Abstract
BACKGROUND
We examined the associations of epithelial sodium channel (ENaC) genes with blood pressure (BP) changes and hypertension incidence in a longitudinal family study.
METHODS
A total of 2,755 Han Chinese participants of the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) baseline examination were eligible for this study. The associations of 43 tag single nucleotide polymorphisms (SNPs) in ENaC genes with BP changes and hypertension incidence were assessed using mixed models to account for the correlations of repeated measures among individuals and within families. A genotype by time interaction term was used to model differences in longitudinal BP change according to genotype over time. Gene-based analyses were conducted using the truncated product method. The Bonferroni method was used to adjust for multiple testing in all analyses.
RESULTS
During an average of 7.4 years follow-up, systolic BP (SBP) and diastolic BP (DBP) increased, and approximately 33% of participants developed hypertension. SCNN1A SNP rs11064153 and SCNN1G SNP rs4401050 were significantly associated with longitudinal changes in SBP after adjustment for multiple testing (P interaction = 5.8×10–4 and 0.001, respectively). Similar but nonsignificant trends were observed for the associations between both rs11064153 and rs4401050 and DBP changes (P interaction = 0.024 and 0.005, respectively) and between rs11604153 and hypertension incidence (P = 0.02). Gene-based analyses also supported the overall association of SCNN1G with longitudinal changes in SBP (P = 2.0×10–4).
CONCLUSIONS
Our findings indicated that SCNN1A and SCNN1G may contribute to BP changes over time in the Han Chinese population. Replication of these findings is warranted.
Keywords: blood pressure, blood pressure changes, epithelial sodium channel (ENaC), hypertension, single nucleotide polymorphisms.
High blood pressure (BP) is the leading risk factor for cardiovascular disease and mortality worldwide.1–3 Even small incremental elevations in BP are associated with an increased risk of cardiovascular events.4 Genetic background contributes to inter-individual variation in BP, with heritability estimates generally ranging 31%–68%.5 Genetic studies of BP and hypertension have implicated more than 50 genes in pathways affecting renal sodium balance and other physiological functions.6
The renal epithelial sodium channel (ENaC) mediates renal reabsorption of sodium, playing a pivotal role in the control of sodium balance, blood volume, and BP.7,8 ENaC consists of 3 homologous subunits (α, β, and γ), which are colocalized in the distal tubules and the collecting ducts of the kidney’s nephrons, where the fine control of sodium reabsorption is carried out.9 The α subunit of ENaC, encoded by SCNN1A on 12p13, supports sodium conductance when expressed alone, whereas the β and γ subunits encoded by SCNN1B and SCNN1G on 16p12 appear to have a structural and/or regulatory role when coexpressed with the α subunit.9,10 Multiple mutations in the ENaC β and γ subunits have been related to Liddle syndrome, a monogenic disorder characterized by severe hypertension and low plasma potassium concentrations.11–14 ENaC’s physiological relevance, combined with findings from monogenic study, make the genes encoding ENaC particularly attractive candidates for genetic study of the complex hypertension phenotype. Although numerous studies have reported the association of common variants in ENaC with BP or hypertension,15–17 very few of these candidate gene studies have been conducted in the Han Chinese population.18 Furthermore, none have examined whether variants in ENaC genes can predict BP change or hypertension incidence in longitudinal study. Herein, we aimed to use both single marker–based and gene-based analyses to assess the relationship of ENaC genes with BP change and incident hypertension in 2,755 Han Chinese participants of the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) follow-up study.
METHODS
Study population
GenSalt is a family-based, dietary feeding study that was conducted among 3,142 study participants in rural north China from 2003 to 2005. Details of the study design and methods have been described previously.19 Briefly, a community-based BP screening was carried out among persons aged 18–60 years in the study villages to identify potential probands. Those with mean systolic BP (SBP) of 130–160mm Hg and/or diastolic BP (DBP) of 85–100mm Hg and no use of antihypertensive medication were recruited, as well as their parents, siblings, spouses, and offspring. Persons who had stage 2 hypertension, secondary hypertension, a history of clinical cardiovascular disease or diabetes, or were pregnant, heavy alcohol drinkers, or currently on a low-sodium diet were excluded from the study.
Institutional review boards at all of the participating institutions approved the GenSalt study. Written informed consents for the program were obtained from each participant.
GenSalt baseline data collection
A standard questionnaire was administered by trained staff to collect information on family pedigrees, demographic characteristics, personal and family medical history, and lifestyle risk factors. Three morning BP measurements were obtained according to a standard protocol on each of the 3 days of baseline observation. All BP readings were measured by trained and certified technicians using a random-zero sphygmomanometer.20 BP was measured with the participant in the sitting position after 5 minutes of rest. In addition, participants were advised to avoid alcohol, cigarette smoking, coffee/tea, and exercise for at least 30 minutes before their BP measurements. Mean BP at baseline was calculated as the mean of 9 measurements from the 3-day baseline observation. Body weight and height were measured twice in light indoor clothing without shoes. Body mass index (BMI) was calculated as kilograms per square meter (kg/m2).
GenSalt follow-up
The GenSalt follow-up study included 2 visits, which were conducted from 2008 to 2009 (visit 1) and from 2011 to 2012 (visit 2). During each follow-up visit, a 3-day clinical examination was conducted using the same standardized protocol as that of the baseline examination. Mean BP was calculated as the average of 9 BP measurements during each of the 2 3-day follow-up visits. Hypertension was defined as SBP ≥140mm Hg or DBP ≥90mm Hg or use of antihypertensive medications.
Of 3,142 individuals participating in the initial screening, 341 subjects were lost to follow-up, and an additional 46 were missing genotype data. The remaining 2,755 participants (87.7%) were eligible for our analysis.
Genotype data and quality control
Three ENaC genes (SCNN1A, SCNN1G, and SCNN1B) were selected based on their potential biological effect on BP regulation. Tag single nucleotide polymorphisms (SNPs) from these genes with pairwise r 2 thresholds <0.9 were identified based on linkage disequilibrium structure in the sample using Tagger software (Dr Paul de Bakker, Broad Institute, Cambridge, MA). We also included SNPs that were previously reported to be associated with BP or hypertension. These 17 SNPs were genotyped using SNPlex assays (Applied Biosystems, Foster City, CA), based on oligonucleotide ligation assay for capillary electrophoresis on ABI 3700 DNA Analyzers (Applied Biosystems). To provide better coverage of these candidate genes, we included an additional 42 SNPs genotyped in a subsample of GenSalt follow-up study participants (n = 1,881) on the Affymetrix 6.0 platform (Affymetrix, Santa Clara, CA).19
Quality control, including checks of Mendelian consistency, genotyping call rate, minor allele frequency, and Hardy–Weinberg equilibrium was performed using PLINK software (version 1.05; Dr Sean Purcell, http://pngu.mgh.harvard.edu/~purcell/plink/).21 We excluded SNPs with minor allele frequency <1%, a low genotyping call rate (< 95%), and deviation from Hardy–Weinberg equilibrium after Bonferroni correction for multiple testing. After data quality control, a total of 43 tag SNPs remained in this analysis. Detailed information for these SNPs, including their genomic locations, allele frequencies, call rates, P values for the Hardy–Weinberg equilibrium test, sample sizes, and genotyping platforms, is presented in Supplementary Table S1. Linkage disequilibrium was calculated using the pairwise r 2 correlation between SNPs, as implemented in Haploview (Dr Mark Daly, MIT/Harvard Broad Institute, Cambridge, MA).22
Statistical analysis
Single marker–based association analysis for longitudinal BP changes.
The additive association between genotyped SNPs and longitudinal BP changes were analyzed using a mixed linear regression model to account for the correlations of repeated measures among individuals as well as the correlations of individuals within families.23,24 To examine the association between genotype and longitudinal BP change, a genotype by follow-up time interaction term was included in the model along with the main effects of these variables. Models were additionally adjusted for the fixed effects of age, sex, BMI, and use of antihypertensive medication. For SNPs that showed significant SNP by follow-up time interactions after Bonferroni correction for multiple testing (α threshold = 0.05/43 = 0.001), the average change in BP per year was calculated for each genotype group after adjusting for age, sex, BMI, and use of antihypertensive medication. To determine whether the method of adjustment for antihypertensive medication use may have influenced study findings, we carried out sensitivity analyses using imputed BP levels for participants taking antihypertensive medication based on a recently published equation from 147 randomized trials rather than directly adjusting for medication use.25 Analyses were implemented using the Proc Mixed procedure in SAS (version 9.2; SAS Institute, Cary, NC).
Single marker–based association analysis for incident hypertension.
For the analyses of incident hypertension, 625 participants already diagnosed with hypertension at baseline were excluded. We examined the additive association between each SNP and incident hypertension using a generalized linear mixed model, which permits multilevel modelling when the response variable follows a binary distribution (e.g., incident hypertension).26 Age, sex, BMI, and follow-up time were adjusted in multivariable analyses. A generalized linear mixed model was implemented using the Proc Glimmix procedure in SAS.
Gene-based association analysis for BP changes and incident hypertension.
The truncated product method, which combines P values from single SNP association analyses, was used to evaluate the overall association of a candidate gene with BP changes over time and hypertension incidence.27,28 For BP changes over time, the P value for the genotype by follow-up time interaction term was used, whereas for hypertension incidence the P value of the genotype term was used. The truncation point was set as τ = 0.05, and the P value for the truncated product method was estimated by 10,000 simulations. Sensitivity analyses were conducted using the truncated product method after excluding significant SNPs within a gene to examine their influence on the gene-based analysis. Bonferroni correction was applied to account for multiple testing among the 3 ENaC genes (α threshold = 0.05/3 = 0.017). Gene-based analysis was performed using R software (version 2.15.2; http://www.r-project.org).
RESULTS
The baseline characteristics of study participants are summarized in Table 1. On average, subjects were 48.7 years old and had a mean BMI of 23.2kg/m2, mean SBP of 123.4mm Hg, and mean DBP of 74.0mm Hg at the baseline examination. In addition, approximately 50% of participants were men, 23% had hypertension, and 7% were taking antihypertensive medication. Mean SBP and DBP increased over follow-up. Furthermore, among those 2,130 participants free from hypertension at baseline, approximately 33% developed hypertension during the average 7.4 years of follow-up.
Table 1.
Characteristics of 2,755 Genetic Epidemiology Network of Salt Sensitivity follow-up study participants
| Baseline | Visit 1 | Visit 2 | |
|---|---|---|---|
| (n = 2,755) | (n = 2,650) | (n = 2,461) | |
| Duration of follow-up, y | — | 4.6±0.7 | 7.4±0.5 |
| Age, y | 48.7±15.9 | 53.7±15.8 | 55.6±15.3 |
| Male, % | 50.3 | 50.1 | 49.6 |
| BMI, kg/m2 | 23.2±3.2 | 23.8±3.6 | 24.4±3.6 |
| Blood pressure, mm Hg | |||
| SBP | 123.4±20.1 | 129.2±21.8 | 134.2±20.0 |
| DBP | 74.0±10.7 | 78.7±11.4 | 81.1±11.2 |
| Antihypertensive medication, % | 7.0 | 11.1 | 16.9 |
| Hypertension, % | 22.7 | 39.2 | 56.2 |
| Hypertension incidencea, % | — | 20.3 | 33.4 |
Data are mean ± SD or percentages.
Abbreviations: BMI, body mass index; DBP, diastolic blood pressure; SBP, systolic blood pressure.
aNewly developed hypertension among 2,130 participants who were free from hypertension at baseline.
Figure 1 shows the association of each of the 43 SNPs with BP changes and incident hypertension. SCNN1A SNP rs11064153 and SCNN1G SNP rs4401050 were significantly associated with longitudinal changes in SBP after correcting for multiple comparisons (P interaction = 5.8×10–4 and 0.001, respectively) (Table 2). Although no SNPs were associated with longitudinal changes in DBP and hypertension incidence after adjustment for multiple testing, marker rs11064153 achieved nominal significance for both phenotypes, and rs4401050 achieved nominal significance for longitudinal changes in DBP (Tables 2 and 3). Results of sensitivity analyses using imputed BP for participants taking antihypertensive medication were consistent with the primary study findings. (P interaction for SBP changes = 1.1×10–4 and 5.7×10–4 for rs11064153 and rs4401050, respectively) (data not shown).
Figure 1.
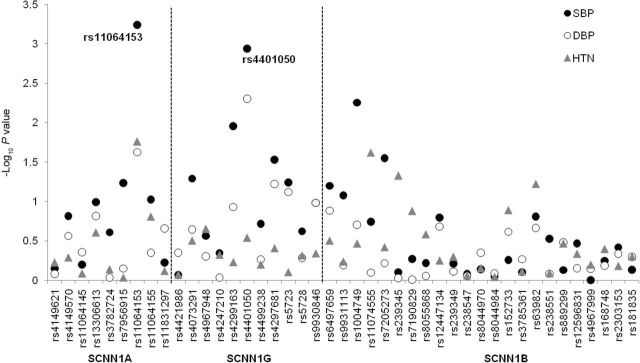
–Log10 P values for the 43 single nucleotide polymorphisms (SNPs) in SCNN1A, SCNN1G, and SCNN1B with longitudinal changes in systolic blood pressure (SBP), diastolic blood pressure (DBP), and hypertension (HTN) incidence. The black and white circles indicate P values for the testing of genotype by follow-up time interactions for SBP and DBP, respectively. The gray triangles indicate P values for the testing of the SNP effect on hypertension incidence. Two labeled SNPs were significantly associated with longitudinal changes in SBP after Bonferroni correction for multiple testing (α threshold = 0.05/43 = 0.001).
Table 2.
Association of variants in epithelial sodium channel genes with blood pressure changes among Genetic Epidemiology Network of Salt Sensitivity participants
| Gene | SNP | Chr | Position | Genotype | SBP | DBP | ||
|---|---|---|---|---|---|---|---|---|
| β (SE) | P interaction value | β (SE) | P interaction value | |||||
| SCNN1A | rs11064153 | 12 | 6488450 | C/C (n = 242) | 1.49 (0.12) | 5.8×10 –4 | 1.00 (0.10) | 0.02 |
| T/C (n = 776) | 1.44 (0.08) | 0.98 (0.06) | ||||||
| T/T (n = 757) | 1.14 (0.07) | 0.82 (0.05) | ||||||
| SCNN1G | rs4401050 | 16 | 23217402 | T/T (n = 28) | 1.64 (0.42) | 0.001 | 1.28 (0.25) | 0.005 |
| C/T (n = 435) | 1.48 (0.12) | 0.89 (0.08) | ||||||
| C/C (n = 2090) | 1.16 (0.06) | 0.77 (0.04) | ||||||
Position was GRCh37.p8. β indicates the average change in BP per year according to genotypes. P interaction is for genotype by follow-up time interactions. P values in boldface indicate statistical significance after Bonferroni correction (0.05/43 = 0.001).
Abbreviations: BP, blood pressure; Chr, chromosome; DBP, diastolic blood pressure; SBP, systolic blood pressure; SNP, single nucleotide polymorphism.
Table 3.
Association of variants in epithelial sodium channel genes with hypertension incidence among Genetic Epidemiology Network of Salt Sensitivity participants
| Gene | SNP | Chr | Position | Maj/Min | MAF | Hypertension incidence | |
|---|---|---|---|---|---|---|---|
| RR (95% CI) | P value | ||||||
| SCNN1A | rs11064153 | 12 | 6488450 | T/C | 0.36 | 1.23 (1.04–1.46) | 0.02 |
| SCNN1G | rs4401050 | 16 | 23217402 | C/T | 0.10 | 1.13 (0.90–1.42) | 0.29 |
Abbreviations: Chr, chromosome; CI, confidence interval; MAF, minor allele frequency; Maj/Min, major allele/minor allele; Position, GRCh37.p8; RR, relative risk: increased risk of hypertension incidence as per minor allele increase; SNP, single nucleotide polymorphism.
Table 2 shows the average change in BP per year according to genotypes of significant SNPs rs11064153 and rs4401050. Compared with those homozygous for the major T allele of SCNN1A marker rs11064153, average increases in SBP over time were larger for those with ≥1 copies of the minor C allele. Mean SBP increases were 1.14, 1.44, and 1.49mm Hg per year for those with rs11064153 genotypes T/T, T/C, and C/C, respectively. Similarly, for SCNN1G marker rs4401050, each copy of the minor T allele was associated with larger increases in SBP, with mean SBP increases of 1.16, 1.48, and1.64mm Hg for genotypes C/C, C/T, and T/T, respectively.
In addition, gene-based analysis found that SCNN1G was significantly associated with longitudinal changes in SBP (P = 2.0×10–4) after Bonferroni correction for multiple testing. SCNN1A also showed nominal significance with SBP changes (P = 0.048). None of the 3 genes showed overall associations with DBP changes or incident hypertension (Table 4). In sensitivity analyses removing the significant markers (rs4401050 in SCNN1G and rs11064153 in SCNN1A), the overall associations of gene-based analyses with SBP changes were attenuated (P = 0.047 and 0.27 for SCNN1G and SCNN1A, respectively).
Table 4.
Gene-based associations of SCNN1A, SCNN1G, and SCNN1B with blood pressure changes and hypertension incidence
| SCNN1A | SCNN1G | SCNN1B | |
|---|---|---|---|
| SBP change | 0.048 | 2.0×10 –4 | 0.14 |
| DBP change | 0.213 | 0.12 | 0.46 |
| HTN incidence | 0.171 | 0.20 | 0.25 |
P values in boldface indicate statistical significance after Bonferroni correction (0.05/3 = 0.02).
Abbreviations: DBP, diastolic blood pressure; HTN, hypertension; SBP, systolic blood pressure.
The pairwise linkage disequilibrium structure of genotyped SNPs in each of the ENaC genes can be found in Supplementary Figure S1.
DISCUSSION
To our knowledge, this is the first investigation to examine the associations of ENaC genes with BP changes and hypertension incidence in the Chinese population. Our study identified 2 novel common variants in the SCNN1A and SCNN1G genes that significantly associated with longitudinal BP changes. Compared with their corresponding major alleles, the SCNN1A rs11604153 and SCNN1G rs4401050 minor alleles predicted larger SBP increases over time. In concordance with findings for SBP change, nominally significant associations were identified for markers rs11604153 and rs4401050 with DBP change and rs11604153 with hypertension incidence. In addition, the gene-based analysis revealed that SCNN1G and SCNN1A were significantly and nominally significantly associated with SBP changes over time, respectively. These findings highlight potentially important contributions of renal sodium-handling genes to long-term BP regulation. Moreover, it adds to our understanding of the genetic architecture of BP progression and hypertension.
Novel SCNN1A marker rs11064153 was associated with longitudinal BP phenotypes in the current analysis. Located in the 5’ flanking region of SCNN1A, marker rs11064153 is a predicted transcription factor binding site29 and may be involved in the regulation of SCNN1A expression. Although rs11064153 or a proxy (r 2 > 0.8) has never previously been associated with BP phenotypes, several other SCNN1A variants have associated with BP traits.15,30,31 For example, similar to our findings, Iwai and colleagues reported the association of a 5’ SCNN1A marker (rs3759324) with cross-sectional BP and hypertension in a Japanese population.15 Unfortunately, neither rs3759324 nor its proxy was genotyped by our study. Using a relatively novel gene-based analysis, we also found a nominally significant association of SCNN1A with SBP changes. However, sensitivity analysis suggested the gene-based association was driven by marker rs11064153. Our findings cumulatively support a role of the SCNN1A gene in BP regulation.
This report provides the first evidence of association of SCNN1G marker rs4401050 with BP changes over time. This marker is located in an intronic region of SCNN1G, with no known function relevance. A significant gene-based association of SCNN1G with SBP change was also identified but could mainly be explained by marker rs4401050. Although our study represents the first report of novel SCNN1G marker rs4401050, previous studies have identified the association of SCNN1G variants with BP phenotypes. For example, we previously reported an association of the rs4299163 minor C allele with increased SBP salt sensitivity in the Han Chinese population.18 Interestingly, this analysis also found the rs4299163 C allele was nominally associated with larger SBP increases over time (P = 0.01). In addition, marker rs4073291 (or a proxy) has been associated with BP and hypertension in a Korean sample,30 as well as with BP salt sensitivity in the Chinese.18 In our report, rs4073291 was also marginally associated with SBP changes over time (P = 0.052). Although our single marker–based and gene-based findings are of some interest, resequencing followed by functional studies will be warranted to determine the causal variant(s) in SCNN1G that predict long-term BP regulation and hypertension.
The SCNN1B gene is located approximately 85kb downstream of SCNN1G. Neither single marker–based analysis nor the gene-based analysis showed significant associations of SCNN1B with BP changes or hypertension incidence. However, some SCNN1B variants have been reported for BP previously. For example, marker rs7205273 was associated with BP in a general Korean population30 and in a Chinese sample with high levels of physical activity,32 suggesting that rs7205273 might influence BP regulation. It is of interest to note that rs7205273 was associated with longitudinal changes in SBP in our analysis (P = 0.03), although it did not maintain significance after adjustment for multiple testing. Further genetic and functional research is still needed to delineate the role of SCNN1B in BP regulation.
There are several strengths of our study. It is the first investigation to examine associations of ENaC genes with BP changes and hypertension incidence. Study attributes, including the recruitment of all Han Chinese participants, should make the analysis robust to population stratification.33 In addition, our study had a high follow-up rate (87.7%), with stringent quality control procedures used during genotyping and measurement of BP and other covariables at each study visit. The power to detect association was further enhanced by using the average of 9 BP measures that were collected at baseline and each follow-up examination, which should have reduced measurement error. However, potential limitations should be addressed. Although our genotyping platform should provide good coverage of common genetic variants,34 some important rare and structural variants in ENaC genes may be missed by our study.
In summary, we provide the first evidence that common variants in SCNN1A and SCNN1G genes were significantly associated with longitudinal changes in BP among a Chinese sample. In concordance with single-marker findings, gene-based analysis revealed potential effects of SCNN1A and SCNN1G on SBP changes. The cumulative evidence highlights that common variants in ENaC genes may play a critical role in long-term BP progression. Further studies are required to replicate these associations and to identify the causal, functional variants in ENaC genes.
SUPPLEMENTARY MATERIAL
Supplementary materials are available at American Journal of Hypertension (http://ajh.oxfordjournals.org).
DISCLOSURE
The authors declared no conflict of interest.
Supplementary Material
ACKNOWELDGMENTS
GenSalt is supported by a cooperative agreement project grant (U01HL072507, R01HL087263, and R01HL090682) from the National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, Maryland. X. Yang is supported by a research training grant (D43TW009107) from the National Institutes of Health John E Fogarty International Center, Bethesda, Maryland.
Dr Shaffer from the Department of Bioinformatics and Biostatistics at Tulane University SPHTM provided statistical consultancy for modeling longitudinal family data.
REFERENCES
- 1. Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, Amann M, Anderson HR, Andrews KG, Aryee M, Atkinson C, Bacchus LJ, Bahalim AN, Balakrishnan K, Balmes J, Barker-Collo S, Baxter A, Bell ML, Blore JD, Blyth F, Bonner C, Borges G, Bourne R, Boussinesq M, Brauer M, Brooks P, Bruce NG, Brunekreef B, Bryan-Hancock C, Bucello C, Buchbinder R, Bull F, Burnett RT, Byers TE, Calabria B, Carapetis J, Carnahan E, Chafe Z, Charlson F, Chen H, Chen JS, Cheng AT, Child JC, Cohen A, Colson KE, Cowie BC, Darby S, Darling S, Davis A, Degenhardt L, Dentener F, Des Jarlais DC, Devries K, Dherani M, Ding EL, Dorsey ER, Driscoll T, Edmond K, Ali SE, Engell RE, Erwin PJ, Fahimi S, Falder G, Farzadfar F, Ferrari A, Finucane MM, Flaxman S, Fowkes FG, Freedman G, Freeman MK, Gakidou E, Ghosh S, Giovannucci E, Gmel G, Graham K, Grainger R, Grant B, Gunnell D, Gutierrez HR, Hall W, Hoek HW, Hogan A, Hosgood HD, 3rd, Hoy D, Hu H, Hubbell BJ, Hutchings SJ, Ibeanusi SE, Jacklyn GL, Jasrasaria R, Jonas JB, Kan H, Kanis JA, Kassebaum N, Kawakami N, Khang YH, Khatibzadeh S, Khoo JP, Kok C, Laden F, Lalloo R, Lan Q, Lathlean T, Leasher JL, Leigh J, Li Y, Lin JK, Lipshultz SE, London S, Lozano R, Lu Y, Mak J, Malekzadeh R, Mallinger L, Marcenes W, March L, Marks R, Martin R, McGale P, McGrath J, Mehta S, Mensah GA, Merriman TR, Micha R, Michaud C, Mishra V, Mohd Hanafiah K, Mokdad AA, Morawska L, Mozaffarian D, Murphy T, Naghavi M, Neal B, Nelson PK, Nolla JM, Norman R, Olives C, Omer SB, Orchard J, Osborne R, Ostro B, Page A, Pandey KD, Parry CD, Passmore E, Patra J, Pearce N, Pelizzari PM, Petzold M, Phillips MR, Pope D, Pope CA, 3rd, Powles J, Rao M, Razavi H, Rehfuess EA, Rehm JT, Ritz B, Rivara FP, Roberts T, Robinson C, Rodriguez-Portales JA, Romieu I, Room R, Rosenfeld LC, Roy A, Rushton L, Salomon JA, Sampson U, Sanchez-Riera L, Sanman E, Sapkota A, Seedat S, Shi P, Shield K, Shivakoti R, Singh GM, Sleet DA, Smith E, Smith KR, Stapelberg NJ, Steenland K, Stockl H, Stovner LJ, Straif K, Straney L, Thurston GD, Tran JH, Van Dingenen R, van Donkelaar A, Veerman JL, Vijayakumar L, Weintraub R, Weissman MM, White RA, Whiteford H, Wiersma ST, Wilkinson JD, Williams HC, Williams W, Wilson N, Woolf AD, Yip P, Zielinski JM, Lopez AD, Murray CJ, Ezzati M, AlMazroa MA, Memish ZA. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012; 380:2224–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. He J, Gu D, Chen J, Wu X, Kelly TN, Huang JF, Chen JC, Chen CS, Bazzano LA, Reynolds K, Whelton PK, Klag MJ. Premature deaths attributable to blood pressure in China: a prospective cohort study. Lancet 2009; 374:1765–1772. [DOI] [PubMed] [Google Scholar]
- 3. Bromfield S, Muntner P. High blood pressure: the leading global burden of disease risk factor and the need for worldwide prevention programs. Curr Hypertens Rep 2013; 15:134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet 2002; 360:1903–1913. [DOI] [PubMed] [Google Scholar]
- 5. Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep 2010; 12:17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basson J, Simino J, Rao DC. Between candidate genes and whole genomes: time for alternative approaches in blood pressure genetics. Curr Hypertens Rep 2012; 14:46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hummler E. Epithelial sodium channel, salt intake, and hypertension. Curr Hypertens Rep 2003; 5:11–18. [DOI] [PubMed] [Google Scholar]
- 8. Rossier BC, Pradervand S, Schild L, Hummler E. Epithelial sodium channel and the control of sodium balance: interaction between genetic and environmental factors. Annu Rev Physiol 2002; 64:877–897. [DOI] [PubMed] [Google Scholar]
- 9. Su YR, Menon AG. Epithelial sodium channels and hypertension. Drug Metab Dispos 2001; 29:553–556. [PubMed] [Google Scholar]
- 10. Burton TJ, O’Shaughnessy KM, Brown MJ. The Epithelial Na Channel as a Determinant of Blood Pressure. Current Hypertension Reviews 2007; 3:45–49. [Google Scholar]
- 11. Bubien JK. Epithelial Na+ channel (ENaC), hormones, and hypertension. J Biol Chem 2010; 285:23527–23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP. Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome. Nat Genet 1995; 11:76–82. [DOI] [PubMed] [Google Scholar]
- 13. Rossi E, Farnetti E, Debonneville A, Nicoli D, Grasselli C, Regolisti G, Negro A, Perazzoli F, Casali B, Mantero F, Staub O. Liddle’s syndrome caused by a novel missense mutation (P617L) of the epithelial sodium channel beta subunit. J Hypertens 2008; 26:921–927. [DOI] [PubMed] [Google Scholar]
- 14. Wang LP, Gao LG, Zhou XL, Wu HY, Zhang L, Wen D, Li YH, Liu YX, Tian T, Fan XH, Jiang XJ, Zhang HM, Hui RT. Genetic diagnosis of Liddle’s syndrome by mutation analysis of SCNN1B and SCNN1G in a Chinese family. Chin Med J (Engl) 2012; 125:1401–1404. [PubMed] [Google Scholar]
- 15. Iwai N, Baba S, Mannami T, Ogihara T, Ogata J. Association of a sodium channel alpha subunit promoter variant with blood pressure. J Am Soc Nephrol 2002; 13:80–85. [DOI] [PubMed] [Google Scholar]
- 16. Hollier JM, Martin DF, Bell DM, Li JL, Chirachanchai MG, Menon DV, Leonard D, Wu X, Cooper RS, McKenzie C, Victor RG, Auchus RJ. Epithelial sodium channel allele T594M is not associated with blood pressure or blood pressure response to amiloride. Hypertension 2006; 47:428–433. [DOI] [PubMed] [Google Scholar]
- 17. Busst CJ, Scurrah KJ, Ellis JA, Harrap SB. Selective genotyping reveals association between the epithelial sodium channel gamma-subunit and systolic blood pressure. Hypertension 2007; 50:672–678. [DOI] [PubMed] [Google Scholar]
- 18. Zhao Q, Gu D, Hixson JE, Liu DP, Rao DC, Jaquish CE, Kelly TN, Lu F, Ma J, Mu J, Shimmin LC, Chen J, Mei H, Hamm LL, He J. Common variants in epithelial sodium channel genes contribute to salt sensitivity of blood pressure: the GenSalt study. Circ Cardiovasc Genet 2011; 4:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The GenSalt Collaborative Research Group. GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens 2007; 21:639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perloff D, Grim C, Flack J, Frohlich ED, Hill M, McDonald M, Morgenstern BZ. Human blood pressure determination by sphygmomanometry. Circulation 1993; 88: 2460–2470. [DOI] [PubMed] [Google Scholar]
- 21. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- 23. Shi G, Rice TK, Gu CC, Rao DC. Application of three-level linear mixed-effects model incorporating gene-age interactions for association analysis of longitudinal family data. BMC Proc 2009; 3:S89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singer JD. Using SAS PROC MIXED to fit multilevel models, hierarchical models, and individual growth models. J Educ Behav Stat 1998; 23:323–355. [Google Scholar]
- 25. Law MR, Morris JK, Wald NJ. Use of blood pressure lowering drugs in the prevention of cardiovascular disease: meta-analysis of 147 randomised trials in the context of expectations from prospective epidemiological studies. BMJ 2009; 338:b1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schabenberger O. Introducing the GLIMMIX procedure for generalized linear mixed models. SUGI 30:Paper 196–30. [Google Scholar]
- 27. Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol 2002; 22:170–185. [DOI] [PubMed] [Google Scholar]
- 28. Sheng X, Yang J. Panel unit root test by combining dependent p-values: a comparative study. J Prob Stat 2011; 2011:617652. [Google Scholar]
- 29. Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 2009; 37:W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin HS, Hong KW, Lim JE, Hwang SY, Lee SH, Shin C, Park HK, Oh B. Genetic variations in the sodium balance-regulating genes ENaC, NEDD4L, NDFIP2 and USP2 influence blood pressure and hypertension. Kidney Blood Press Res 2010; 33:15–23. [DOI] [PubMed] [Google Scholar]
- 31. Xu H, Li NF, Hong J, Zhang L, Zhou L, Li T, Ou Yang WJ, Cheng QY. Relationship between four single nucleotide polymorphisms of epithelial sodium channel alpha subunit gene and essential hypertension of Kazakhs in Xinjiang. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2009; 31:740–745. [DOI] [PubMed] [Google Scholar]
- 32. Montasser ME, Gu D, Chen J, Shimmin LC, Gu C, Kelly TN, Jaquish CE, Rice T, Rao DC, Cao J, Liu DP, Whelton PK, He J, Hixson JE. Interactions of genetic variants with physical activity are associated with blood pressure in Chinese: the GenSalt study. Am J Hypertens 2011; 24:1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gu D, Kelly TN, Hixson JE, Chen J, Liu D, Chen JC, Rao DC, Mu J, Ma J, Jaquish CE, Rice TK, Gu C, Hamm LL, Whelton PK, He J. Genetic variants in the renin-angiotensin-aldosterone system and salt sensitivity of blood pressure. J Hypertens 2010; 28:1210–1220. [PMC free article] [PubMed] [Google Scholar]
- 34. Nishida N, Koike A, Tajima A, Ogasawara Y, Ishibashi Y, Uehara Y, Inoue I, Tokunaga K. Evaluating the performance of Affymetrix SNP Array 6.0 platform with 400 Japanese individuals. BMC Genomics 2008; 9:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


