Key Points
BCR activation enhances eIF4A m7GTP cap-binding.
The 5′UTR of CARD11 suppresses protein translation.
Abstract
Human diffuse large B-cell lymphomas (DLBCLs) often aberrantly express oncogenes that generally contain complex secondary structures in their 5′ untranslated region (UTR). Oncogenes with complex 5′UTRs require enhanced eIF4A RNA helicase activity for translation. PDCD4 inhibits eIF4A, and PDCD4 knockout mice have a high penetrance for B-cell lymphomas. Here, we show that on B-cell receptor (BCR)-mediated p70s6K activation, PDCD4 is degraded, and eIF4A activity is greatly enhanced. We identified a subset of genes involved in BCR signaling, including CARD11, BCL10, and MALT1, that have complex 5′UTRs and encode proteins with short half-lives. Expression of these known oncogenic proteins is enhanced on BCR activation and is attenuated by the eIF4A inhibitor Silvestrol. Antigen-experienced immunoglobulin (Ig)G+ splenic B cells, from which most DLBCLs are derived, have higher levels of eIF4A cap-binding activity and protein translation than IgM+ B cells. Our results suggest that eIF4A-mediated enhancement of oncogene translation may be a critical component for lymphoma progression, and specific targeting of eIF4A may be an attractive therapeutic approach in the management of human B-cell lymphomas.
Introduction
During B-lymphocyte development, antigen engagement of the B-cell receptor (BCR) propagates signals promoting cell growth, survival, and maturation. Naïve follicular B cells express membrane-bound immunoglobulins of the immunoglobulin (Ig)M and IgD isotype, which rely on the associated Igα and Igβ for signal transduction.1,2 On T cell-dependent antigen-induced activation within the germinal center (GC), most B cells undergo isotype switching, mainly to IgG, and differentiate into Ig-secreting plasma cells or memory B cells. T-cell help is often mediated through CD40, which serves as a costimulatory signal for B-cell growth, somatic hypermutation, and isotype switching.3 GC-B and memory B cells that express IgG, which has a much longer cytoplasmic tail than IgM and IgD, display more robust signaling on BCR activation, including higher levels of mitogen-activated protein kinase and calcium signaling, than naïve IgM-expressing B cells,1,2,4 although the precise mechanistic differences between IgG- and IgM-mediated signals are still debated.5 Diffuse large B-cell lymphomas (DLBCLs), representing the most common lymphoid malignancy in adults,6 derive from antigen-experienced B cells. DLBCLs with a more aggressive phenotype often commandeer BCR signaling in an antigen-independent fashion,7 and the BCR isotype is well correlated with B-cell lymphoma subtypes and disease progression.8 Although several oncogenes are often dysregulated in DLBCL, the mechanism that links BCR signaling, elevated oncoprotein expression, and lymphoma progression remains unknown.
Signaling through the BCR triggers the activation of downstream pathways including Ras/Raf/MEK/Erk, AKT/MTOR, and p70s6K.9,10 MTORC1 phosphorylates and releases eIF4E-binding proteins (4EBPs), allowing eIF4E to bind eIF4G and the mRNA cap.11 Another substrate of MTORC1 is p70s6K, which enhances the RNA helicase activity of eIF4A by phosphorylating EIF4B, allowing it to subsequently bind eIF4A.12-14 p70s6K also phosphorylates the eIF4A inhibitor PDCD4,15 which results in its βTRCP-mediated ubiquitination and degradation, freeing eIF4A for mRNA binding and cap-complex formation.16 Significantly, 86% of PDCD4−/− mice succumb to B-cell lymphomas.17 Furthermore, gene expression analysis of different B-cell compartments demonstrated that GC B cells have markedly repressed PDCD4 expression.18 A recent study demonstrated that the eIF4A cofactor, EIF4B, is overexpressed in DLBCL and results in the selective upregulation of mRNAs encoding antiapoptotic and DNA repair proteins.19 Conversely, the eIF4A inhibitors Hippuristanol and Silvestrol have shown a large degree of B-cell specificity and promise in treating B-cell malignancies.20-22
Genes involved in proliferation, including oncogenes c-Myc and CCND2, are enriched for complex 5′ untranslated regions (UTRs), whereas housekeeping genes generally harbor less complex 5′UTRs. Interestingly, oncogenes containing complex 5′UTRs, CARD11, BCL10, and MALT1 (referred to collectively as the CBM complex), are predicted to have ΔG (Gibb's free energy) values of −174, −258, and −156, respectively.23 These values have been reported to suppress translation initiation.24 Given that the CBM complex is required for the coupling of BCR signaling to nuclear factor (NF)-κB signaling after antigen exposure, we explored whether these genes are subject to regulation at the level of translation and, specifically, whether they are regulated by eIF4A, which is required for translation of mRNA with such highly complex 5′UTR structures.25-28
Here we demonstrate a hitherto unrecognized link between BCR activation and eIF4A-dependent upregulation of translation. We also show that oncogenes implicated in DLBCL development, such as CARD11, BCL10, and MALT1, contain complex 5′UTRs. Expression of these short-lived proteins is sensitive to eIF4A inhibition in both primary and malignant B cells. Our data provide a compelling rationale for the use of eIF4A pharmacological agents in B-cell lymphoma and other lymphoid diseases, characterized by unrestricted BCR activation and enhanced dependency on the CBM complex and NF-κB activation.
Methods
Human splenic B-cell isolation and cell culture
Human splenic B cells from otherwise healthy motor vehicle collision patients were provided by the R. Adams Cowley Shock Trauma Center and University of Maryland Greenebaum Cancer Center Pathology Biorepository and Research Core in accordance with the guidelines of the University of Maryland Medical School Institutional Review Board and conform to the Declaration of Helsinki. Spleens were minced on ice, and lymphocytes were separated by Ficoll and viably frozen in 95% fetal bovine serum (FBS) and 5% dimethylsulfoxide. B-cell populations were isolated through negative selection using a B-cell Isolation Kit (MiltenyiBiotec) per the manufacturer’s protocol. Purified lymphocytes were washed in cold Hanks balanced salt solution containing 5% FBS and stained with CD19-allophycocyanin (APC) and CD3-fluorescein isothiocyanate (15 minutes, on ice; BD Biosciences and Biolegend). Cells were analyzed using a FACSCanto II (BD Biosciences) flow cytometer and determined to be CD19+/CD3− with purity >95%. Human splenic B cells were cultured in RPMI containing 10% FBS, 1% penicillin/streptomycin (Pen/Strep), and 1% amphotericin-B. SUDHL4 and Toledo cells were purchased from ATCC and cultured in RPMI containing 10% FBS and 1% Pen/Strep. OCI-Ly10 cells were a kind gift from Dr Pasqualucci and cultured in Iscove modified Dulbecco medium containing 20% human serum and 1% Pen/Strep.
B-cell activation assay
After negative selection, 2.5 × 106 cells/mL human splenic B cells were treated with mouse IgG1κ (isotype control; eBioscience), anti-human IgA+IgG+IgM(H+L) (anti-BCR; Jackson ImmunoResearch), anti-CD40 clone 5C3 (anti-CD40; eBioscience), or anti-BCR + anti-CD40 for 24 hours.
Drug treatment
Human splenic B cells were treated with isotype control or anti-BCR antibodies as described under B-cell activation assay. B cells were then simultaneously treated with either vehicle control (dimethylsulfoxide) or 10 nM Silvestrol (MedChem Express) for 24 hours.
Purification of human B cells with flow cell sorting
Viably frozen human splenic lymphocytes from patients between the ages of 41 and 88 years were thawed and stained with CD19-APC (BD Biosciences), IgG-Brilliant Violet (BD Biosciences), and IgM-phycoerythrin (eBioscience) in Hanks balanced salt solution containing 5% FBS. Cell sorting was performed as described in the supplemental Methods available on the Blood Web site.
Fluorescence-activated cell sorter analysis for B-cell activation and viability
For B-cell activation, human splenic B cells were stained with CD69-fluorescein isothiocyanate (eBioscience, 15 minutes, on ice) 24 hours after activation. For viability, SUDHL4 cells were stained with propidium iodide (PI) (Sigma-Aldrich, 15 minutes, on ice) 24 hours after activation and treatment. Acquisition was performed on a FACSCantoII (BD Biosciences) flow cytometer, and data were analyzed with FlowJo (TreeStar, Inc.).
Isotype specific B-cell activation assay
After flow sorting, 2.5 × 106 cells/mL of IgG+/IgM− and IgG−/IgM+ B cells were treated with the following conditions: anti-goat control, 2 μg/mL F(ab′)2 donkey anti-goat IgG (H+L), goat anti-human IgG + anti-goat: 2.5 μg/mL goat anti-human IgG Fcγ, or goat anti-human IgM + anti-goat: 2.5 μg/mL goat anti-human IgM Fc5μ (Jackson ImmunoResearch). Initial viability of sorted B cells was 82%, and viability of activated cells was 75% to 80% in all treatment regimens (supplemental Figure 4B).
Western blotting
Western blotting was performed as described in the supplemental Methods.
7-Methyl-GTP Sepharose 4B pull-down
7-methyl-GTP Sepharose 4B pull-downs were performed as described in the supplemental Methods.
Lentiviral 5′UTR reporter generation, viral production, and luciferase assay
5′UTR assays were performed as previously described29,30 (supplemental Methods).
Polysome preparation
Polysomes were performed as previously described31 (supplemental Methods).
Trichloroacetic acid precipitation
Trichloracetic acid precipitation was performed as described in the supplemental Methods.
Protein stability assay
Protein stability was performed as described in the supplemental Methods.
RNA isolation and reverse transcription
Quantitative reverse transcription-polymerase chain reaction was performed as described in the supplemental Methods. using primers shown in supplemental Table 1.
In vitro assessment of global protein translation
Analysis of de novo translation was carried out as previously described32 (supplemental Methods).
Bioinformatics analysis of 5′UTR sequences
BCR-related genes (supplemental Table 2) were evaluated for the complexity of their 5′UTRs using DINAMelt as previously described.23
Statistical analysis
Results were analyzed using Microsoft Excel. P values were calculated using a paired Student t test. Statistical significance was determined when P < .05 was attained.
Results
BCR activation enhances protein translation in human splenic B cells
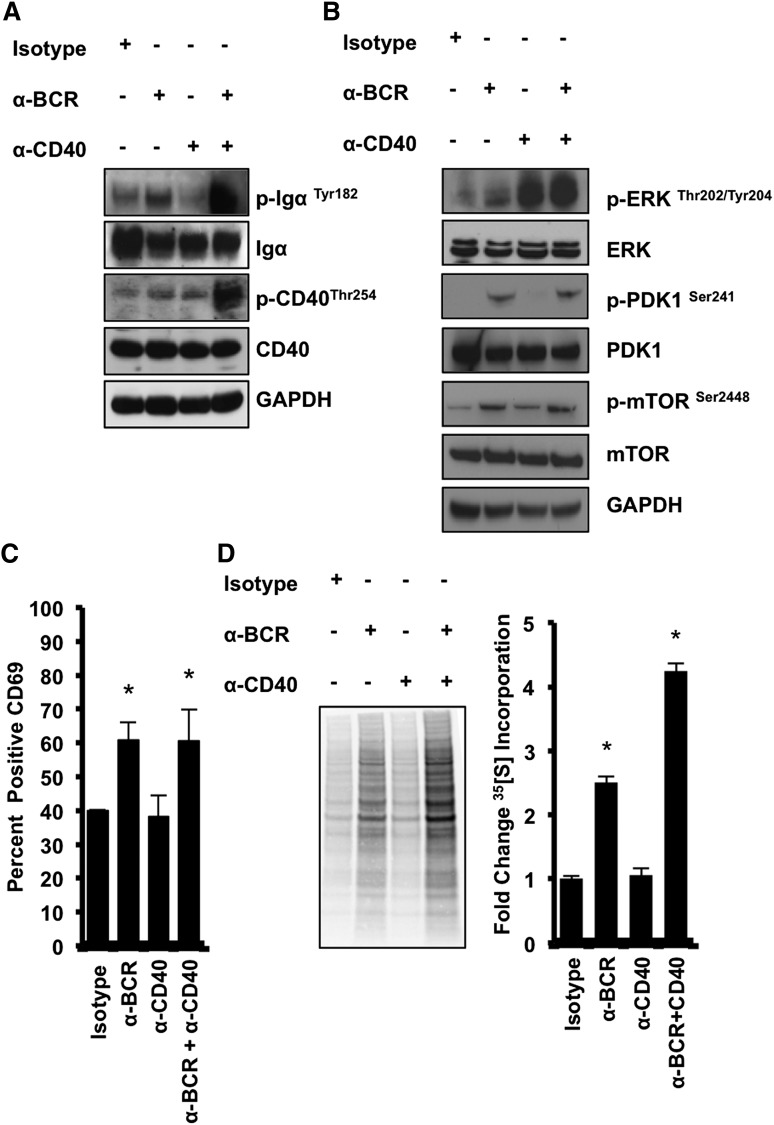
We activated human splenic B cells with antibodies against BCR, CD40, or BCR and CD40 together. Anti-BCR treatment mimics antigen-specific stimulation, whereas CD40 engagement serves as a costimulatory signal typical for T cell-dependent B-cell activation.3 On activation by anti-BCR, with or without anti-CD40, we observed enhanced phosphorylation of Igα at Tyr182, whereas we only observed an increase in CD40 phosphorylation at Thr254 under anti-BCR+anti-CD40 conditions (Figure 1A; supplemental Figure 1A). Regulators of protein translation were analyzed for Erk phosphorylation at Thr202/Tyr204, PDK1 phosphorylation at Ser241, and MTOR phosphorylation at Ser2448 (Figure 1B), as these are activating phosphorylation events, and collectively, these kinases result in full activation of p70s6K.33 CD40 activation enhances extracellular signal-regulated kinase (ERK) activation (Figure 1B; supplemental Figure 1B), which is consistent with previous reports,34 whereas anti-BCR induces phosphorylation of both PDK1 and MTOR (Figure 1B). The activation marker CD69 is upregulated under anti-BCR conditions (Figure 1C).35 A significant increase in global protein synthesis was observed in B cells activated by BCR stimulation relative to control cultures and B cells stimulated by anti-CD40 alone, as demonstrated by 35S-radiolabeled amino acid incorporation (Figure 1D), whereas polysome distribution profiles showed enhanced polysome formation on stimulation by anti-BCR or anti-CD40 (supplemental Figure 1C-E). Together, these results demonstrate that BCR activation enhances global protein translation.
Figure 1.
BCR activation enhances protein translation in human splenic B cells. (A) Protein lysates from human splenic B cells were harvested 30 minutes after treatment with either Isotype, anti-BCR, anti-CD40, or anti-BCR+CD40 and then analyzed by western blot analysis for p- Igα (Tyr182), total Igα, p-CD40 (Thr254), and total CD40. (B) Lysates were harvested and analyzed for p-ERK (Thr202/Tyr204), total ERK, p-PDK1 (Ser241), total PDK1, p-MTOR (Ser2448), and total MTOR. GAPDH served as a loading control, and data are representative of 3 independent experiments. (C) Human splenic B cells were harvested, washed, and stained for CD69 24 hours after activation. Fluorescence-activated cell sorter analysis was used to quantify the surface CD69 expression. Data are means ± standard error of the mean (SEM) of 3 independent experiments. (D) Cells were incubated with 1 mCi L-[35S]methionine and l-[35S]cysteine 24 hours after treatment, and lysates were harvested and analyzed for radiolabel incorporation. Densitometry was used to quantify radiolabel incorporation, and data are means ± SEM of 3 different experiments (3 individual spleens; *P < .05).
BCR activation significantly enhances eIF4A cap-binding activity in human splenic B cells
To determine how BCR activation affects the p70s6K-eIF4A axis, lysates were harvested after B-cell activation and analyzed by western blot. Increased p70s6K phosphorylation at Thr389, which is carried out by MTOR, as well as phosphorylation of p70s6K substrates RPS6, PDCD4, and EIF4B, suggest that the p70s6K axis is activated under anti-BCR conditions, consistent with previous studies10 (Figure 2A). Additionally, we observed a reduction in PDCD4 under anti-BCR conditions, suggesting that, on phosphorylation, PDCD4 is ubiquitinated and degraded as previously reported16 (Figure 2A). Phosphorylation of the inhibitor of eIF4E, 4EBP1, at Thr36/37 was enhanced when B cells were stimulated with both anti-BCR and anti-CD40 (Figure 2A, lane 4). This may be due to increased MTORC1 activity on BCR stimulation (Figure 1B), although in addition to MTOR, PIM2, which is expressed in B cells regardless of activation status (Figure 2A), can also phosphorylate 4EBP1. Protein lysates were incubated with m7GTP Sepharose beads and analyzed for eIF4E, eIF4G, and eIF4A cap-binding activity. We observed enhanced eIF4A cap-binding activity specifically under anti-BCR conditions, with no change in eIF4E cap-binding activity. Cap-binding by eIF4G was enhanced both in anti-BCR and anti-CD40 conditions (Figure 2B). Total eIF4E protein expression (Figure 2B; supplemental Figure 2) increased only slightly under anti-BCR conditions. This observation is consistent with previous studies demonstrating increased eIF4E protein expression on lymphocyte activation.36 Together, these data demonstrate that eIF4A cap-binding activity and p70s6K signaling is rapidly enhanced on BCR activation.
Figure 2.
BCR activation significantly enhances eIF4A cap-binding activity in human splenic B cells. (A) Protein lysates from human splenic B cells were harvested 24 hours after treatment with Isotype, anti-BCR, anti-CD40, or anti-BCR+CD40 and then analyzed by western blot analysis for p-4EBP1 (Thr36/37), total 4EBP1, PIM2, p-p70s6K (Thr389), total p70s6K, p-EIF4B (Ser406), total EIF4B, p-RPS6 (Ser235/236), RPS6, p-PDCD4 (Ser67), and total PDCD4. GAPDH served as a loading control, and data are representative of 3 independent experiments. (B) Lysates were incubated with m7GTP Sepharose beads overnight at 4°C. Western blot analysis was used to observe eIF4A, eIF4G, and eIF4E binding to m7GTP. Data are representative of 3 independent experiments (3 individual spleens).
Silvestrol reduces eIF4A cap-binding activity, protein translation, and oncoprotein expression in activated human splenic B cells
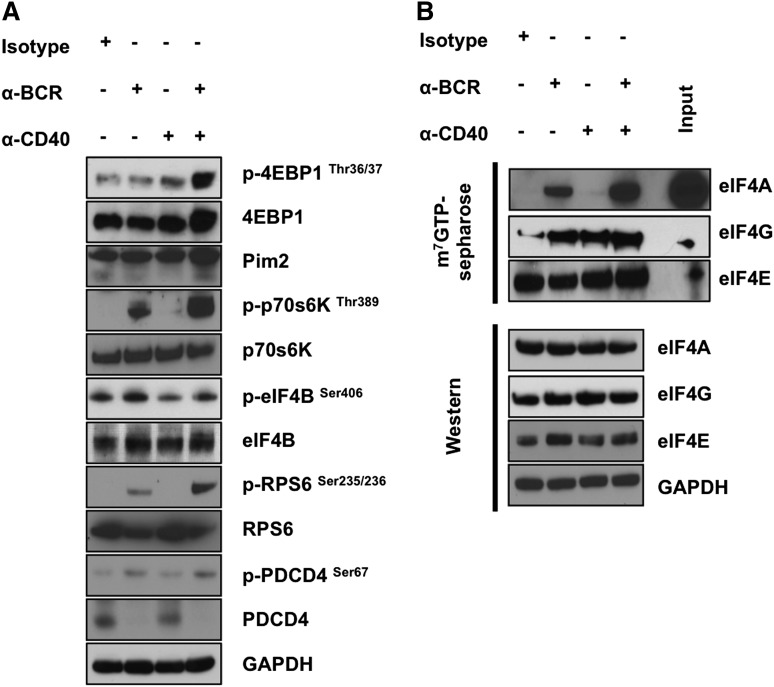
To explore the therapeutic potential for eIF4A inhibition in the context of BCR activation, we treated human splenic B cells with either an isotype control or anti-BCR antibody and simultaneously treated cells in both conditions with either vehicle or 10 nM of the eIF4A inhibitor Silvestrol. We used a dose of 10 nM of Silvestrol for 24 hours, which is a previously established dose and time point for inhibiting protein translation without significantly inducing apoptosis in primary chronic lymphocytic leukemia cells.22,37 Silvestrol reduced eIF4A cap-binding under anti-BCR conditions (Figure 3A), as well as eIF4E total protein levels and eIF4E cap-binding under all conditions. Silvestrol also attenuated global protein translation on BCR activation, shown either through mRNA polysomal loading and 80S formation (supplemental Figure 3B-C) or by 35S-radiolabeled amino acid incorporation into nascent proteins (Figure 3B). Treatment with Silvestrol effectively reduced BCR-induced radiolabel incorporation back to levels comparable to isotype plus vehicle treatment (Figure 3B).
Figure 3.
Silvestrol reduces eIF4A cap-binding activity, protein translation, and oncoprotein expression in activated human splenic B cells. (A) Protein lysates from human splenic B cells were harvested 24 hours after treatment with Isotype + Vehicle, Isotype + 10 nM Silvestrol, anti-BCR + Vehicle, or anti-BCR + 10 nM Silvestrol and incubated with m7GTP Sepharose beads overnight at 4°C. Western blot analysis was used to observe eIF4A and eIF4E binding to m7GTP. Data are representative of 3e independent experiments. (B) Twenty-four hours after treatment, cells were incubated with 1 mCi L-[35S]methionine and l-[35S]cysteine, and lysates were harvested and analyzed for radiolabel incorporation. Densitometry was used to quantify radiolabel incorporation, and data are means ± SEM of 3 different experiments. (C) Protein levels of CARD11, BCL10, MALT1, c-Myc, and CCND2 were measured by western blotting. GAPDH served as a loading control. (D) Densitometry was used to quantify protein expression, and data are means ± SEM of 3 independent experiments (3 individual spleens). (E) Cells were fractionated through sucrose gradients, and the relative distribution of CARD11 (normalized to GAPDH) was analyzed by quantitative reverse transcription-polymerase chain reaction analysis of RNA in each of the 10 gradient fractions. Data are representative of 2 independent experiments (2 individual spleens; *P < .05).
Throughout the course of our studies, we monitored the expression of several components of the BCR signaling pathway and found that CARD11, BCL10, and MALT1, which are considered proto-oncogenes, are enhanced at the protein level on BCR activation (Figure 3C). Similar to other proto-oncogenes such as CCND2 and c-Myc, CBM complex protein levels are enhanced on BCR activation and are more sensitive to eIF4A inhibition than housekeeping genes such as GAPDH (Figure 3C-D). For CARD11, the change in protein expression could not be accounted for by enhanced mRNA expression, as total mRNA levels of CARD11 did not change on activation and actually increased in splenic B cells treated with Silvestrol (supplemental Figure 3A). Instead, we observed enhanced loading of CARD11 mRNA to the higher molecular weight polysomal fractions (numbers 8-10) on activation, which could be reduced on treatment with Silvestrol (Figure 3E). Together, these data suggest that pharmacological inhibition of eIF4A-mediated translation on BCR activation potently reduces the expression of oncoproteins including CARD11, BCL10, and MALT1; specifically, CARD11 translation is enhanced on BCR activation.
eIF4A cap-binding activity and protein translation is higher in IgG-expressing and IgG-activated B cells
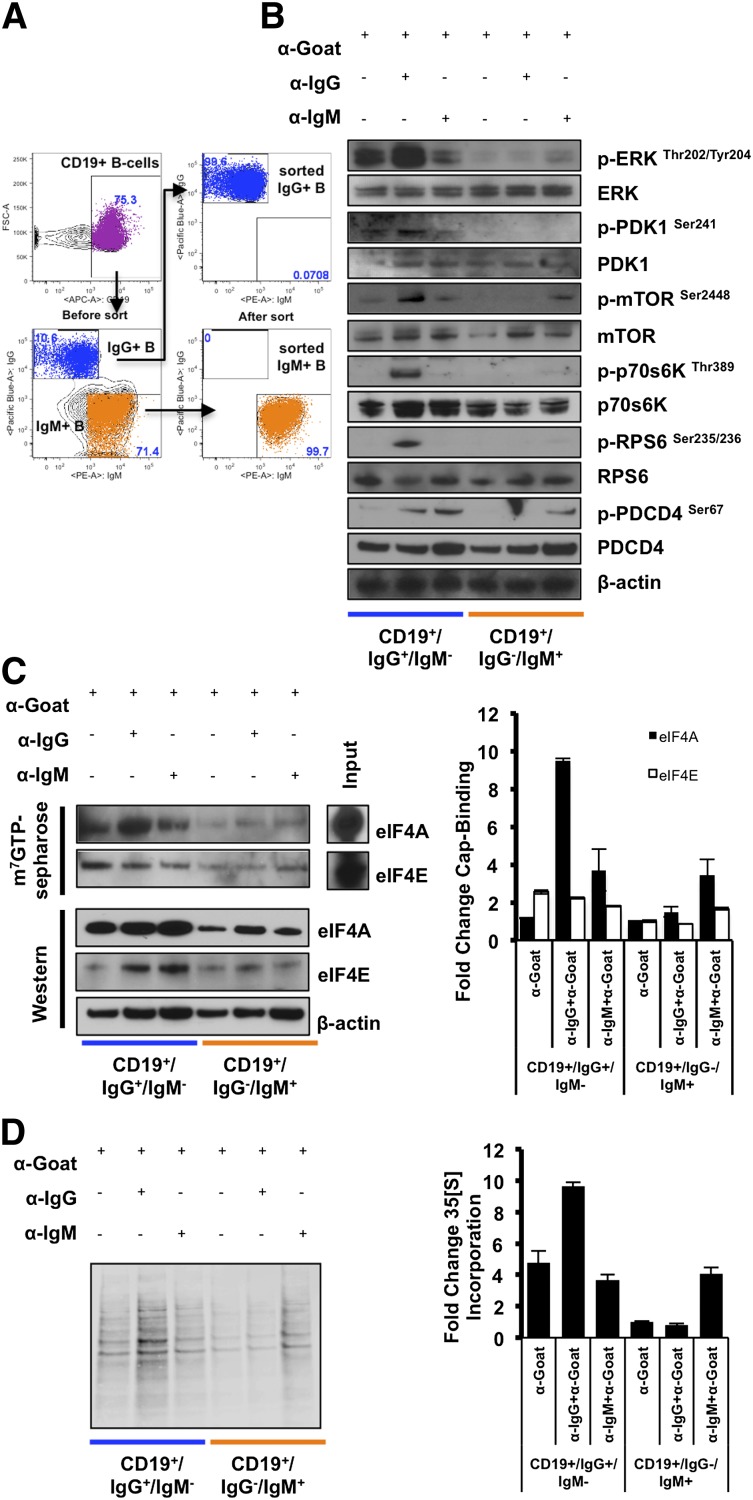
Stimulation through IgG and IgM produces qualitatively different BCR signals. IgG is expressed on memory and a subset of GC B cells, whereas IgM is mainly expressed on naïve and marginal zone B cells.1 To determine whether the different signaling capacities of IgG and IgM are also reflected in different translational activation, we sorted human splenic B cells for IgG+/IgM− or IgG−/IgM+ subpopulations (Figure 4A). Neither the sorting nor the staining procedure appears to activate p70s6K signaling in splenic lymphocytes (supplemental Figure 4A, lanes 1-4). IgG+/IgM− and IgG−/IgM+ B cells were sorted with similar initial viabilities (82 ± 2.5% and 76 ± 3%, respectively), as assessed by trypan blue exclusion. Equal numbers of cells of each population were treated with either anti-IgG Fcγ-specific or anti-IgM Fcμ-specific antibodies followed by secondary antibody cross-linking 30 minutes after primary antibody incubation. After 24 hours of treatment, cell viability remained comparable under all conditions (supplemental Figure 4B). We observed enhanced ERK phosphorylation in IgG+ B cells under both basal conditions (Figure 4C, lanes 1 and 3) and anti-IgG activation (lane 2) compared with IgM+ B cells (lanes 4-6), consistent with previous reports.1 Additionally, we observed substantially more enhanced phosphorylation of PDK1, MTOR, p70s6K, and RPS6 in activated IgG+ B cells compared with activated IgM+ B cells (Figure 4B, lane 2), although we only observed slightly enhanced phosphorylation of PDCD4 and did not observe any significant downregulation of total PDCD4 in stimulated cells. Furthermore, eIF4A binding to the m7GTP cap was more enhanced in IgG+ B cells under both basal conditions (Figure 4C, lanes 1 and 3) and activation (lane 2) compared with IgM+ B cells (lanes 4-6), whereas IgG+ B cells also had higher protein expression of eIF4A and eIF4E than IgM+ B cells. Consistent with these findings, we observed that IgG+ B cells are roughly fivefold more translationally active under basal conditions compared with IgM+ B cells, as demonstrated by 35S-radiolabeled amino acid incorporation (Figure 4D, lanes 1 and 4), and global protein synthesis further increased in activated IgG+ B cells (Figure 4D, lane 2) and in activated IgM+ B cells (lane 6). Importantly, Ig isotype-specific BCR activation of IgG+ B cells enhanced CARD11 expression, in a manner that can be attenuated by treatment with Silvestrol, whereas in IgM+ B cells, CARD11 expression was only reduced by Silvestrol in resting but not activated B cells (supplemental Figure 4C). Together, these results demonstrate that resting IgG+ human splenic B cells have more eIF4A activity and protein translation compared with IgM+ B cells, which is further enhanced on BCR activation, inducing higher activity of the p70s6K-eIF4A axis in IgG+ B cells compared with IgM+ B cells.
Figure 4.
eIF4A cap-binding activity and protein translation is higher in IgG-expressing and IgG-activated B cells. (A) Human splenic lymphocytes isolated from motor vehicle collision patients between the ages of 41 and 88 years were sorted for IgG high/IgM low or IgG low/IgM high CD19+ B-cell populations. (B) Each population was treated with no treatment, goat anti-human IgG Fcγ, or goat anti-human IgM Fc5μ. Isotype-specific activation was then induced 30 minutes after primary antibody incubation by treating all conditions with anti-goat IgG. Twenty-four hours after activation protein lysates were harvested and analyzed for p-ERK (Thr202/Tyr204), total ERK, p-PDK1 (Ser241), total PDK1, p-MTOR (Ser2448), total MTOR, p-p70s6K (Thr389), total p70s6K, p-RPS6 (Ser235/236), RPS6, p-PDCD4 (Ser67), and total PDCD4. β-Actin served as a loading control, and data are representative of 2 independent experiments. (C) Twenty-four hours after activation, protein lysates were incubated with m7GTP Sepharose beads overnight at 4°C, and western blot was used for analysis of eIF4E and eIF4A cap-binding activity. Densitometry was used to quantify protein expression, and data are means ± SEM of 3 independent experiments. (D) Twenty-four hours after isotype-specific activation, cells were incubated with 1 mCi L-[35S]methionine and l-[35S]cysteine, and lysates were harvested and analyzed for radiolabel incorporation. Densitometry was used to quantify radiolabel incorporation, and data are means ± SEM of 3 different experiments.
CARD11, BCL10, and MALT1 have short protein half-lives, complex 5′UTRs, and are sensitive to eIF4A inhibition
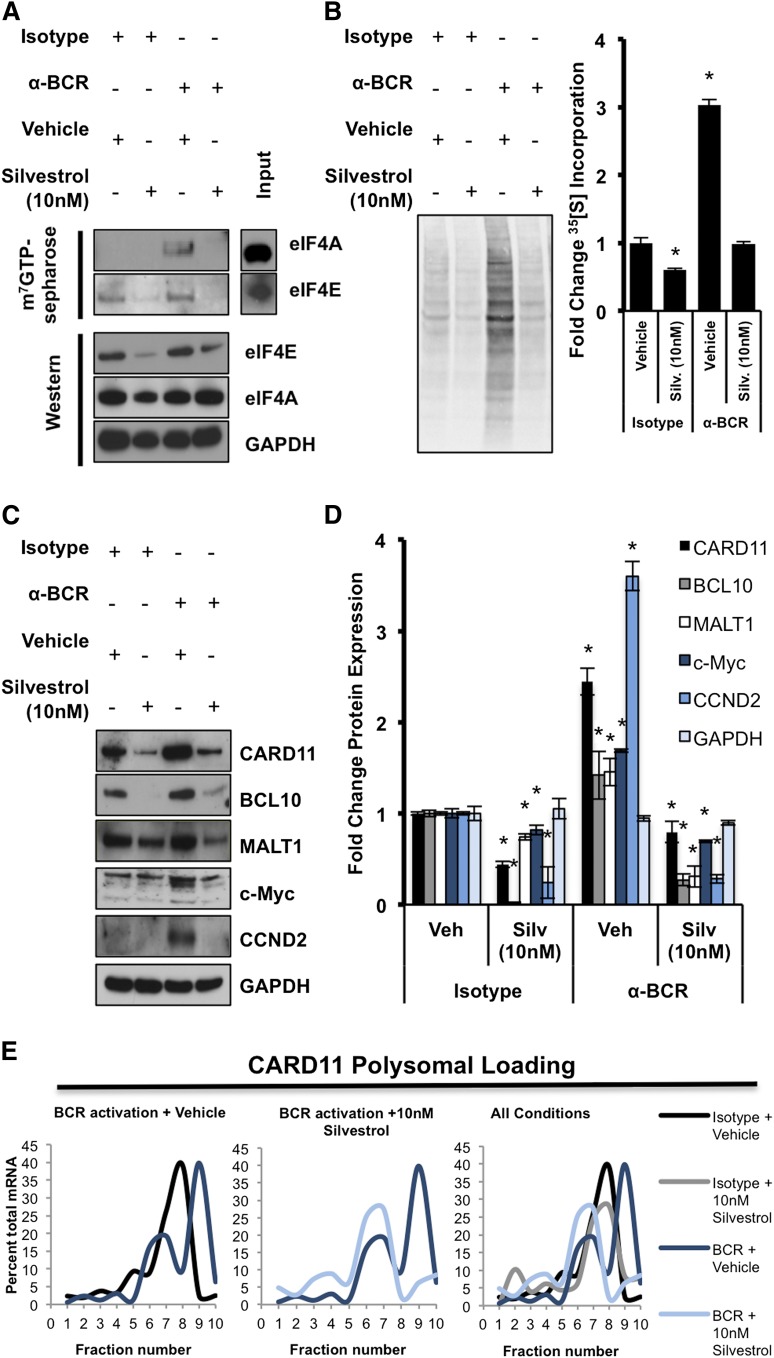
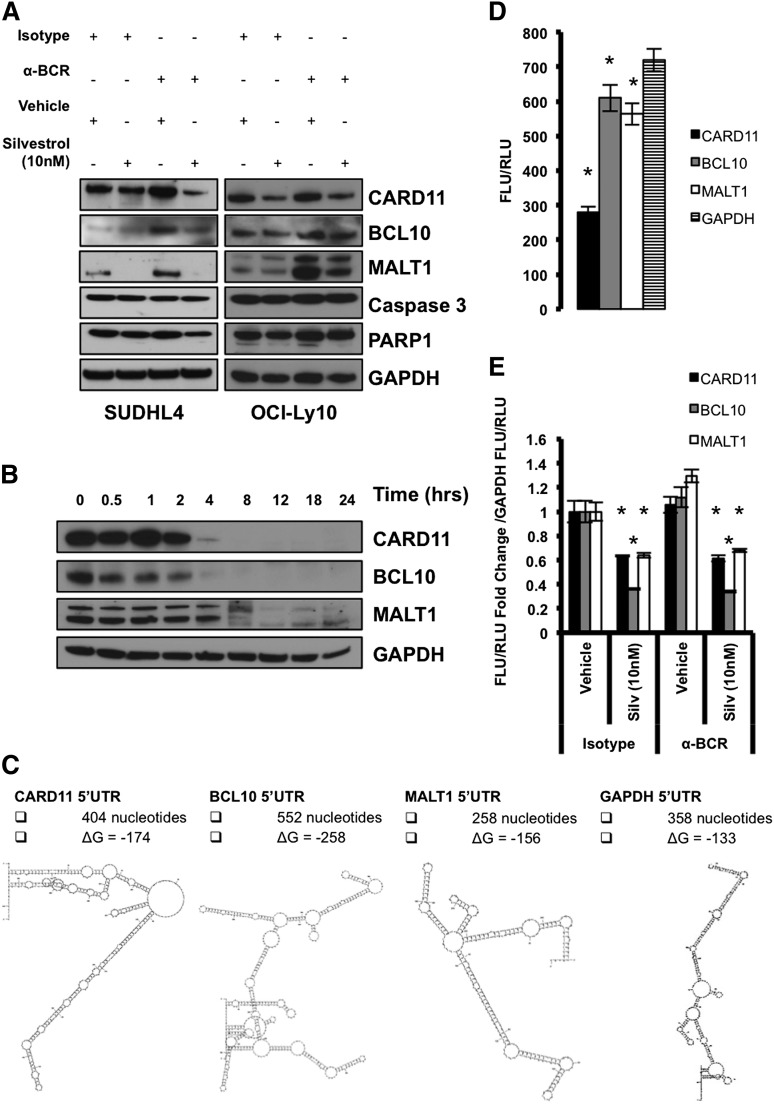
Because the 5′UTRs of CARD11, BCL10, and MALT1 are highly complex (Figure 5C) and their translation is dependent on enhanced eIF4A activity, their role in regulation of protein translation initiation was tested directly using luciferase reporters. Due to the limitations of transient transfection of human splenic B cells, we instead used the DLBCL cell lines SUDHL4 and OCI-Ly10, which respond to antigen stimulation, as well as the DLBCL cell line Toledo. We treated DLBCL cells with either isotype control or anti-BCR antibody and then simultaneously treated both conditions with vehicle or Silvestrol. Similar to splenic B cells, we found elevated levels of CARD11, BCL10, and MALT1 24 hours after BCR activation, whereas expression was reduced with Silvestrol treatment (Figure 5A; supplemental Figure 5A). These changes were not due to reduced viability, as we observed no significant cell death 24 hours after activation or treatment, as assessed by PI staining (supplemental Figure 5C) and western blot analysis of caspase-3 and PARP1 cleavage (Figure 5A; supplemental Figure 5A). We did observe a significant increase in cell death 48 hours after Silvestrol treatment in both SUDHL4 and OCI-Ly10 but not Toledo cells as assessed by PI staining (supplemental Figure 5D). Cycloheximide blockade of protein translation showed that CARD11, BCL10, and MALT1 all have much shorter half-lives of roughly 3, 2, and 6 hours, respectively, compared with the low-turnover housekeeping protein GAPDH (Figure 5B; supplemental Figure 5B). These findings are consistent with the notion that the CARD11, BCL10, and MALT1 oncogenes, like many other oncogenes, are tightly regulated at the level of protein translation initiation owing to their complex 5′UTRs and short half-lives.
Figure 5.
The 5′UTR of CARD11 represses protein translation. SUDHL4 and OCI-Ly10 cells were harvested 24 hours after treatment with Isotype + Vehicle, Isotype + 10 nM Silvestrol, anti-BCR + Vehicle, or anti-BCR + 10 nM Silvestrol. (A) Protein levels of CARD11, BCL10, and MALT1 were measured by western blotting. GAPDH served as a loading control, and data are representative of 2 independent experiments. (B) Cells were treated with 100 μg/mL of cycloheximide, and lysates were collected at the indicated time points. Protein levels of CARD11, BCL10, and MALT1 were measured by western blotting. GAPDH served as a loading control, and data are representative of 2 independent experiments. (C) Predicted 5′UTR structures and ΔG of respective genes. (D) A graph of the ratio between Firefly luciferase units (FLU) and renilla luciferase units (RLU) demonstrating the impact each respective 5′UTR has on cap-dependent translation. Data are means ± SEM from triplicates and are representative of 3 different experiments. (E) A graph of the fold change from Isotype + Vehicle for CARD11, BCL10, and MALT1 5′UTR FLU/RLU ratios normalized to GAPDH’s 5′UTR FLU/RLU ratio. Data are means ± SEM from triplicates and are representative of 3 different experiments (*P < .05).
To directly test the function of the 5′UTRs of CARD11, BCL10, and MALT1, that have predicted ΔG values of −174, −258, and −156, respectively (Figure 5C), we cloned them into dual luciferase plasmid constructs. In these plasmids, Firefly luciferase expression is controlled by cap-dependent translation and the respective 5′UTR element, whereas renilla luciferase expression is controlled by the eIF4A-independent hepatitis C virus (HCV)–internal ribosomal entry site element.38 The GAPDH 5′UTR from the longest and most complex transcript (NM_001256799.2, predicted ΔG of −133), as opposed to the 5′UTR from the shortest and least complex transcript (NM_002046.5, predicted ΔG of −70), was also cloned in and served as a control (Figure 5C). SUDHL4 cells that were stably transduced with each construct were treated with either isotype control or anti-BCR antibodies and simultaneously treated with either vehicle or Silvestrol. When normalized to the GAPDH ratio between cap-dependent and cap-independent translation, all components of the CBM complex were more sensitive to Silvestrol treatment than GAPDH (Figure 5E). Also, under basal conditions, the ratio between cap-dependent and cap-independent translation was greatly reduced in the CARD11 construct compared with GAPDH, demonstrating that the 5′UTR of CARD11 effectively suppresses translation initiation (Figure 5D). Together, these data demonstrate that CARD11, BCL10, and MALT1 have short protein half-lives, complex 5′UTRs, and are sensitive to eIF4A inhibition.
The 5′UTR of eIF4E represses protein translation
The reduction in total eIF4E protein expression on Silvestrol treatment was surprising and suggested that eIF4E protein translation may be highly dependent on eIF4A activity. We analyzed the 5′UTRs of the major eIF4E1 and eIF4A1 mRNA transcripts using DINAMelt,23 which varied in complexity (supplemental Figure 6A). These observations suggest that eIF4E1 might require enhanced eIF4A helicase activity to augment protein expression of eIF4E1 on lymphocyte activation. Using the dual luciferase approach described above, surprisingly, we found that, although the complex form of the eIF4E 5′UTR greatly suppressed translation (supplemental Figure 6B), translation was not enhanced on BCR activation, whereas the noncomplex form of the eIF4E 5′UTR, which did not suppress translation, responded to BCR activation (supplemental Figure 6C). Furthermore, neither the 5′UTRs of eIF4A nor of eIF4E responded to Silvestrol treatment under any condition (supplemental Figure 6C). Further studies will be required to determine the significance of the highly complex 5′UTR of eIF4E and its interactions with eIF4A.
Discussion
It has been established that eIF4A activity is directly correlated to the degree of mRNA 5′UTR secondary structure.39 A significant role for augmented eIF4F activity and cap-dependent translation during lymphomagenesis has been established through both mouse models and in vitro pharmacological studies.40-42 Furthermore, PDCD4−/− mice have a high penetrance for B-cell lymphomas.17 Moreover, Silvestrol has demonstrated chemosensitization properties in refractory murine lymphoma models40 and multiple tumor types characterized by persistent formation of the eIF4F complex through multiple mechanisms.43 Interestingly, in an NCI 60-cell line screen, Silvestrol produced a pattern of roughly 10-fold greater total growth inhibition in B-cell malignancies than the average of all other cell lines tested.22,44-46 Most recently, it has been reported that a unique RNA G-quadruplex structure increases the 5′UTR structure complexity of some oncogenic messages, which were shown to be highly dependent on eIF4A activity.47 The advent of pharmacological inhibitors of eIF4A such as Silvestrol and Hippuristanol combined with these compounds’ general specificity and high efficacy in B-lymphoid tumors suggests that the role for eIF4A in B-cell malignancies should be further elucidated.
PDCD4 has been shown to be downregulated by miR-21, a target of NF-κB transcription, and is correlated with activated B-cell (ABC) DLBCLs.48 Furthermore, gene expression analysis of different B-cell compartments demonstrated that GC B cells have markedly repressed PDCD4 expression.18 Although rapid induction of NF-κB-dependent transcription and enhanced p70s6K signaling10 on BCR activation are well characterized, much less is known about the potential involvement of MTOR and p70s6K in the regulation of translation initiation during the early phases of B-cell activation. To our surprise, we observed that eIF4E cap-binding activity was not highly modulated on BCR activation despite enhanced eIF4E protein expression (Figure 2B). Instead, we observed a significant increase in p70s6K signaling and eIF4A cap-binding activity (Figure 2A-B). p70s6K is subject to regulation by ERK, PDK1, and MTORC1, which phosphorylate p70s6K on its autoinhibitory domain, activation loop, and hydrophobic motif, respectively.33 Consistent with previous studies, we found that these kinases, as well as PDCD4 and RPS6, substrates of p70s6K, are phosphorylated and activated on BCR activation (Figures 1B and 2A). Additionally, total PDCD4 protein levels were reduced on BCR activation, which frees eIF4A to bind the cap-binding complex (Figure 2A-B). Collectively, these results demonstrate that BCR activation greatly enhances eIF4A cap-binding activity through upstream p70s6K and PDCD4 signaling.
The pharmacological inhibitor of eIF4A, Silvestrol, was used to investigate the impact of eIF4A on global translation on BCR activation. Treatment with Silvestrol was able to reverse BCR-induced eIF4A cap-binding (Figure 3A). Additionally, eIF4E cap-binding and total eIF4E protein levels were reduced on Silvestrol treatment under both isotype treatment and anti-BCR stimulation (Figure 3A). Although the 5′UTRs of some eIF4E transcripts are highly complex, thus far we have not found evidence that eIF4A would directly regulate eIF4E translation either at basal conditions or upon activation (supplemental Figure 6A-C). Silvestrol treatment also resulted in a drastic reduction in BCR-induced protein translation as assessed by both 80S formation/polysomal mRNA recruitment (supplemental Figure 3B) and 35S-radiolabeled amino acid incorporation (Figure 3B). On BCR activation of human splenic B cells, we observe that protein levels of CARD11, BCL10, and MALT1 are upregulated and are sensitive to Silvestrol treatment (Figure 3C-D).
It is important to note that quantitative increases in protein translation can impact cell growth during cellular transformation, representing a first hit for cancer formation, whereas enhanced translation initiation through increased activity of eIFs, such as eIF4A, may also qualitatively enhance the translation efficiency of the poorly translated mRNAs involved in cell growth and survival.42 We observed that IgG+ human splenic B cells display higher levels of p70s6K signaling on activation than IgM+ B cells (Figure 4B). Consequently, IgG+ B cells also have enhanced eIF4A cap-binding activity (Figure 4C) and are more translationally active (Figure 4D) under both basal and activated conditions than IgM+ B cells. Despite the fact that IgM+ B cells exhibit lower levels of eIF4E and eIF4A cap-binding, global protein translation is as much or even more increased on BCR activation than in IgG+ B cells. Multiple factors can also augment protein synthesis including enhanced transcription and mRNA capping, which are likely promoted on BCR activation. Furthermore, we demonstrated that IgG+ B cells enhance CARD11 expression on BCR activation unlike IgM+ B cells, which may be the result of enhanced eIF4A activity (supplemental Figure 4C). Additionally, this increase in CARD11 expression in IgG+ B cells can be attenuated by treatment with Silvestrol, whereas CARD11 expression was only reduced in IgM+ B cells that were not stimulated and treated with Silvestrol, which may be the result of not only differential eIF4E and eIF4A cap-binding capabilities but basal expression of these proteins (supplemental Figure 4C). Clearly, IgG+ and IgM+ B cells are qualitatively different with respect to eIF4A-dependent regulation of protein translation, and this difference may also contribute to the fact that antigen-experienced B cells are typically the cell of origin for many lymphomas, including DLBCL.
The CBM complex is required for the coupling of BCR signaling to NF-κB signaling.25-27 After antigen exposure, CARD11 serves as a scaffolding protein for BCL10 and MALT1 that promotes the activation of the IκB kinase β.28 Normal B cells display antigen-independent tonic BCR signaling, and NF-κB activation is required for the survival and differentiation of specific B-cell subpopulations.49-54 We used the DLBCL cell lines SUDHL4, OCI-Ly10, and Toledo to both confirm our results obtained in splenic B cells and to further investigate the translational regulation of CBM components (Figure 5A; supplemental Figure 5A). We observed that CARD11, BCL10, and MALT1 are relatively short-lived proteins (Figure 5B; supplemental Figure 5B), suggesting that translational inhibition may be a rational therapy for malignancies dependent on these oncogenes. Furthermore, we observed that the 5′UTRs of CARD11, BCL10, and MALT1 suppress translation initiation (Figure 5D), and all CBM components were sensitive to pharmacological inhibition of eIF4A (Figure 5E). Studies have demonstrated that CARD11 expression is required for constitutive activation of NF-κB in ABC-DLBCL. RNA interference approaches demonstrated a strong dependence of ABC-DLBCL cells on CARD11, BCL10, and MALT1 expression.27 Furthermore, roughly 10% of ABC-DLBCL cases harbor activating CARD11 mutations, thus rendering them refractory to Bruton's tyrosine kinase inhibition.26
In our study, we found that eIF4A cap-binding activity is increased on BCR activation, whereas eIF4E cap-binding activity remained relatively unaltered. Additionally, CARD11, BCL10, and MALT1 protein expression were found to be upregulated on BCR activation and sensitive to eIF4A inhibition. Furthermore, we identified that antigen-experienced IgG+ B cells are more translationally active and highly modulate the translation of the CARD11 oncogene on activation and pharmacological inhibition of eIF4A. Finally, we observed that CARD11, BCL10, and MALT1 harbor 5′UTRs that are sensitive to inhibition of eIF4A/cap-dependent translation and are also short-lived proteins, suggesting that pharmacological inhibition of eIF4A and cap-dependent translation may be rational targets for malignancies dependent on CBM expression such as ABC-DLBCLs.26,27
Acknowledgments
The authors thank Dr Phil Sharp for the pFR_HCV_xb construct and the staff and nurses at the R. Adams Cowley Shock Trauma Center, the staff of the Pathology Core, and the staff of the Translational Laboratory Shared Service of the University of Maryland Medical Center. Flow cytometry analysis and cell sorting were performed at the Marlene & Stewart Greenebaum Cancer Center Flow Cytometry Shared Service.
This work was supported in part by a Merit Review Award from the Department of Veterans Affairs (to R.B.G.) and National Institutes of Health grants R01AA017972 (National Institute on Alcohol Abuse and Alcoholism, to R.B.G.) and R01CA164311 (National Cancer Institute, to R.B.G).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.J.S. and F.L. conceived of and designed the research, performed research, analyzed data, and wrote the paper; R.J.P., K.M.-M., Q.C., S.H., B.D., and A.L.L. performed research, analyzed data, and wrote the paper; C.R., R.N.B., J.D., B.B., R.T., D.S., R.F., N.H., J.P., C.R., M.D.K., M.B., J.R., S.S., and T.S. provided samples; and R.B.G. conceived of and designed the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ronald B. Gartenhaus, Marlene & Stewart Greenebaum Cancer Center, Department of Medicine, University of Maryland Medical Center, 655 West Baltimore St, BRB 9-011, Baltimore, MD 21201; e-mail: rgartenhaus@som.umaryland.edu.
References
- 1.Wakabayashi C, Adachi T, Wienands J, Tsubata T. A distinct signaling pathway used by the IgG-containing B cell antigen receptor. Science. 2002;298(5602):2392–2395. doi: 10.1126/science.1076963. [DOI] [PubMed] [Google Scholar]
- 2.Dogan I, Bertocci B, Vilmont V, et al. Multiple layers of B cell memory with different effector functions. Nat Immunol. 2009;10(12):1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 3.Kehry MR. CD40-mediated signaling in B cells. Balancing cell survival, growth, and death. J Immunol. 1996;156(7):2345–2348. [PubMed] [Google Scholar]
- 4.Monroe JG. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol. 2006;6(4):283–294. doi: 10.1038/nri1808. [DOI] [PubMed] [Google Scholar]
- 5.Horikawa K, Martin SW, Pogue SL, et al. Enhancement and suppression of signaling by the conserved tail of IgG memory-type B cell antigen receptors. J Exp Med. 2007;204(4):759–769. doi: 10.1084/jem.20061923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 7.Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov. 2013;12(3):229–243. doi: 10.1038/nrd3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blenk S, Engelmann J, Weniger M, et al. Germinal center B cell-like (GCB) and activated B cell-like (ABC) type of diffuse large B cell lymphoma (DLBCL): analysis of molecular predictors, signatures, cell cycle state and patient survival. Cancer Inform. 2007;3:399–420. [PMC free article] [PubMed] [Google Scholar]
- 9.Gauld SB, Dal Porto JM, Cambier JC. B cell antigen receptor signaling: roles in cell development and disease. Science. 2002;296(5573):1641–1642. doi: 10.1126/science.1071546. [DOI] [PubMed] [Google Scholar]
- 10.Li HL, Forman MS, Kurosaki T, Puré E. Syk is required for BCR-mediated activation of p90Rsk, but not p70S6k, via a mitogen-activated protein kinase-independent pathway in B cells. J Biol Chem. 1997;272(29):18200–18208. doi: 10.1074/jbc.272.29.18200. [DOI] [PubMed] [Google Scholar]
- 11.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 12.Bi X, Ren J, Goss DJ. Wheat germ translation initiation factor eIF4B affects eIF4A and eIFiso4F helicase activity by increasing the ATP binding affinity of eIF4A. Biochemistry. 2000;39(19):5758–5765. doi: 10.1021/bi992322p. [DOI] [PubMed] [Google Scholar]
- 13.Dennis MD, Jefferson LS, Kimball SR. Role of p70S6K1-mediated phosphorylation of eIF4B and PDCD4 proteins in the regulation of protein synthesis. J Biol Chem. 2012;287(51):42890–42899. doi: 10.1074/jbc.M112.404822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers GWJ, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274(18):12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 15.Yang HS, Jansen AP, Komar AA, et al. The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol. 2003;23(1):26–37. doi: 10.1128/MCB.23.1.26-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314(5798):467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- 17.Hilliard A, Hilliard B, Zheng SJ, et al. Translational regulation of autoimmune inflammation and lymphoma genesis by programmed cell death 4. J Immunol. 2006;177(11):8095–8102. doi: 10.4049/jimmunol.177.11.8095. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Iqbal J, Xiao L, et al. Distinct gene expression profiles in different B-cell compartments in human peripheral lymphoid organs. BMC Immunol. 2004;5(20):20. doi: 10.1186/1471-2172-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvilleur E, Sbarrato T, Hill K, et al. A role for eukaryotic initiation factor 4B overexpression in the pathogenesis of diffuse large B-cell lymphoma. Leukemia. 2014;28(5):1092–1102. doi: 10.1038/leu.2013.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang BY, Su BN, Chai H, et al. Silvestrol and episilvestrol, potential anticancer rocaglate derivatives from Aglaia silvestris. J Org Chem. 2004;69(10):3350–3358. doi: 10.1021/jo040120f. [DOI] [PubMed] [Google Scholar]
- 21.Bordeleau ME, Mori A, Oberer M, et al. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2(4):213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- 22.Lucas DM, Edwards RB, Lozanski G, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113(19):4656–4666. doi: 10.1182/blood-2008-09-175430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33(Web Server Issue):W577-W581. [DOI] [PMC free article] [PubMed]
- 24.Willis AE. Translational control of growth factor and proto-oncogene expression. Int J Biochem Cell Biol. 1999;31(1):73–86. doi: 10.1016/s1357-2725(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 25.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4(5):348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 26.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870):1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 27.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rawlings DJ, Sommer K, Moreno-García ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6(11):799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 29.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21(4):533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 30.Kutner RH, Zhang XY, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4(4):495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 31.Mazan-Mamczarz K, Zhao XF, Dai B, et al. Down-regulation of eIF4GII by miR-520c-3p represses diffuse large B cell lymphoma development. PLoS Genet. 2014;10(1):e1004105. doi: 10.1371/journal.pgen.1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazan-Mamczarz K, Lal A, Martindale JL, Kawai T, Gorospe M. Translational repression by RNA-binding protein TIAR. Mol Cell Biol. 2006;26(7):2716–2727. doi: 10.1128/MCB.26.7.2716-2727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Zhong C, Wang F, Qu F, Ding J. Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif. Biochem J. 2013;454(1):39–47. doi: 10.1042/BJ20121863. [DOI] [PubMed] [Google Scholar]
- 34.Mizuno T, Rothstein TL. B cell receptor (BCR) cross-talk: CD40 engagement enhances BCR-induced ERK activation. J Immunol. 2005;174(6):3369–3376. doi: 10.4049/jimmunol.174.6.3369. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez-Mateos P, Cebrián M, Acevedo A, López-Botet M, De Landázuri MO, Sánchez-Madrid F. Expression of a gp33/27,000 MW activation inducer molecule (AIM) on human lymphoid tissues. Induction of cell proliferation on thymocytes and B lymphocytes by anti-AIM antibodies. Immunology. 1989;68(1):72–79. [PMC free article] [PubMed] [Google Scholar]
- 36.Bjur E, Larsson O, Yurchenko E, et al. Distinct translational control in CD4+ T cell subsets. PLoS Genet. 2013;9(5):e1003494. doi: 10.1371/journal.pgen.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin C, Rajabi H, Rodrigo CM, Porco JAJ, Jr, Kufe D. Targeting the eIF4A RNA helicase blocks translation of the MUC1-C oncoprotein. Oncogene. 2013;32(17):2179–2188. doi: 10.1038/onc.2012.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12(1):67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svitkin YV, Pause A, Haghighat A, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7(3):382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schatz JH, Oricchio E, Wolfe AL, et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med. 2011;208(9):1799–1807. doi: 10.1084/jem.20110846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86(2):178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 42.Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol. 2013;5(2):a012336. doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boussemart L, Malka-Mahieu H, Girault I, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513(7516):105–109. doi: 10.1038/nature13572. [DOI] [PubMed] [Google Scholar]
- 44.Monks A, Scudiero D, Skehan P, et al. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J Natl Cancer Inst. 1991;83(11):757–766. doi: 10.1093/jnci/83.11.757. [DOI] [PubMed] [Google Scholar]
- 45.Saradhi UV, Gupta SV, Chiu M, et al. Characterization of silvestrol pharmacokinetics in mice using liquid chromatography-tandem mass spectrometry. AAPS J. 2011;13(3):347–356. doi: 10.1208/s12248-011-9273-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. National Cancer Institute. COMPARE methodology. http://dtp.nci.nih.gov/docs/compare/compare_methodology.html. Accessed April 12, 2014.
- 47.Wolfe AL, Singh K, Zhong Y, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513(7516):65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gu L, Song G, Chen L, et al. Inhibition of miR-21 induces biological and behavioral alterations in diffuse large B-cell lymphoma. Acta Haematol. 2013;130(2):87–94. doi: 10.1159/000346441. [DOI] [PubMed] [Google Scholar]
- 49.Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 50.Pasparakis M, Schmidt-Supprian M, Rajewsky K. IkappaB kinase signaling is essential for maintenance of mature B cells. J Exp Med. 2002;196(6):743–752. doi: 10.1084/jem.20020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egawa T, Albrecht B, Favier B, et al. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr Biol. 2003;13(14):1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 52.Hara H, Wada T, Bakal C, et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18(6):763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 53.Jun JE, Wilson LE, Vinuesa CG, et al. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity. 2003;18(6):751–762. doi: 10.1016/s1074-7613(03)00141-9. [DOI] [PubMed] [Google Scholar]
- 54.Newton K, Dixit VM. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr Biol. 2003;13(14):1247–1251. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]