Speirs V., Thorne J. L., Thygesen H. H., Zougman A., Hull M. A., Hanby A. M., Hughes T. A. (2013) miR-26b is down-regulated in carcinoma-associated fibroblasts from ER-positive breast cancers leading to enhanced cell migration and invasion. J. Pathol. 231, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu N., Zhao X., Liu M., Liu H., Yao W., Zhang Y., Cao S., Lin X. (2011) Role of microRNA-26b in glioma development and its mediated regulation on EphA2. PLoS One 6, e16264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang S., He X. (2011) The role of microRNAs in liver cancer progression. Br. J. Cancer 104, 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yue X., Wang H., Zhao F., Liu S., Wu J., Ren W., Zhu Y. (2012) Hepatitis B virus-induced calreticulin protein is involved in IFN resistance. J. Immunol. 189, 279–286 [DOI] [PubMed] [Google Scholar]
- 29. Arora H., Qureshi R., Park A. K., Park W. Y. (2011) Coordinated regulation of ATF2 by miR-26b in γ-irradiated lung cancer cells. PLoS One 6, e23802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu W., Wang X., Ding X., Li Y., Zhang X., Xie P., Yang J., Wang S. (2012) MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One 7, e34165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paddison P. J., Caudy A. A., Bernstein E., Hannon G. J., Conklin D. S. (2002) Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu Y., Lu Y., Zhang Q., Liu J. J., Li T. J., Yang J. R., Zeng C., Zhuang S. M. (2012) MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 40, 4615–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thiagalingam S., Foy R. L., Cheng K. H., Lee H. J., Thiagalingam A., Ponte J. F. (2002) Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: molecular basis of its occurrence. Curr. Opin. Oncol. 14, 65–72 [DOI] [PubMed] [Google Scholar]
- 34. Li S. P., Wang H. Y., Li J. Q., Zhang C. Q., Feng Q. S., Huang P., Yu X. J., Huang L. X., Liang Q. W., Zeng Y. X. (2001) Genome-wide analyses on loss of heterozygosity in hepatocellular carcinoma in Southern China. J. Hepatol. 34, 840–849 [DOI] [PubMed] [Google Scholar]
- 35. Fabian M. R., Sonenberg N., Filipowicz W. (2010) Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 [DOI] [PubMed] [Google Scholar]
- 36. Shirasu K., Lahaye T., Tan M. W., Zhou F., Azevedo C., Schulze-Lefert P. (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366 [DOI] [PubMed] [Google Scholar]
- 37. Brancaccio M., Menini N., Bongioanni D., Ferretti R., De Acetis M., Silengo L., Tarone G. (2003) Chp-1 and melusin, two CHORD containing proteins in vertebrates. FEBS Lett. 551, 47–52 [DOI] [PubMed] [Google Scholar]
- 38. Ferretti R., Palumbo V., Di Savino A., Velasco S., Sbroggiò M., Sportoletti P., Micale L., Turco E., Silengo L., Palumbo G., Hirsch E., Teruya-Feldstein J., Bonaccorsi S., Pandolfi P. P., Gatti M., Tarone G., Brancaccio M. (2010) Morgana/chp-1, a ROCK inhibitor involved in centrosome duplication and tumorigenesis. Dev. Cell 18, 486–495 [DOI] [PubMed] [Google Scholar]
- 39. Gano J. J., Simon J. A. (2010) A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol. Cell. Proteomics 9, 255–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu J., Luo S., Jiang H., Li H. (2005) Mammalian CHORD-containing protein 1 is a novel heat shock protein 90-interacting protein. FEBS Lett. 579, 421–426 [DOI] [PubMed] [Google Scholar]
- 41. Michowski W., Ferretti R., Wisniewska M. B., Ambrozkiewicz M., Beresewicz M., Fusella F., Skibinska-Kijek A., Zablocka B., Brancaccio M., Tarone G., Kuznicki J. (2010) Morgana/CHP-1 is a novel chaperone able to protect cells from stress. Biochim. Biophys. Acta 1803, 1043–1049 [DOI] [PubMed] [Google Scholar]
- 42. Johnston A. M., Naselli G., Gonez L. J., Martin R. M., Harrison L. C., DeAizpurua H. J. (2000) SPAK, a STE20/SPS1-related kinase that activates the p38 pathway. Oncogene 19, 4290–4297 [DOI] [PubMed] [Google Scholar]
- 43. Hoppe-Seyler K., Sauer P., Lohrey C., Hoppe-Seyler F. (2012) The inhibitors of nucleotide biosynthesis leflunomide, FK778, and mycophenolic acid activate hepatitis B virus replication in vitro. Hepatology 56, 9–16 [DOI] [PubMed] [Google Scholar]
- 44. Liu S. C., Bassi D. E., Zhang S. Y., Holoran D., Conti C. J., Klein-Szanto A. J. (2002) Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. Mol. Carcinog. 34, 131–139 [DOI] [PubMed] [Google Scholar]
- 45. Hong T. J., Kim S., Wi A. R., Lee P., Kang M., Jeong J. H., Hahn J. S. (2013) Dynamic nucleotide-dependent interactions of cysteine- and histidine-rich domain (CHORD)-containing Hsp90 cochaperones Chp-1 and melusin with cochaperones PP5 and Sgt1. J. Biol. Chem. 288, 215–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shang L., Tomasi T. B. (2006) The heat shock protein 90-CDC37 chaperone complex is required for signaling by types I and II interferons. J. Biol. Chem. 281, 1876–1884 [DOI] [PubMed] [Google Scholar]