Abstract
Retinoblastoma (RB) is a children's ocular cancer caused by mutated retinoblastoma 1 (Rb1) gene on both alleles. Rb1 and other related genes could be regulated by microRNAs (miRNA) via complementarily pairing with their target sites. MicroRNA-21 (miR-21) possesses the oncogenic potential to target several tumor suppressor genes, including PDCD4, and regulates tumor progression and metastasis. However, the mechanism of how miR-21 regulates PDCD4 is poorly understood in RB. We investigated the expression of miRNAs in RB cell lines and identified that miR-21 is one of the most deregulated miRNAs in RB. Using qRT-PCR, we verified the expression level of several miRNAs identified by independent microarray assays, and analyzed miRNA expression patterns in three RB cell lines, including Weri-Rb1, Y79 and RB355. We found that miR-19b, -21, -26a, -195 and -222 were highly expressed in all three cell lines, suggesting their potential role in RB tumorigenesis. Using the TargetScan program, we identified a list of potential target genes of these miRNAs, of which PDCD4 is one the targets of miR-21. In this study, we focused on the regulatory mechanism of miR-21 on PDCD4 in RB. We demonstrated an inverse correlation between miR-21 and PDCD4 expression in Weri-Rb1 and Y79 cells. These data suggest that miR-21 down-regulates Rb1 by targeting PDCD4 tumor suppressor. Therefore, miR-21 could serve as a therapeutic target for retinoblastoma.
Keywords: Retinoblastoma, miRNA, miR-21, PDCD4.
Introduction
Retinoblastoma (RB) is a common pediatric intraocular tumor represented by a mutation of the retinoblastoma gene (Rb1) on both alleles. There are about 250-350 newly diagnosed RB cases every year in the United States 1, 2. Retinal tumors develop rapidly in early childhood, usually before the age of five. Common treatments now include chemotherapy, focal therapy such as laser or cryotherapy, and enucleation 3. Metastasis of RB is often fatal, despite the high cure rate in children 4. Later in life, survivors with bilateral RB, an inheritable form, will face increased risk of developing other RB-related nonocular cancers 5, 6, and a trilateral RB, known as pineoblastoma, a tumor in pineal gland 7. Previous report indicates that 30-40% of all RBs were found to be bilateral 8, 9. The metastasis could be seen in both heritable and inheritable cases, as tumor cells frequently invade the optic nerve, orbit and choroid 10, 11. Thus, methods to minimize the requirement of enucleation, decrease the risk of tumor metastasis and the development of secondary cancers, preserve vision and prevent systemic toxicity are crucially needed 1.
The critical role of Rb1 in cell proliferation attracted much attention after it was first discovered in RB. The Rb1 gene is located at chromosome 13q14 and functions as a tumor suppressor, which regulates the cell cycle, specifically cell proliferation and apoptosis. The retinoblastoma protein (pRb) binds to the transcription factor of the E2F family to mediate the succession of the G1/S phase 12. As described in Knudson's “two-hit theory” model of tumorigenesis 13, the heritable form of RB is derived from one germ line mutation and one somatic mutation. On the other hand, the non-heritable or sporadic form is due to two somatic mutations. Loss of the Rb1 gene in RB is necessary for tumorigenic initiation as well as subsequent genetic and epigenetic alterations causing tumor metastasis 1, 12, 14, 15.
miRNA is a small non-coding single-stranded RNA (18-24nt), widespread throughout the human genome. It is well documented for their post-transcriptional modifications on gene expression 16-18. According to miRBase Release 21 (June 2014), 2,588 mature human miRNAs have been identified 19, 20. Primary miRNA (pri-miRNA) is transcribed by RNA polymerase II and then cleaved by endonuclease RNase III Drosha in the nucleus to release stem-loop shaped pre-miRNA, which are approximately 70 nucleotides in length 14, 21. After being exported into the cytoplasm and processed by another endonuclease, Dicer, pre-miRNA becomes mature double-strand miRNA. One strand of mature miRNA is integrated into the RNA-induced silencing complex (RISC) to regulate target genes 16, 22. Imperfectly or perfectly base pairing the 3' UTR of a target mRNA with the conserved seed region (~5 nucleotides) in the 5' end of a miRNA caused genomic instability and transcriptional degradation, as well as translational repression 18, 21, 23, 24. Furthermore, due to important features of miRNAs, a single miRNA can target more than one mRNA molecule and single target mRNA can be recognized by multiple miRNAs. The interaction between miRNAs and its targets creates a complex regulatory network 18, 22, 25.
miRNA transcripts are often located at fragile genomic sites related to cancer and are expressed abnormally because of alterations in cancer genome 26, 27. Several cancer-related miRNAs and their target genes in tumor cells have been identified 14, 25 and their disposition clarified as either towards tumor suppression or oncogenesis 16, 18. For instance, loss of miRNA-15/-16 in chronic lymphocytic leukemia 16, 18, 27, 28, miRNA let-7 in lung cancer 16, 18, 27, 29, and miRNA-143/ miRNA-145 in colon cancer 30, miR-638 in breast cancer 31 provided some evidences towards the tumor suppressing capabilities of miRNA. Similarly, over-expressed miRNA-21/ miRNA-155 in human breast cancer 16, 18, 32 showed the oncogenic potential of miRNAs. Recently, we found that a number of miRNAs, including miR-21, are involved in early breast cancer progression using microarray assay on microdissected formalin-fixed paraffin-embedded (FFPE) samples 33.
Targeting miRNAs could be a novel therapeutic option due to its regulatory role during cancer development and progression. miRNA-34a was reported as a tumor suppressor in RB cell lines 14. The inverse correlation between down-regulated hsa-let-7b and overexpressed HMGA-1 and HMGA-2 was found in FFPE RB tumor samples 15. By profiling miRNA expression in RB cell line HXO-RB44 under normal and hypoxic conditions, researchers identified miR-181 as a hypoxia-related miRNA and that the inhibition of miR-181b impeded cell growth in RB cells under hypoxia conditions 34.
In this study, we first tried to determine the expression of a set of miRNAs related to RB in three RB cell lines, Weri-Rb1, Y79 and RB 355. We found a list of five miRNAs: miR-19b, miR-21, miR-26a, miR-195 and miR-222, which might be involved in the proliferation and metastasis in RB. Then we analyzed the predicted target genes of these miRNA in RB pathologies by miRNA and mRNA interactions. Among them, miR-21 is well studied and is a known onco-miR that is highly expressed in cancers, such as breast cancer 35-37, glioblastoma 38, 39, glioma 40, colorectal cancer 41, prostate cancer 42, 43, hepatocellular cancer 44, non-small cell lung cancer 45, papillary thyroid carcinoma 46, gastric cancer 47, osteosarcoma 48, diffuse large B-cell lymphoma 49 and laryngeal carcinoma 50. However, to the best of our knowledge, there is no report on the role of miR-21 in RB initiation and development. In this study, we focused on elucidating the role of miR-21 in regulating one of its target genes, the programmed cell death 4 (PDCD4), a tumor suppressor involving cell cycle regulation.
Materials and Methods
Cell culture
The human RB cell lines Y79, Weri-RB1 and RB355 were provided by Dr. Hakim Djaballah at the Memorial Sloan-Kettering Cancer Center. They were cultured in RPMI-1640 (Lonza) with 1% penicillin-streptomycin, 2 mM glutamine and 20% fetal bovine serum (HyClone, Thermo Scientific) 51. All cell lines were grown in suspension at a concentration of 105 - 106 cells/ml and incubated at 37 ˚C with 5% CO2.
Isolation of total RNAs and miRNAs
Total RNAs and miRNAs from the cell lines were isolated using TRIzol® Reagent (Invitrogen) and the mirVana miRNA Isolation Kit (Ambion) respectively. Concentration and purity of the RNA samples were assayed using the NanoDrop ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE).
qRT-PCR for miRNAs and mRNAs
The Taqman MiRNA Reverse Transcript Kit (Applied Biosystems), wherein a stem-loop RT primer specifically hybridizes with a miRNA molecule and a MultiScribe™ Reverse Transcriptase, was used for reverse transcription (RT). The RT reaction mixture was incubated at 16˚C for 30 minutes, 42˚C for 30 minutes, 85˚C for 5 minutes, and then stored at 4˚C. The RT products were subsequently amplified with the mature miRNA-specific Taqman® MiRNA Assay Kit (Applied Biosystems) using the ABI 7300 Real-Time PCR system. The PCR was performed at 95 ˚C for 10 minutes, followed by 40 cycles of 95 ˚C for 15 seconds and 60 ˚C for 60 seconds. U6 small nuclear RNA was used as an internal control for normalization. PDCD4 primers were synthesized by the Integrated DNA Technologies (IDT) and the sequences were as follows: Forward, 5'-GGC CTC CAA GGA GTA AGA CC-3'; Reverse, 5'-AGG GGT CTA CAT GGC AAC TG-3'. The cDNAs were synthesized via iScript cDNA Synthesis Kit (Bio-Rad Laboratories) and amplified with SYBR® GreenER™ qPCR Supermix Universal (Invitrogen) using the ABI 7300 Real-Time PCR system. The RT reactions were carried out at 25˚C for 5 minutes, 42˚C for 30 minutes, 85˚C for 5 minutes and then held on 4˚C. The PCR reactions were performed under the following conditions, 50˚C for 2 minutes, 95˚C for 10 minutes and then 40 cycles of 95˚C for 15 seconds and 60˚C for 60 seconds. 18S rRNA gene was used for normalization. The qRT-PCR analyses for each miRNA and mRNA in three cell lines were performed in quadruplicates and triplicates respectively. Relative quantitation was calculated using the 2-Δ(ΔCt) method where fold change is equal to 2-[(Mean ΔCt Target Gene) - (Mean ΔCt Internal Control)].
Transfection of miR-21 mimic and inhibitor in RB cell lines
The miR-21 mimic is a small double-strand RNA that mimics endogenous precursor miRNA, and the miR-21 inhibitor is a single-strand RNA that is designed to bind to and inhibit endogenous miRNA. Transfection of Weri-Rb1, Y79 and RB355 cell lines was executed with Lipofectamine™ 2000 (Invirtrogen) according to the manufacturer's protocol. Briefly, cells were plated in 6-well plates at 30-50% confluence in antibiotics-free growth medium one day before transfection. To make the transfection complex, we diluted 70pmole of Pre-miR™ miRNA precursor, Anti- miR™ miRNA inhibitors (hsa-miR-21) or miRNA negative control (Ambion) with 5µl Lipofectamine™ 2000 (Invirtrogen) in 2ml of Opti-MEM® I Reduced Medium (Invitrogen). Medium were changed to complete growth medium (20%FBS) after 4-6 hours. After 48 h transfection, total RNA and protein were isolated.
Western blot assays
Proteins were extracted from transfected cells using 10X RIPA lysis buffer (Millipore). Protein concentration was measured using the Pierce® BCA protein assay kit (Thermo). The primary anti-PDCD4 (rabbit) antibody was purchased from Rockland (Gilbertsville, PA) and anti-β-actin (mouse) antibody was obtained from Sigma-Aldrich. Protein with 1X Laemmli buffer was denatured at 95˚C for 5 minutes before electrophoresis using the 10% Mini-PROTEAN® TGX Precast Gels (BIO-RAD), under the condition of 120V, for 1 - 1.5 hours. Protein was transferred onto the nitrocellulose membrane. The membrane was blocked with 5% non-fat milk/ PBST for one hour at RT and incubated with the primary antibody (1:5,000 dilution) at 4 ˚C overnight. The secondary antibody (1:100,000 dilution) was applied for 1-1.5h at RT. The blot signals were detected using the SuperSignal West Dura Chemiluminescent Substrate (Thermo) and visualized via Image Lab™ system software (BIO-RAD). β-actin was used as an internal control. The band intensity was quantified using the Image J software.
Statistical analysis
Data were presented as the mean ± standard deviation (S.D.). The one-way analysis of variance (ANOVA) was performed for the miRNA differential expression in cell lines. The Student's t-test was applied to compare two transfected groups. P-value ≤ .05 or .01 was considered statistically significant and presented with one and two asterisks respectively.
Results and Discussion
Differentially expressed miRNAs in three RB cell lines
miRNAs, such as miR-19b, 93, -141 and 181c, miR-181a, -183, -222 and let-7a were identified earlier as RB-related miRNAs using a Human miRNA 2k custom array (Agilent) 24. A separate study identified 11 over-expressed miRNAs in RB compared to normal retina, most notably a fourfold increase in miR-320, miR-373 and miR-503 using the miRCURYTM array microarray (Exiqon) 27. To explore the relative expression of miRNAs in three RB cell lines, Weri-Rb1, Y79 and RB355, we selected a list of 23 candidate miRNAs (miR-19b, -21, -26a, -29b, -93, -96, -141, -181a, -181c, -183, -187,-195, -200b, -200c, -205, -214, -222, -381,-556-3p, -557, -644, -1287 and let-7a) from aforementioned studies for qRT-PCR analysis.
Interestingly, those miRNAs, such as miR-19, -21, -26, -29, -93, -96, -141, -181, -183, -195, -200b, -205 and -222, were predicted to target one or more of the 320 retinal genes, which are known to be involved in visual functions or related to inherited retinal diseases 52. These target genes were identified by a combination of the RetinaCentral, the Retinal Information Network databases, and the comparison of retinal/ RPE dbEST libraries between five tissue-specific libraries 52.
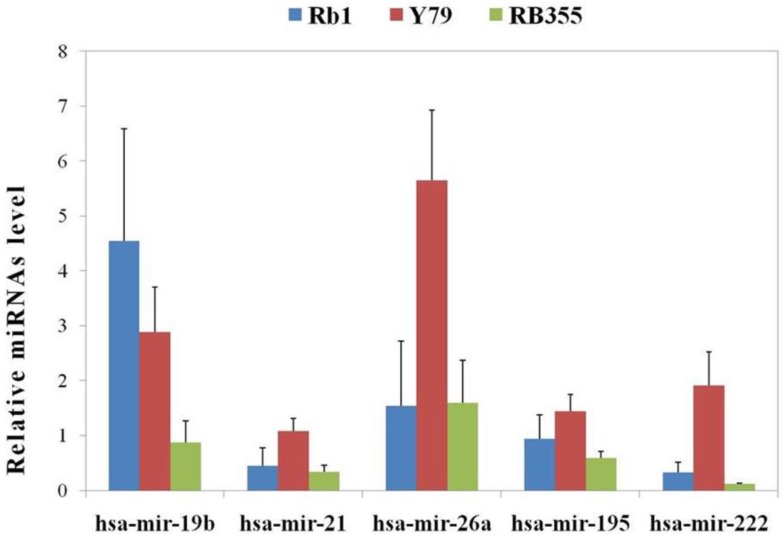
In addition, we found that the expression of some miRNAs, such as miR-141, -187, -200b, 200c, -214 and -381, was very low in all three RB cell lines (data not shown). Others, such as miR-195, miR-19b, miR-26a, miR-21 and miR-222 were extensively over-expressed in one cell line as compared to the other two (Figure 1). miR-19b was deregulated as previously reported 24 in RB cell lines, with greater expression in Weri-Rb1 than in Y79 cells and low levels in RB355 cells. miR-26a, which is expressed in normal retina 52, appeared with significantly different expression levels among the three cell lines, with the highest expression being found in Y79 cells and a comparable expression level in both Wer-Rb1 and RB355 cells. Others, such as miR-21 and miR-222, were well documented by independent studies in regards to their regulatory activity within a number of cancers and also showed differential expression in RB cells. It should be noted that four of the five miRNAs were commonly overexpressed in Y-79 cells as compared to Rb-1 and Rb-355 cells. Alongside our previous observation of Y-79 cells as having increased potency in metastasis and proliferation, we suggest those four miRNAs might have the ability to predict metastasis and proliferation of the RB. However, miR-19b was expressed higher exclusively in Rb-1 cells and required further investigation.
Figure 1.
The differential expression patterns of miRNAs in three RB cells. Five miRNAs were identified by ANOVA tests to be overexpressed in one cell line compared to the other two. Four of the five miRNAs had a common increase in expression level in Y-79 cells, whereas miR-19b had a higher expression level in Weri-Rb1 cells (Rb1). All data were measured in triplicate and presented as the mean ± standard deviation (S.D.).
Target gene prediction of the candidate miRNAs
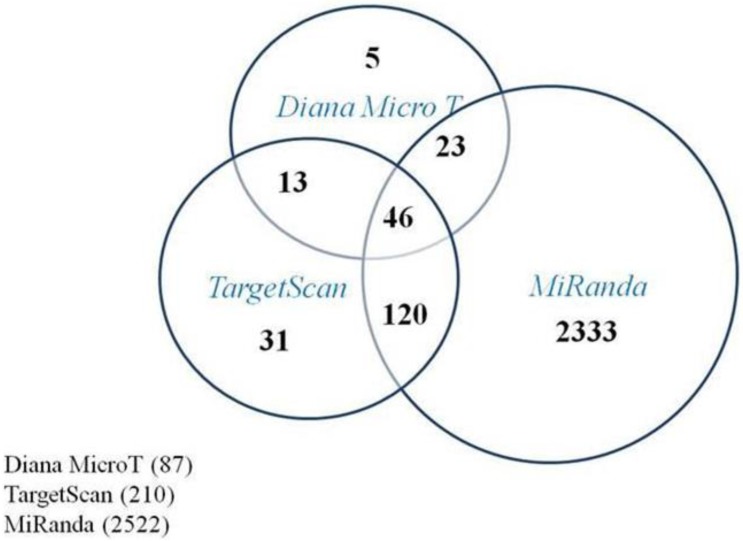
In order to understand how these five miRNAs function in RB tumor development and progression, predicted target genes of those five miRNAs were obtained via three computational algorithms, miRanda (http://www.microrna.org/), TargetScan (http://www.targetscan.org/), and DianaMicroT (http://diana.cslab.ece.ntua.gr/microT/). For each miRNA, predicted target genes were displayed if at least predicted by two algorithms. The predicted miR-21 target genes are presented in Figure 2.
Figure 2.
Program prediction of target genes of miR-21. The Venn-diagram displays the putative targets genes of miR-21 as predicted by Diana MicroT, TargetScan, and Miranda. There were 202 candidates predicted by at least two programs and 46 candidates by all three algorithms.
miR-21 and one of its target genes, PDCD4 in RB
To study the biological significance of the differentially expressed miRNAs, we predicted putative target genes, which were documented in previous studies. PDCD4 is one of the well reported targets of miR-21. Reduction or loss of PDCD4 expression was often seen in various cancers, including lung cancer 53, heptocellular carcinoma 54, ductal breast cancer 55, oral squamous cell carcinoma 56, and colorectal cancer 57. PDCD4 protein levels were found to be inversely correlated with miR-21 levels in human glioblastoma cell lines 58, colorectal samples 41 and cell lines 59, and breast tumor specimens 35.
Furthermore, the conserved target site for miR-21 in the 3'-UTR of PDCD4 was perfectly matched through bioinformatics predictions. The suppression of reporter gene activity with the 3'-UTR of PDCD4 verified the down-regulatory effect of miR-21 in colorectal cells 59, cervical carcinoma cells 60, glioblastoma cells 58, breast cancer cells 61, and myelogenous leukemia cells 62-64. The TargetScan program was applied to confirm the miR-21 binding site in the 3'-UTR of PDCD4 and the sequence for the binding site was well conserved among species as shown in Figure 3.
Figure 3.
miR-21 seed sequence matches the target site in the 3' UTR of PDCD4. The seed sequence of miR-21 was identified by TargetScan and accurately matched to the 3' UTR of PDCD4 in six different species.
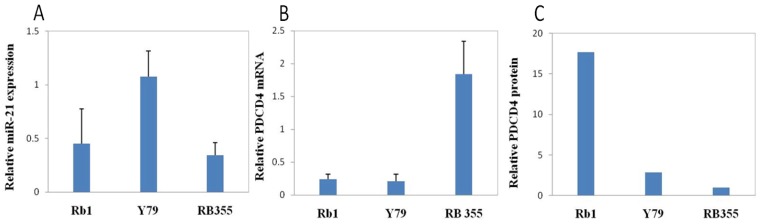
In glioblastoma T98G cells, high levels of miR-21 repressed PDCD4 58. We reasoned that similar interactions might also exist in RB. Thus, we assessed both PDCD4 mRNA and protein levels in RB cell lines, Rb1, Y79 and Rb355. We expected that PDCD4 expression might be inversely correlated with miR-21 expression. The implication being, cell lines with high endogenous miR-21 might contain a low level of PDCD4 protein, and vice versa. When compared between Rb1 and Y79 cells, miR-21 was barely detectable and the most abundant amount of PDCD4 protein was found in Rb1, whereas Y79 cells displayed a higher miR-21 level and low PDCD4 mRNA and protein (Figure 4.). Rb-355 cells have similarly low expression levels of miR-21 and high levels of PDCD4 mRNA. However, Rb-355 rarely expressed the PDCD4 protein, indicating PDCD4 might be subject to more complex post-transcriptional regulation.
Figure 4.
Expression of miR-21 and PDCD in RB cells. Total RNA and total protein were isolated from three RB cell lines by mirVana and RIPA respectively. qRT-PCR was performed to measure the expression level of miR-21 and PDCD4 mRNA. Western blot analysis was performed to measure the protein level of PDCD4. Y79 cells had the highest miR-21 level along with a low PDCD4 mRNA and protein level. Rb1 had a lower miR-21 expression level, but the highest PDCD4 protein level. RB355 cells had a low miR-21 expression level along with the highest PDCD4 mRNA level, but barely expressed any PDCD4 protein.
Overexpression of PDCD4 could inhibit cell invasion in MDA-MB-231 breast cancer cells 35 as well as OSCC cells 56. We hypothesize that higher miR-21 levels accompanied by attenuated PDCD4 protein levels in Y79 cell lines might be closely related to its more aggressive nature in tumor growth and metastasis. Meanwhile, lower miR-21 and up-regulated expression of PDCD4 protein in Rb1 cells denotes its non-metastasis feature. As for RB355 cells, knowledge is limited on the invasive ability and metastatic propensity.
Previous studies also implied that the Y79 cell line represents more metastatic potential as compared to the Rb1 cell line. The metastatic characteristics of Y79 cells were observed after intravitreous cavity injections in immune-deficient mice, while Rb1 cells represented a less-metastatic model 65. Tumors formed in Y79-injected mice aggressively invaded subretinal and choroidal space, and then migrated to the optic nerve as well as the brain and contralateral optic nerve. On the other hand, the mice injected with Rb1 cells developed localized tumors located in the anterior uveal tissue. In vitro, suspended Y79 cells adhered to monkey choroid and rat C6 glioma cells, but not to human embryonic kidney cells. The binding of Y79 RB cells to the choroid and glioma cells, which were derived from target sites of invasion, were confirmed as true cell-cell interactions since trypsin treatment interrupted the binding of Y79 cells, but neuraminidase did not. Few Rb1 cells interacted with choroidal and glioma cells. It was believed that the distinct ability and inclination to metastasize in Rb1 and Y79 RB cells might result from differences in membrane protein structure. Moreover, the invasive Y79 cells lost expression of N-cadherin and cadherin-11 proteins, contributing to RB tumor progression 66, 67. Rb1 cells relied on cadherin-mediated cell-cell interaction 66, which is calcium-dependent 67. Ectopic expression of N-cadherin and cadherin-11 in Y79 cells transplanted into the vitreous of mice showed a less extensive optic nerve invasion, which was similar to Rb1 cell xenografts as compared to Y79 cell controls.
In a previous study, Dalgard et al. discovered that precursor and mature miR-34a in RB cell lines were expressed in different levels: they were higher in Rb1 cells and lower in Y79 cells 14. In Y79 cells, the level of miR-34a rose after adding p53-activating agents. Overexpression of miR-34a diminished cell growth and increased caspase-3/7 activity in both RB cells, particularly in Y79 cells. It is likely that miR-34a expression levels represent the aggressiveness of both cells, due to the miRNAs' epigenetic manipulation on RB cell lines contributing to invasion variability. Thus, Y79 seems to be more aggressive when compared to Weri-Rb1 cells, and higher expression level of miR-21 along with diminished PDCD4 level might serve as an indicator of its aggressive nature.
Inhibition or overexpression of miR-21 in RB cell lines
We sought to explore whether or not miR-21 regulates endogenous PDCD4 in RB cell lines Rb1 and Y79, as well as to observe the miR-21 and PDCD4 expression levels after transient transfections. The negative regulation of miR-21 on its target gene, PDCD4, has been established in other cancers. The protein expression level of PDCD4 could be down- and up-regulated by a miR-21 inhibitor and precursor respectively in OSCC cells 56 and cervical carcinoma Hela cells 60. A similar study showed that anti-miR-21 transfected in leukemia HL60 and K562 cells increased the mRNA and protein expression of PDCD4 62-64.
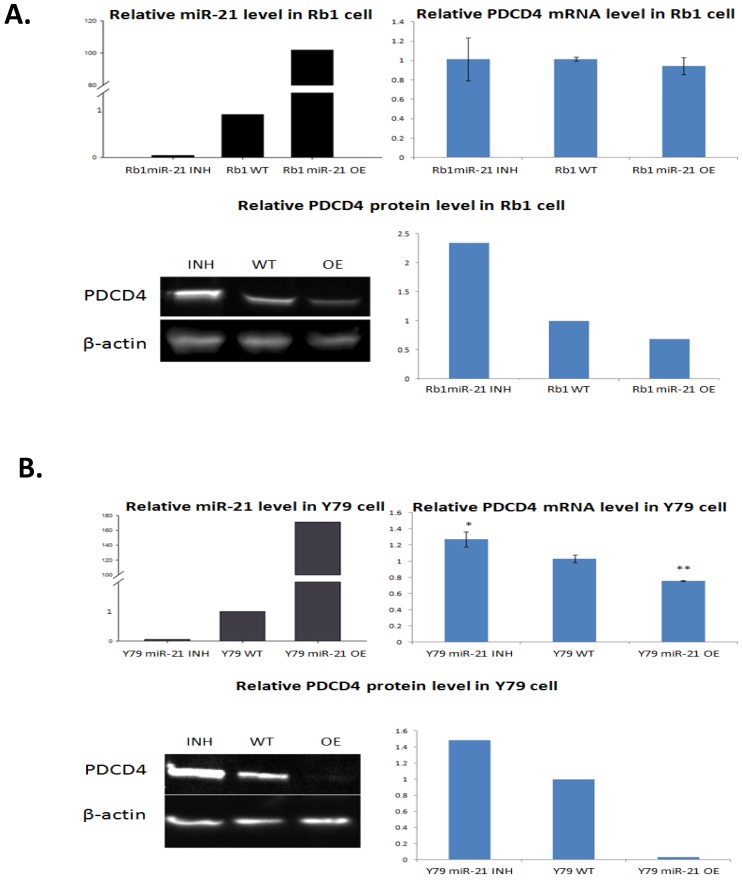
To validate the similar alterations of miR-21 and PDCD4, we performed qRT-PCR and Western blotting to compare RB cells with miR-21 inhabitation and overexpression to negative controls. The transfection with the miR-21 inhibitor and mimic were efficient and could elevate miR-21 to more than 100-fold levels in cell lines when compared to negative controls in both Rb1 and Y79 cells (Figure 5).
Figure 5.
Expression of PDCD4 mRNA and protein in Rb1 and Y79 cells when transfected by miR-21 mimic or inhibiotor. Rb1 and Y79 cells were transfected with miR-21 mimic and inhibitor respectively. Two days after transfection, total RNA and protein were isolated. qRT-PCR showed the inhibition and overexpression were successful in comparison with the wild type control. PDCD4 mRNA levels in Rb1 cells did not show a significant change, but a sigifnicant fold change was present in Y79 cell data. Western blotting revealed PDCD4 proteins in both cells as having an inverse correlation with the miR-21 level, in which miR-21 overexpression reduced the PDCD4 protein while miR-21 inhibition restored the PDCD4 protein level.
The PDCD4 protein level significantly decreased in Rb-1 and Y-79 cells when miR-21 is overexpressed. Meanwhile, inhibition of miR-21 successfully restored PDCD4 protein levels; eliminating the possibility that miR-21 overexpression silences PDCD4 due to the non-specific off-target binding effect.
By targeting PDCD4, miR-21 could manage tumor growth and invasion. In colorectal RKO cells that endogenously expressed high levels of miR-21, anti-miR-21 transfection caused PDCD4 protein levels to increase and also blocked cell invasion and distal metastasis 59. In a similar vein, pre-miR-21-transfected Colo206f cells that were naturally low in miR-21 displayed decreased PDCD4 protein levels and increased cell invasion. In cervical carcinoma Hela cells, the knockdown of miR-21 restrained cell proliferation and colony formation 60. In parallel, elevated expression of miR-21 in RB via a precursor might consistently lessen PDCD4 expression, a responder to apoptosis in cancer, thus stimulating cell proliferation. In contrast, miR-21 inhibition might contribute to restoring PDCD4 expression in RB cells, resulting in suppression of tumor cell growth and invasion. These imply that miR-21 may serve as a therapeutic target by suppressing tumor growth and triggering apoptosis pathways by regulating PDCD4 gene expression in RB.
Conclusions
Our data suggests that miR-21 may play a significant regulatory role in RB tumor cell migration and invasion. Therefore, it may serve as a therapeutic target in RB as it regulates Rb1 gene expression by directly targeting PDCD4.
Acknowledgments
We are grateful to Dr. Hakim Djaballah (Memorial Sloan-Kettering Cancer Center, US) for providing the Weri-RB1, Y-79 and RB355 cell lines and Dr. Gail M. Seigel (State University of New York at Buffalo, US) for the gift of RB143, Y79 and WERI-RB27 cell lines. This research was supported in part by the Wendy Will Case award (to SWF), the Hao Foundation Award (to SWF) and the Elaine H. Snyder Cancer Research Award (to SWF).
References
- 1.Schefler AC, Abramson DH. Retinoblastoma: what is new in 2007-2008. Curr Opin Ophthalmol. 2008;19:526–34. doi: 10.1097/ICU.0b013e328312975b. doi:10.1097/ICU.0b013e328312975b 00055735-200811000-00013 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Abramson DH. Retinoblastoma in the 20th century: past success and future challenges the Weisenfeld lecture. Invest Ophthalmol Vis Sci. 2005;46:2683–91. doi: 10.1167/iovs.04-1462. doi:46/8/2684 [pii] 10.1167/iovs.04-1462. [DOI] [PubMed] [Google Scholar]
- 3.Melamud A, Palekar R, Singh A. Retinoblastoma. Am Fam Physician. 2006;73:1039–44. [PubMed] [Google Scholar]
- 4.Theodoropoulou S, Kolovou PE, Morizane Y, Kayama M, Nicolaou F, Miller JW. et al. Retinoblastoma cells are inhibited by aminoimidazole carboxamide ribonucleotide (AICAR) partially through activation of AMP-dependent kinase. FASEB J. 2010;24:2620–30. doi: 10.1096/fj.09-152546. doi:fj.09-152546 [pii] 10.1096/fj.09-152546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abramson DH, Ronner HJ, Ellsworth RM. Second tumors in nonirradiated bilateral retinoblastoma. Am J Ophthalmol. 1979;87:624–7. doi: 10.1016/0002-9394(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 6.Eng C, Li FP, Abramson DH, Ellsworth RM, Wong FL, Goldman MB. et al. Mortality from second tumors among long-term survivors of retinoblastoma. J Natl Cancer Inst. 1993;85:1121–8. doi: 10.1093/jnci/85.14.1121. [DOI] [PubMed] [Google Scholar]
- 7.Bader JL, Miller RW, Meadows AT, Zimmerman LE, Champion LA, Voute PA. Trilateral retinoblastoma. Lancet. 1980;2:582–3. doi: 10.1016/s0140-6736(80)92009-7. [DOI] [PubMed] [Google Scholar]
- 8.Draper GJ, Sanders BM, Brownbill PA, Hawkins MM. Patterns of risk of hereditary retinoblastoma and applications to genetic counselling. Br J Cancer. 1992;66:211–9. doi: 10.1038/bjc.1992.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moll AC, Imhof SM, Bouter LM, Kuik DJ, Den Otter W, Bezemer PD. et al. Second primary tumors in patients with hereditary retinoblastoma: a register-based follow-up study, 1945-1994. Int J Cancer. 1996;67:515–9. doi: 10.1002/(SICI)1097-0215(19960807)67:4<515::AID-IJC9>3.0.CO;2-V. doi:10.1002/(SICI)1097-0215(19960807)67:4<515::AID-IJC9>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 10.Shields CL, Shields JA, Baez K, Cater JR, De Potter P. Optic nerve invasion of retinoblastoma. Metastatic potential and clinical risk factors. Cancer. 1994;73:692–8. doi: 10.1002/1097-0142(19940201)73:3<692::aid-cncr2820730331>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 11.Honavar SG, Singh AD, Shields CL, Meadows AT, Demirci H, Cater J. et al. Postenucleation adjuvant therapy in high-risk retinoblastoma. Arch Ophthalmol. 2002;120:923–31. doi: 10.1001/archopht.120.7.923. doi:ecs10156 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Ganguly A, Shields CL. Differential gene expression profile of retinoblastoma compared to normal retina. Mol Vis. 2010;16:1292–303. [PMC free article] [PubMed] [Google Scholar]
- 13.Knudson AG. Mutation and cancer: statistical study of retinoblastoma. Proceedings of the National Academy of Sciences of the United States of America. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalgard CL, Gonzalez M, deNiro JE, O'Brien JM. Differential microRNA-34a expression and tumor suppressor function in retinoblastoma cells. Invest Ophthalmol Vis Sci. 2009;50:4542–51. doi: 10.1167/iovs.09-3520. doi:iovs.09-3520 [pii] 10.1167/iovs.09-3520. [DOI] [PubMed] [Google Scholar]
- 15.Mu G, Liu H, Zhou F, Xu X, Jiang H, Wang Y. et al. Correlation of overexpression of HMGA1 and HMGA2 with poor tumor differentiation, invasion, and proliferation associated with let-7 down-regulation in retinoblastomas. Hum Pathol. 2010;41:493–502. doi: 10.1016/j.humpath.2009.08.022. doi:S0046-8177(09)00335-9 [pii] 10.1016/j.humpath.2009.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–96. doi: 10.1038/sj.onc.1209913. doi:1209913 [pii] 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 17.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O. et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. doi:ng1590 [pii] 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 18.Sioud M, Cekaite L. Profiling of miRNA expression and prediction of target genes. Methods Mol Biol. 2010;629:257–71. doi: 10.1007/978-1-60761-657-3_16. doi:10.1007/978-1-60761-657-3_16. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Grocock R, van Dongen S, Bateman A, Enright A. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. doi:gkm952 [pii] 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. doi:nrg2843 [pii] 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 22.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. doi:nrm1644 [pii] 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. doi:10.1038/nature02871 nature02871 [pii] [DOI] [PubMed] [Google Scholar]
- 24.Huang JC, Babak T, Corson TW, Chua G, Khan S, Gallie BL. et al. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–9. doi: 10.1038/nmeth1130. doi:nmeth1130 [pii] 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 25.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–55. doi: 10.1038/cgt.2008.8. doi:cgt20088 [pii] 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 26.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S. et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. doi:10.1073/pnas.0307323101 0307323101 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao JJ, Yang J, Lin J, Yao N, Zhu Y, Zheng J. et al. Identification of miRNAs associated with tumorigenesis of retinoblastoma by miRNA microarray analysis. Childs Nerv Syst. 2009;25:13–20. doi: 10.1007/s00381-008-0701-x. doi:10.1007/s00381-008-0701-x. [DOI] [PubMed] [Google Scholar]
- 28.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. doi:10.1073/pnas.242606799242606799 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H. et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. doi:10.1158/0008-5472.CAN-04-063764/11/3753 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ Jr. et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer research. 2009;69:8157–65. doi: 10.1158/0008-5472.CAN-09-1996. doi:0008-5472.CAN-09-1996 [pii] 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan X, Peng J, Fu Y, An S, Rezaei K, Tabbara S. et al. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple negative breast cancer. Breast cancer research: BCR. 2014;16:435.. doi: 10.1186/s13058-014-0435-5. doi:10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. doi:65/16/7065 [pii] 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 33.Chen L, Li Y, Fu Y, Peng J, Mo MH, Stamatakos M. et al. Role of deregulated microRNAs in breast cancer progression using FFPE tissue. PloS one. 2013;8:e54213.. doi: 10.1371/journal.pone.0054213. doi:10.1371/journal.pone.0054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X, Jia R, Zhou Y, Song X, Wang J, Qian G. et al. Microarray-based analysis: identification of hypoxia-regulated microRNAs in retinoblastoma cells. Int J Oncol. 2011;38:1385–93. doi: 10.3892/ijo.2011.961. doi:10.3892/ijo.2011.961. [DOI] [PubMed] [Google Scholar]
- 35.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–9. doi: 10.1038/cr.2008.24. doi:cr200824 [pii] 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 36.Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 13: R2. doi:bcr2803 [pii] 10.1186/bcr2803. [DOI] [PMC free article] [PubMed]
- 37.Song B, Wang C, Liu J, Wang X, Lv L, Wei L, MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res. 29: 29. doi:1756-9966-29-29 [pii] 10.1186/1756-9966-29-29. [DOI] [PMC free article] [PubMed]
- 38.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 90: 144-55. doi:labinvest2009126 [pii] 10.1038/labinvest. 2009. 126. [DOI] [PubMed]
- 39.Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P. et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55. doi: 10.1038/labinvest.2009.126. doi:labinvest2009126 [pii] 10.1038/labinvest.2009.126. [DOI] [PubMed] [Google Scholar]
- 40.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS. et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80. doi: 10.1128/MCB.00479-08. doi:10.1128/MCB.00479-08 MCB.00479-08 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang KH, Miller N, Kheirelseid EA, Ingoldsby H, Hennessy E, Curran CE. et al. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol. 2011;37:597–603. doi: 10.1016/j.ejso.2011.04.001. doi:S0748-7983(11)00247-2 [pii] 10.1016/j.ejso.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–5. doi: 10.1016/j.bbrc.2009.03.077. doi:S0006-291X(09)00543-9 [pii] 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 43.Schramedei K, Morbt N, Pfeifer G, Lauter J, Rosolowski M, Tomm JM, MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene. doi:onc201115 [pii] 10.1038/onc. 2011. 15. [DOI] [PMC free article] [PubMed]
- 44.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. doi:S0016-5085(07)01002-5 [pii] 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 411: 846-52. doi:S0009-8981(10)00169-5 [pii] 10.1016/j.cca. 2010. 02.074. [DOI] [PubMed]
- 46.Jazdzewski K, Boguslawska J, Jendrzejewski J, Liyanarachchi S, Pachucki J, Wardyn KA, Thyroid Hormone Receptor {beta} (THRB) Is a Major Target Gene for MicroRNAs Deregulated in Papillary Thyroid Carcinoma (PTC) J Clin Endocrinol Metab. 96: E546-53. doi:jc.2010-1594 [pii] 10.1210/jc.2010-1594. [DOI] [PMC free article] [PubMed]
- 47.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W. et al. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–66. doi: 10.1038/labinvest.2008.94. doi:labinvest200894 [pii] 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 48.Ziyan W, Shuhua Y, Xiufang W, Xiaoyun L. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med Oncol. doi:10.1007/s12032-010-9563-7. [DOI] [PubMed]
- 49.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC. et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–61. doi: 10.1002/ijc.22800. doi:10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 50.Liu M, Wu H, Liu T, Li Y, Wang F, Wan H. et al. Regulation of the cell cycle gene, BTG2, by miR-21 in human laryngeal carcinoma. Cell Res. 2009;19:828–37. doi: 10.1038/cr.2009.72. doi:cr200972 [pii] 10.1038/cr.2009.72. [DOI] [PubMed] [Google Scholar]
- 51.Antczak C, Kloepping C, Radu C, Genski T, Muller-Kuhrt L, Siems K. et al. Revisiting old drugs as novel agents for retinoblastoma: in vitro and in vivo antitumor activity of cardenolides. Invest Ophthalmol Vis Sci. 2009;50:3065–73. doi: 10.1167/iovs.08-3158. doi:iovs.08-3158 [pii] 10.1167/iovs.08-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48:3962–7. doi: 10.1167/iovs.06-1221. doi:48/9/3962 [pii] 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Knosel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S. et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200:640–6. doi: 10.1002/path.1378. doi:10.1002/path.1378. [DOI] [PubMed] [Google Scholar]
- 54.Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y. et al. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–12. doi: 10.1038/sj.onc.1209634. doi:1209634 [pii] 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 55.Wen YH, Shi X, Chiriboga L, Matsahashi S, Yee H, Afonja O. Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep. 2007;18:1387–93. [PubMed] [Google Scholar]
- 56.Reis PP, Tomenson M, Cervigne NK, Machado J, Jurisica I, Pintilie M. et al. Programmed cell death 4 loss increases tumor cell invasion and is regulated by miR-21 in oral squamous cell carcinoma. Mol Cancer. 2010;9:238.. doi: 10.1186/1476-4598-9-238. doi:1476-4598-9-238 [pii] 10.1186/1476-4598-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mudduluru G, Medved F, Grobholz R, Jost C, Gruber A, Leupold JH. et al. Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer. 2007;110:1697–707. doi: 10.1002/cncr.22983. doi:10.1002/cncr.22983. [DOI] [PubMed] [Google Scholar]
- 58.Chen Y, Liu W, Chao T, Zhang Y, Yan X, Gong Y. et al. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272:197–205. doi: 10.1016/j.canlet.2008.06.034. doi:S0304-3835(08)00526-0 [pii] 10.1016/j.canlet.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 59.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S. et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. doi:1210856 [pii] 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 60.Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388:539–42. doi: 10.1016/j.bbrc.2009.08.044. doi:S0006-291X(09)01592-7 [pii] 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 61.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–95. doi: 10.1093/nar/gkp117. doi:gkp117 [pii] 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu J, Zhu X, Li Y, Dong D, Yao J, Lin C. et al. miRNA-21 regulates arsenic-induced anti-leukemia activity in myelogenous cell lines. Med Oncol. 2011;28:211–8. doi: 10.1007/s12032-009-9413-7. doi:10.1007/s12032-009-9413-7. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Zhu X, Gu J, Dong D, Yao J, Lin C. et al. Anti-miR-21 oligonucleotide sensitizes leukemic K562 cells to arsenic trioxide by inducing apoptosis. Cancer Sci. 2010;101:948–54. doi: 10.1111/j.1349-7006.2010.01489.x. doi:CAS1489 [pii] 10.1111/j.1349-7006.2010.01489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Y, Zhu X, Gu J, Hu H, Dong D, Yao J. et al. Anti-miR-21 oligonucleotide enhances chemosensitivity of leukemic HL60 cells to arabinosylcytosine by inducing apoptosis. Hematology. 2010;15:215–21. doi: 10.1179/102453310X12647083620840. doi:10.1179/102453310X12647083620840. [DOI] [PubMed] [Google Scholar]
- 65.Chevez-Barrios P, Hurwitz MY, Louie K, Marcus KT, Holcombe VN, Schafer P. et al. Metastatic and nonmetastatic models of retinoblastoma. Am J Pathol. 2000;157:1405–12. doi: 10.1016/S0002-9440(10)64653-6. doi:S0002-9440(10)64653-6 [pii] 10.1016/S0002-9440(10)64653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laurie N, Mohan A, McEvoy J, Reed D, Zhang J, Schweers B. et al. Changes in retinoblastoma cell adhesion associated with optic nerve invasion. Mol Cell Biol. 2009;29:6268–82. doi: 10.1128/MCB.00374-09. doi:MCB.00374-09 [pii] 10.1128/MCB.00374-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schiffman JS, Grunwald GB. Differential cell adhesion and expression of N-cadherin among retinoblastoma cell lines. Invest Ophthalmol Vis Sci. 1992;33:1568–74. [PubMed] [Google Scholar]