Abstract
Typhoid fever poses significant burden on healthcare systems in Southeast Asia and other endemic countries. Several epidemiological and genomic studies have attributed pseudogenisation to be the major driving force for the evolution of Salmonella Typhi although its real potential remains elusive. In the present study, we analyzed genomes of S. Typhi from different parts of Southeast Asia and Oceania, comprising of isolates from outbreak, sporadic and carrier cases. The genomes showed high genetic relatedness with limited opportunity for gene acquisition as evident from pan-genome structure. Given that pseudogenisation is an active process in S. Typhi, we further investigated core and pan-genome profiles of functional and pseudogenes separately. We observed a decline in core functional gene content and a significant increase in accessory pseudogene content. Upon functional classification, genes encoding metabolic functions formed a major constituent of pseudogenes as well as core functional gene clusters with SNPs. Further, an in-depth analysis of accessory pseudogene content revealed the existence of heterogeneous complements of functional and pseudogenes among the strains. In addition, these polymorphic genes were also enriched in metabolism related functions. Thus, the study highlights the existence of heterogeneous strains in a population with varying metabolic potential and that S. Typhi possibly resorts to metabolic fine tuning for its adaptation.
Salmonella are enteric bacteria that can infect a broad range of host species causing various infectious diseases. Presently, there are two well recognized species of Salmonella - S. bongori and S. enterica. Further, based on Kauffman-White classification scheme, S. enterica is divided into six distinct subspecies and more than 2500 serovars1. However, serovar S. Typhi of Salmonella enterica subspecies enterica infects only humans resulting in a systemic infection, typhoid fever2. This infection is of immense concern to public health worldwide as it is responsible for about 21.6 million cases of which 1% become fatal, on an average, per year3. About 90% of this morbidity and mortality stems from Asia due to high endemicity of typhoid fever in developing countries where drinking water quality and sewage treatment facilities are poor4. The control and prevention strategies are also severely hampered due to the emergence of antibiotic resistant strains in these regions, which are responsible for periodic outbreaks and sporadic cases causing severe complications and mortality5. Further, the presence of these antimicrobial resistance genes carried on mobile elements such as integrons and self-transmissible plasmids like that of IncH1, which was reported to be associated with many strains from endemic zones such as Vietnam, pose a constant threat to public health world-wide6,7.
For a host adapted strain like S. Typhi, survival in the host and dissemination are vital for establishing persistent infections. Some infected individuals even serve as asymptomatic reservoirs who continue to shed the bacterium in stools for a long period of time8,9. The studies on transmission dynamics of salmonellae have emphasized on monitoring of the carrier isolates for effective epidemiological tracking and surveillance10. The carrier isolates have also been shown to exhibit similar pulsed field gel electrophoresis (PFGE) profiles with other S. Typhi isolates from various regions of Southeast Asia. Therefore, it appears that the spread of S. Typhi occurs mostly through carrier individuals11.
In the past, various genomic studies have attributed signatures like pseudogenisation (loss of gene function) or gene deletion for host restriction in pathogenic bacteria12. It was also reported that in human-restricted serovar S. Typhi, pseudogenisation is an active process compared to other generalist serovars like S. Typhimurium13. Further, the extent of this pseudogenisation also varies considerably even among host restricted serovars14,15,16. Pseudogenes constitute up to 4.5% of S. Typhi gene pool, making them an important driver of genome re-assortment over time17. However the potential role of pseudogenisation in persistence and adaptation of S. Typhi still remains elusive.
Given this, it is important to characterize S. Typhi's pan-genome, more importantly with respect to functional and pseudogene complements and investigate their gene-frequency distributions among various strains. The pan-genome of a species is the complete inventory of genes in the population and is always significantly greater than the gene content of an individual18. The pan genome is composed of both ‘core genome’ and ‘accessory genome’ where accessory part is comprised of genes shared by some but not all strains. This accessory or dispensable part confers various selective advantages such as antibiotic resistance, niche adaptation, pathogenicity and host specificity19,20. However, the residual core part of genome that keeps a very high sequence similarity of about 95% ANI (Average Nucleotide Identity), encodes all the fundamental biological processes essential for survival18. Thus, the pan-genome analysis helps us to better understand the population genetic structure and provides cues about the mechanisms underlying adaptation and evolution of bacteria. Studies based on whole genome comparative analyses carried out at the population level involving other S. enterica serovars such as Paratyphi A and Agona have recently provided significant insights into the evolution of these serovars21,22.
The whole genome sequences corresponding to eight strains previously isolated from different endemic regions of Southeast Asia and Oceania were extensively analyzed in this study. These strains were associated with different clinical manifestations - outbreaks, sporadic cases, carrier strains and fatal episodes. The strain BL196 was associated with a large outbreak in Kelantan, Malaysia in 200523. Strains CR0044 and CR0063 were isolated from carrier individuals in 2007 after an outbreak and were reported to share PFGE profiles with the strain BL19624,25. Strain ST0208 was associated with a sporadic case in Kuala Lumpur, Malaysia26. The previous findings have also recorded shared PFGE patterns among the isolates from Southeast Asia11. Strains UJ308A and UJ816A were isolated in Papua New Guinea from fatal and non-fatal cases, respectively, in 199827. Genomes of multi-drug resistant strains, CT18 from Vietnam, and P-stx-12 strain isolated from a carrier individual in India28,29, were also analyzed. Some of the earlier studies based on PFGE observed minimal to moderate diversity among the isolates from Papua New Guinea and elsewhere in Asia30,31, thus verifying the limited observed diversity if not a clonal nature of this organism. Herein, we analyzed genomes of the strains described in some of the above pioneering studies. These genomes, although limited in number, were chosen owing to their being most authentic available representatives of geographically distinct populations from different endemic countries such as India, Vietnam, Papua New Guinea and Malaysia and thus were used for extensive genomic analyses hitherto unreported for these unique strains. Our comprehensive genomic analyses reported herein highlight the possible evolutionary mechanisms and in particular, the impact of pseudogenes on the evolution of S. Typhi in different patient types from the typhoid-endemic countries of the east.
Results
Phylogeny
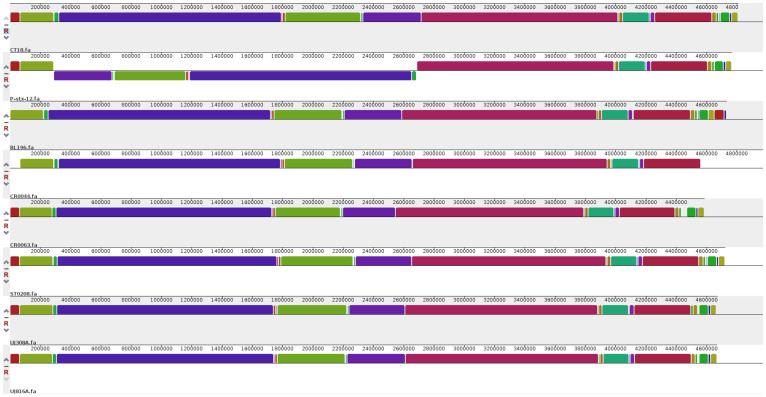
The whole genome based phylogenetic tree allowed us to understand the close genetic relationship among various strains as shown in Figure 1. The strain BL196 isolated during the outbreak, and the carrier strain CR0044 isolated a year later, co-clustered revealing close similarity. This suggests that the strain CR0044 could have emerged due to clonal expansion of BL196, whereas another carrier strain CR0063 might have accumulated enough variations allowing it to cluster separately. The two strains isolated from Papua New Guinea, UJ816A and UJ308A, also clustered together with respect to all other strains. This observation by whole genome based phylogeny corroborates with the PFGE based analysis of Thong et al, where S. Typhi strains from Papua New Guinea showed highly similar PFGE patterns exhibiting limited genetic diversity among the strains30. As typhoid cases were rarely detected in Papua New Guinea before 1985, the limited observed diversity might be due to clonal expansion of a single ancestral strain30. The strains CT18, P-stx-12, ST0208 have shown up independently in the tree. The close similarity of these genomes is also reflected in whole genome alignment as depicted (Figure 2). This analysis once again reinforces the genetically monomorphic nature of this pathogen and our observations are in concurrence with the previous findings based on Multilocus sequence typing and other techniques17,32. A similar co-clustering pattern was also observed with Maximum Likelihood based phylogenetic tree constructed using core gene clusters without paralogs (Supplementary Figure S1).
Figure 1. Phylogenomic Tree.
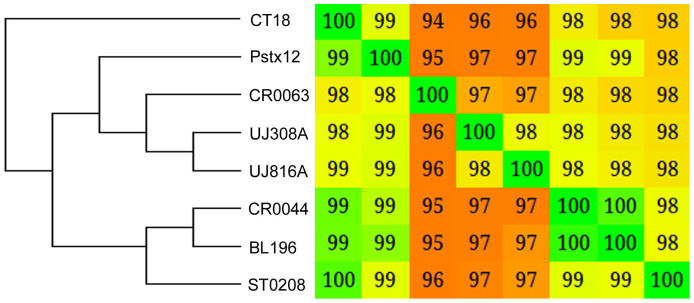
The whole genome information was used to build the distance matrix using Gegenees. The phylogenetic tree was developed using SplitsTree by NJ method. This revealed close similarity among genomes and also co-clustering of strains isolated from the same regions.
Figure 2. Genome alignment.
The whole genome alignment of all eight genomes was generated using progressiveMauve50. Each colored block represents similar sequences in the respective genomes.
Mobile elements
The phages and insertion sequence (IS) elements of the two complete genomes, CT18 and P-stx-12 have been already reported28,29. The IS elements belonging to the family IS200/605, IS3, IS256 were commonly observed in all the other draft genomes we analyzed herein. However, the strain CR0063 also contained copies that belonged to IS1 family. The determination of exact copy number of these IS elements was difficult because of the draft status of the genomes. Further search for putative phage elements revealed presence of 4 intact phage sequences together with various phage remnants in each of the genomes (Supplementary table S1). Gifsy-2 and fels-2 phage sequences were common in most of the genomes. The atypical regions which encode genes mostly associated with virulence are designated as Salmonella pathogenicity islands (SPI). BRIG33 was used to represent the status of major pathogenicity islands in these genomes as shown in Supplementary Figure S2. A list of genomic islands detected in these genomes as well as of the genes encoded by them is provided (Supplementary table S2). The plasmid related genes were not found in any strains other than CT18 and P-stx-12. The characteristics of the plasmids present in these strains along with the orthologous genes shared by them have already been discussed previously28,29.
Pan-genome analysis
The pan-genome content measured up to a total of 5426 genes, 1.07 times higher than the average number of genes per individual strain. The pan-genome extrapolation was carried out in accordance with Heap's law34. The Heap's law can be represented by the following equation:
n = k*N−α, where n is pan-genome size, N is the number of genomes and k, γ are curve specific constants where α = 1 − γ.
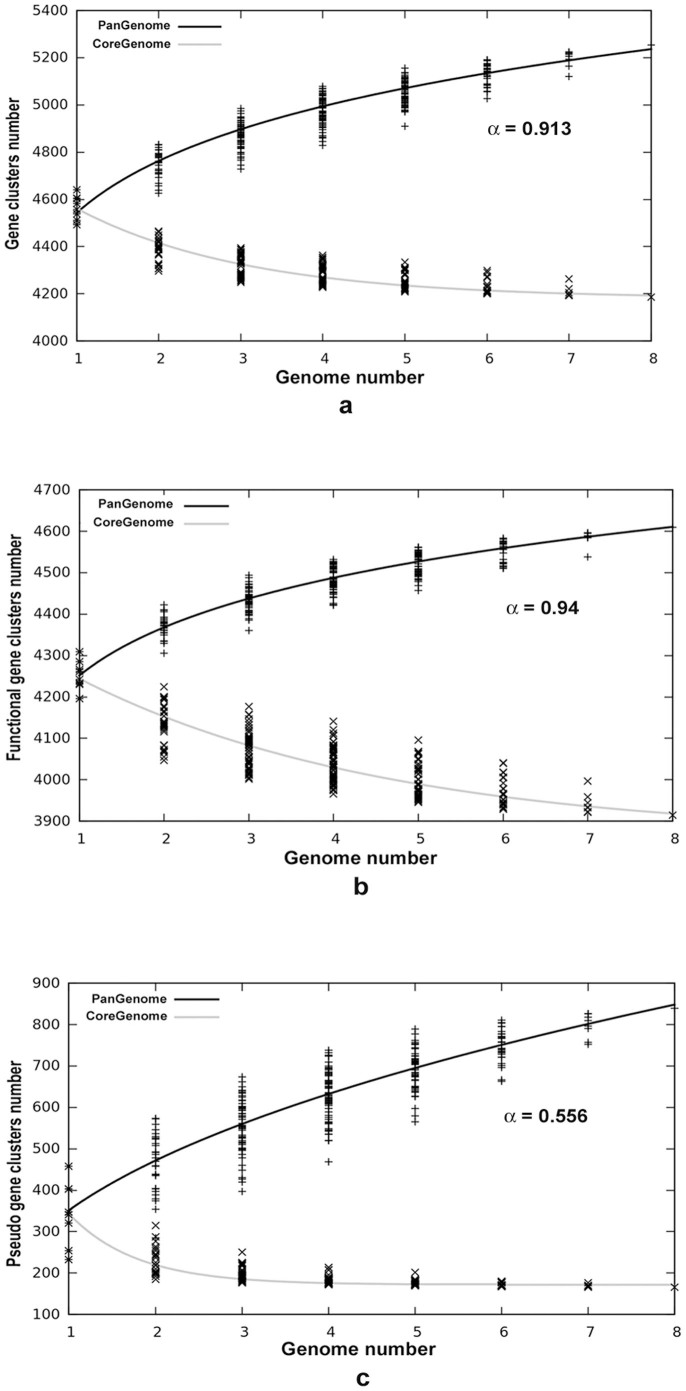
The exponential term α determines whether pan-genome of a bacterial species is closed or open. For α > 1 (γ < 0) the pan-genome is considered closed i.e. sampling more genomes will not affect the pan-genome size, whereas for α < 1 (0 < γ > 1) the pan-genome remains open and addition of more genomes would increase its size. In this study, the k and γ values were determined as 4486 and 0.087 respectively. The pan-genome analysis of S. Typhi strains revealed an α value of 0.913 implying a highly conservative nature of these endemic isolates (Figure 3a).
Figure 3. Pan and Core Genome Distribution.
(a). Pan and core genome developments using median values of the combinations of all eight genomes. (b). Pan and core genome developments of functional genes of these eight isolates. Here it can be observed that core genome is decreasing sharply. (c). Pan and core genome developments of pseudogenes of these eight isolates. It can be seen that pan genome of pseudogenes is highly non conservative with a steep increase in accessory content while the core genome reached convergence.
Further, to investigate the effect of pseudogenisation on gene frequency distributions of functional and pseudogenes, their pan and core genome components were determined separately. The pan-genome of functional genes contained a total of 4632 genes which was 1.03 times the average functional gene content per strain, whereas the pan-genome of pseudogenes contained a total of 857 genes which was 2.49 times the average pseudogene content per strain. This increased proportion of pseudogene content compared to functional pan-genome suggests that pseudogenisation is an active process in S. Typhi and this increase is also reflected in the pan-genome curve of pseudogenes (Figure 3c).
The values k and γ for functional gene clusters after curve fitting were determined as 4054 and 0.06 respectively, with a α value of 0.94. In contrast with what we observed in functional genes scenario, after curve fitting, α value for pseudogenes was 0.556 (Figure 3b, 3c), with k and γ values determined as 342.5 and 0.444 respectively. The α value of 0.94 indicates that the pan-genome of the functional genes is highly restricted in nature to allow any significant intake of foreign DNA and thus corroborates with high collinearity observed among these endemic strains. However the pan-genome of pseudogenes with an α value of 0.556 showed a very non-conservative nature as shown in Figure 3c and thus reemphasizes that pseudogenisation of functional genes is an ongoing process in S. Typhi.
The core genome of S. Typhi
The core genome of a species includes a subset of genes that are shared by all strains. The core genome of our endemic strains contained 4131 genes. This core genome size tended to decrease upon increasing the number of genomes; therefore, the curve fitting and extrapolation was done by least square fit of the exponential regression decay. This equation is written as: 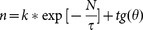 , where n is the expected core genome size, N denotes number of genomes and k, tg(θ) are constants that fit the curve. In this equation, the first term
, where n is the expected core genome size, N denotes number of genomes and k, tg(θ) are constants that fit the curve. In this equation, the first term 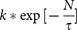 will tend towards zero and the second term tg(θ) tends to converge towards a specific value. The analysis revealed a convergence value of 4124 genes which indicates a minimal genome content retained by the bacteria to perform basic biological processes (Figure 3a).
will tend towards zero and the second term tg(θ) tends to converge towards a specific value. The analysis revealed a convergence value of 4124 genes which indicates a minimal genome content retained by the bacteria to perform basic biological processes (Figure 3a).
The core genome of functional and pseudogenes was also determined separately and these distributions provided some significant pointers. The core genome of functional genes was determined to be around 3558 genes and was still decreasing as shown in core genome curve of functional genes (Figure 3b) with the convergence value tg(θ) as 3495 obtained upon solving the equation. However, in the case of pseudogenes, the core genome has already reached its convergence with a tg(θ) value of 166 genes (Figure 3c). Further, the core genome profiles of functional and pseudogenes (Figure 3b, 3c) imply that core genome of pseudogenes constitutes a minor component of the total pseudogene content unlike that of core genome of functional genes. Moreover, these findings also stress on the need to analyze the role of these high number of accessory pseudogenes which are causing a steep increase in its pan-genome.
The pseudogenes identified in this analysis (Supplementary table S3) included various fimbrial proteins, methyl accepting chemotaxis proteins and certain secreted effector proteins. Some of these pseudogenes were potentially homologous to the genes found to be associated with important cellular functions such as anaerobic metabolism – ethanolamine utilization, being precursors of vitB12 synthesis, or acting as electron donors (formate dehydrogenase, galactarate dehydrogenase, succinyl glutamic semialdehyde dehydrogenase) and acceptors (tetrathionate reductase, trimethylamine-N-oxide reductase, nitric oxide reductase). The affordability to dispense such genes in intracellular bacteria like S. Typhi has already been previously reported and discussed35,36.
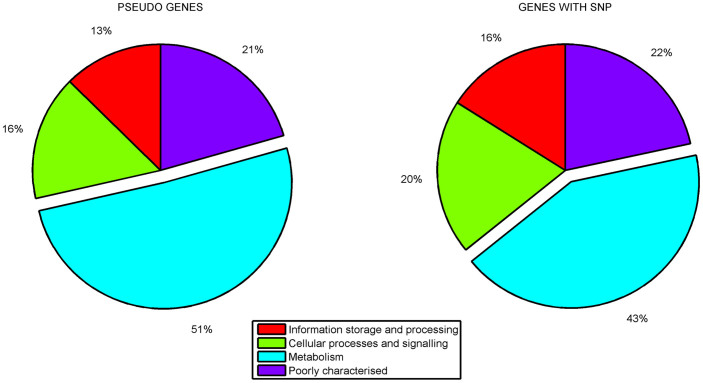
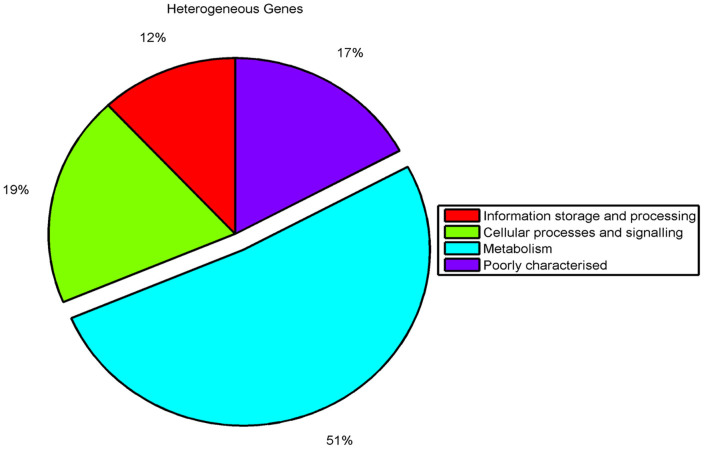
Further to evaluate pseudogene distribution among various functional classes, they were classified into COG functional categories based on RPS BLAST. This analysis showed that, of those functionally classified, majority of the pseudogenes were related to metabolic processes: carbohydrate metabolism, amino acid transport and metabolism, inorganic ion transport etc. (Figure 4a). Further, when core functional gene clusters with SNPs (604clusters out of 3333 core clusters) were assigned COG classification, a higher proportion of functionally classified genes was observed in the same functional categories related to metabolic functions as observed in case of pseudogenes (Figure 4b). This enrichment of pseudogenes observed in the metabolism related genes was also statistically significant according to the proportionality z-test. Thus, from our observations, as depicted (Figure 4a, 4b), it can be inferred that metabolism related gene repertoire is under constant fine tuning and might relate to a rapid adaptation to the immediate local niche.
Figure 4. The proportion of functionally classified pseudogenes and the functional genes with SNPs according to COG classification.
The pie chart represents the proportion of various functional classes among the pseudogenes and the functional genes with SNPs. The figure clearly shows the enrichment of metabolism related genes in pseudogenes and the functional genes with SNPs.
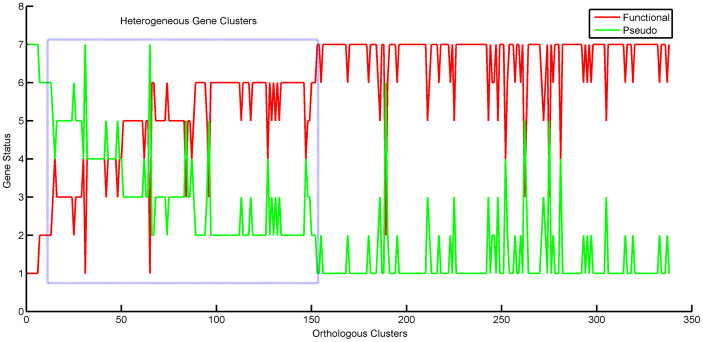
To gain further insights into the differential pseudogene content among various strains, we focused on the accessory pseudogene clusters marked by absence of a corresponding ortholog in at least one or more strains. For this analysis, only those accessory pseudogene clusters which do not have paralogs were considered. The absence of an ortholog in these clusters indicated only two possibilities: either the ortholog is not present in the strain or there exists a functional complement in the strain. Therefore, status of these accessory pseudogenes in each cluster was marked as P for pseudogene, F for functional complement and N for absence of a gene. Finally, we considered only those clusters where the orthologs were present in P or F states and removed those which had N in any of the strains. The plot of these pseudogene clusters along with their respective status in the genomes revealed a mixed profile (Figure 5). This analysis provides evidence for the existence of a heterogeneous mixture of functional and pseudogene complements in the population. Further, COG classification of those pseudogene clusters with P or F status in at least two query strains has shown that these were also enriched in metabolism related functions and this proportion was statistically significant (Figure 6). Thus the comparison points at the existence of heterogeneous strains with varying metabolic potential and might confer an adaptive advantage for the persistence of the pathogen.
Figure 5. Accessory pseudogene clusters analysis.
The status of genes in each accessory pseudogene cluster was marked as P for pseudogene, F for functional complement and N for absence of gene. The clusters where the orthologs were present in P or F states were considered in plot. This shows the heterogeneous existence of functional and pseudogene complements in the population.
Figure 6. Proportion of heterogeneous genes classified according to COG functional categories.
The figure represents the distribution of accessory pseudogenes (those having variable functional and pseudogene status in atleast two strains) among various COG functional categories. The genes related to metabolism were clearly enriched in the accessory pseudogenes.
We also performed pairwise comparisons of the strains of different clinical spectrum in order to determine if there are any state specific genes that entail different clinical level phenotypes of the strains. For this, we considered a total of four different pair-wise comparisons: outbreak versus carrier strains in 2 sets (BL196 & CR0044 and, BL196&CR0063) from Kelantan, Malaysia; a pair of strains (BL196 & ST0208) associated with an outbreak (BL196) and sporadic case (ST0208) from Malaysia; and a pair of strains (UJ308A & UJ816A) associated with fatal (UJ308A) and non-fatal (UJ816A) cases from Papua New Guinea. The core and specific functional gene content as well as pseudogene content were determined for all these strains. Further, after identifying the specific functional and pseudogenes of each strain in comparison, we also checked if a given strain in comparison carried an ortholog in a different functional state (functional or pseudo) than that of its corresponding strain, or vice versa. In this way, gene contents of all the strains were analyzed. Although this analysis helped us develop pairwise inventories of complimentary functional genes and pseudogenes, it did not identify any specific pattern of potential associations that could be attributed to a strain of a certain clinical spectrum conveying an acute or a carrier stage, for example.
Discussion
The previous whole genome based study on S. Typhi by Holt et al17 revealed that these genomes are highly clonal with minimal genomic variation due to SNPs, recombination and horizontal gene acquisition. In the present study also we observed a close genetic relatedness among the strains from different endemic zones of Southeast Asia/Oceania sharing similarity of 94–98% as shown (Figures 1, 2). In the past, comparative genomic studies have proposed that pseudogenisation is the main driving force in evolution of this organism when compared to others like acquiring foreign genetic material through HGT (Horizontal Gene Transfer) or gain of function17,28,35. Therefore, we attempted to understand the pseudogene pool of various isolates in greater detail and cues they could provide about various adaptation and survival mechanisms harnessed by this pathogen. We identified most of the pseudogenes including those caused due to frameshift mutations, as these were not detected by Holt et al because of the low quality of sequence data17.
The pan-genome analysis of these isolates has revealed limited potential for horizontal gene acquisition. This characteristic of the gene pool is a commonly observed phenomenon in case of intracellular organisms as they have limited contact with the potential gene donors37. Moreover, when the same analysis was carried out for functional and pseudogenes separately, it was observed that the core content of functional genes is still declining whereas the pseudogene content recorded steady increase in pan-genome (Figure 3). This decreasing trend of functional core genome and an increase in the pan-pseudogene content indicates that potential loss of functional genes might be a consequence of active pseudogenisation. Though an active pseudogenisation was observed, we could not detect any significant reduction in genome size or gene content indicating that pseudogenisation perhaps does not entail concurrent or consequent gene deletion in case of S. Typhi in contrast to other important human pathogens such as Mycobacterium leprae wherein pseudogenisation is followed by deletion, thus downsizing the genome38. Further, this finding could just be a reflection of different inactivating mutation rates or varying negative or positive selection pressures experienced by different isolates and/or lineages. Another possible explanation could be due to reversion of pseudogenes39 although any gain of function may be rare, or only occurring in a small number of genes through point mutations17. Given this situation, a definitive mechanism can be confirmed only through genetic and functional studies involving serial isolates. Collectively, from these findings, we believe that over the course of evolution, S. Typhi has resorted to maintain its genome size through a fine balance between functional and pseudogenes.
Further, when the core functional gene clusters with SNPs were functionally classified, it was observed that these genes majorly belonged to metabolism related functions. A significant number of pseudogenes also belonged to same functional category as core functional genes with SNPs (Figure 4). This convergence of the core functional genes with SNPs and the pseudogenes indeed emphasizes the stress on the metabolic machinery. In addition, the analysis of accessory pseudogene clusters identified 338 clusters with mixed profile of pseudo and functional gene complements in various strains (Figure 5). Upon functional classification, even these polymorphic genes were found to be enriched in metabolism related functions (Figure 6). This could be an advantageous mechanism for the bacterium to modulate its metabolic repertoire through pseudogenisation depending on its specific local niche40. Similar survival strategy is reported in other pathogenic bacteria where virulence optimization is achieved at the cost of certain metabolic genes41,42. Given this, it can be surmised that this modulation could be one of the major mechanisms underlying the carrier state. Hence, it is important to focus on characterizing the metabolic potential and its implications on virulence of S. Typhi.
The heterogeneity displayed by functional and pseudogene content of these isolates, especially even in those collected from the same region (Kelantan, Malaysia) over a period of time, provides explanation for the interplay between them and supports previous hypothesis that the restoration of function might be occurring through mutation17. However this can be only further proved with functional studies on serial isolates from a single individual.
S. Typhi encounters drastically different environments from its initial point of entry into the small intestine up to its final colonization of internal organs like gall bladder for chronic carriage43. To succeed in these varying environments, it might be very important to optimize its metabolism through loss of function, conferring an advantage within its immediate local niche.
The above observations regarding pan and core genome distributions of functional and pseudogenes lend support to the idea that S. Typhi maintains an efficient balance through various mechanisms, such that its genome is not degraded beyond a certain level. At the same time, a heterogeneous profile of functional and pseudo gene complements could possibly culminate in a more hospitable metabolic environment. Collectively, these orchestrated genome dynamics most likely appear to aid in persistence and host adaptation. Genome analysis of this limited but important collection of strains could provide us some significant pointers regarding adaptation of this organism which appears to be possibly influenced by a conserved nature of its genome. Further, inclusion of more number of genomes in the analysis would possibly enhance the understanding of these observations.
Nevertheless, given these findings, it will be possible to advance the current knowledge of the carrier state in Salmonella pathogens underlying continuous emergence and reemergence of typhoid in endemic regions.
Methods
Sequence information
The S. Typhi strains chosen for the analysis have been isolated from various countries of Southeast Asia/Oceania and were isolated at different time points by different researchers. The genome collection included two complete genomes - CT18, P-stx-12 strains and six draft genomes - UJ308A, UJ816A, BL196, CR0063, CR0044 and ST0208 which were available in public domain through NCBI. Few other strains from Southeast Asia which are available in NCBI could not be included in the analysis because of the low sequence coverage and poor quality of data.
Refinement of assembly and annotation
The contigs from WGS master records of S. Typhi genomes (UJ308A, UJ816A, BL196, CR0044, CR0063, ST0208) were downloaded from NCBI. These contigs were ordered according to a reference using standalone BLAST. The high quality filtered reads of respective strains were mapped to the contigs using BWA alignment tool44. The alignment file was visualized using Tablet alignment viewer45 to sort the scaffolds in correct order based on paired-end read information. The regions with low coverage were manually inspected before including them into final genome.
After finalizing the order of the contigs, they were linked using a linker sequence (NNN NNC ATT CCA TTC ATT AAT TAA TTA ATG AAT GAA TGN NNN N) that encodes start and stop codons in all six frames. The contigs thus obtained was submitted to ISGA pipeline for annotation46. The two complete genomes CT-18 and P-stx-12 were also re-annotated to homogenize the data with a single annotation platform. This annotation pipeline uses Glimmer 3 for prediction of ORFs and BLASTx for searches based on sequence similarity47. The predicted ORFs were scanned to identify protein domains using HMMPfam (http://hmmer.janelia.org/). The tRNAscan-SE and RNAmmer were used respectively for the detection of tRNA and rRNA48,49. The whole genome alignment of all these genomes was generated using progressive Mauve50.
For the identification of pseudogenes, BLASTn was performed for all the nucleotide sequences of query ORFs against the functional proteins of Salmonella strains, which were submitted to NCBI as complete genomes. The corresponding protein sequences of the best five hits of nucleotide BLAST were considered for performing BLASTx against individual query ORFs. Then, to mark the latter as a pseudogene, based on above results, threshold of more than 60% coverage of query length and 98% identity were applied. The above method could detect all the pseudogenes that originated due to a nonsense mutation resulting in early termination of translation. To identify pseudogenes that were formed due to potential frame shifts causing protein fragmentation were identified using an inbuilt module of PanOCT51. The BLASTp result of query ORFs against the functional genes of Salmonella strains was provided as input to PanOCT. The number of BLAST matches needed to confirm a protein fragment/frame-shift was set to 1 and the frame-shift overlap parameter as 1.33. In the case of proteins which are split due to frame-shifts, the major fragment was considered in the final pseudogene list, so that the number did not over-represent.
Phylogenomic analysis
The whole genome based phylogeny was performed for all eight strains using Gegenees (version 1.1.4)52 which employs a fragmented all-against-all comparison of the genomes and builds a distance matrix file suitable to construct a phylogenetic tree and heat map. The phylogenetic tree was built by NJ (Neighbor-joining) method using SplitsTree software (version 1.1.4)53. The detailed methodology of core genome based phylogeny using maximum likelihood is explained in Supplementary information.
Detection of mobile elements
All of these strains included various mobile phages or phage like elements. To identify these elements, PhiSpy54, an algorithm that combines both similarity and composition based strategies, was used. These predictions were compared with results obtained from PHAST (A Fast Phage search tool)55. IS elements were identified using IS finder56. The genomic islands in these strains were identified using IslandViewer57.
Pan-genome analysis
Pan-genome analysis represents the variation in gene content of different strains. The determination of pan and core genome requires correct identification of orthologous clusters of all selected strains. This was done using OrthoMCL58 which is mainly developed for clustering of orthologous protein sequences based on user defined percent match cutoff and minimum protein length.
Further, the pan-genome and core genome of the two strains A and B (AB) were calculated as follows: pan-genome AB is composed of the sum of gene sets of A and B (strain A and non-orthologous genes of strain B) and the core genome AB is composed of orthologous genes that are present in both A and B. Upon addition of more genomes, pan-genome was estimated in an additive manner whereas the core genome was determined in a reductive manner. The median values of all possible combinations of genomes were considered to further examine the patterns of pan- and core genomes. The curve fitting of pan-genome was done using Heap's law whereas that of core genome using least square fit of the exponential regression decay as described previously by Tettelin et al34.
Initially, orthologous clusters of all strains were generated with the percent match threshold of 85% and minimum protein length of 50 amino acids. Later, the functional genes and pseudogenes were analyzed separately as mentioned by Liang et al59, but their respective orthologous clusters were generated using OrthoMCL with same percent match cutoff. However, minimum protein length considered for generating functional gene clusters was 50 amino acids whereas for pseudogenes it was set to 10 amino acids. The extrapolations of pan-genome and core genome of functional and pseudogenes was done as mentioned above, but individually for each of them. The detailed explanation of COG classification using RPS BLAST (NCBI) is given in supplementary information. The statistical two sample z-proportionality test was applied to calculate the significance for the enrichment of various functional classes.
Detection of SNP in core genome
The core functional gene clusters were identified as those which contain only one representative protein from each of the query strain. SNP detection in these core functional gene clusters was done by aligning the corresponding sequences of each cluster using ClustalW60.
Author Contributions
R.B. and N.A. designed and conducted the study. N.K. helped in analysis of data and preparation of the manuscript. A.K.L. and S.S. provided help in writing scripts.
Additional Information
Accession codes CT18 (NC_003198), P-stx-12 (NC_016832), UJ308A (AJTD00000000), UJ816A (AJTE00000000), BL196 (AJGK00000000), CR0063 (AKIC00000000), CR0044 (AKZO00000000) and ST0208 (AJXA00000000).
Supplementary Material
Supplementary information
Table S2
Table S3
Acknowledgments
We would like to thankfully acknowledge Prof. Kwai-Lin Thong for information about some of the strains studied herein and to all those authors who contributed to data about these strains as available in the public domain. We would also like to thank Donepudi RaviTeja for his help with gnuplot and R. We thankfully acknowledge funding received from Department of Biotechnology, Government of India (Ref. No. BT/HRD/NBA/34/01/2011(ix) and BT/PR6921/MED/29/699/2013). RB would like to acknowledge the UGC-RFSMS fellowship.
References
- Coburn B., Grassl G. A. & Finlay B. B. Salmonella, the host and disease: a brief review. Immunol Cell Biol 85, 112–118 (2007). [DOI] [PubMed] [Google Scholar]
- Boyle E. C., Bishop J. L., Grassl G. A. & Finlay B. B. Salmonella: from pathogenesis to therapeutics. J Bacteriol 189, 1489–1495 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J. A., Luby S. P. & Mintz E. D. The global burden of typhoid fever. Bull World Health Organ 82, 346–353 (2004). [PMC free article] [PubMed] [Google Scholar]
- Gopinath S., Carden S. & Monack D. Shedding light on Salmonella carriers. Trends Microbiol 20, 320–327 (2012). [DOI] [PubMed] [Google Scholar]
- Chandel D. S., Chaudhry R., Dhawan B., Pandey A. & Dey A. B. Drug-resistant Salmonella enterica serotype Paratyphi A in India. Emerg Infect Dis 6, 420–421 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E. et al. Temporal fluctuation of multidrug resistant salmonella typhi haplotypes in the Mekong river delta region of Vietnam. PLoS Negl Trop Dis 5, e929 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploy M. C. et al. Integron-associated antibiotic resistance in Salmonella enterica serovar typhi from Asia. Antimicrob Agents Chemother 47, 1427–1429 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby T., McLaughlin L., Gopinath S. & Monack D. Salmonella's long-term relationship with its host. FEMS Microbiol Rev 36, 600–615 (2012). [DOI] [PubMed] [Google Scholar]
- Kalai Chelvam K., Chai L. C. & Thong K. L. Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut Pathog 6, 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzas C. et al. The effect of heterogeneous infectious period and contagiousness on the dynamics of Salmonella transmission in dairy cattle. Epidemiol Infect 136, 1496–1510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thong K. L. et al. Analysis of Salmonella typhi isolates from southeast Asia by pulsed-field gel electrophoresis. J Clin Microbiol 33, 1938–1941 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumler A. & Fang F. C. Host specificity of bacterial pathogens. Cold Spring Harb Perspect Med 3, a010041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 (2001). [DOI] [PubMed] [Google Scholar]
- Holt K. E. et al. Pseudogene accumulation in the evolutionary histories of Salmonella enterica serovars Paratyphi A and Typhi. BMC Genomics 10, 36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C. H. et al. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res 33, 1690–98 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson N. R. et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18, 1624–1637 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E. et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet 40, 987–993 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F. U. D. Is the pan-genome also a pan-selectome? F1000Res 1, 16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial ‘pan-genome’. Proc Natl Acad Sci U S A 102, 16530–16530 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D., Donati C., Tettelin H., Masignani V. & Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev 15, 589–594 (2005). [DOI] [PubMed] [Google Scholar]
- Zhou Z. et al. Transient Darwinian selection in Salmonella enterica serovar Paratyphi A during 450 years of global spread of enteric fever. Proc Natl Acad Sci U S A 111, 12199–12204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. et al. Neutral genomic microevolution of a recently emerged pathogen, Salmonella enterica serovar Agona. PLoS Genet 9, e1003471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddam R. et al. Genetic Fine Structure of a Salmonella enterica Serovar Typhi Strain Associated with the 2005 Outbreak of Typhoid Fever in Kelantan, Malaysia. J Bacteriol 194, 3565–3566 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. P. et al. Genome Sequence and Comparative Pathogenomics Analysis of a Salmonella enterica Serovar Typhi Strain Associated with a Typhoid Carrier in Malaysia. J Bacteriol 194, 5970–5971 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddam R. et al. Genome sequencing and analysis of Salmonella enterica serovar Typhi strain CR0063 representing a carrier individual during an outbreak of typhoid fever in Kelantan, Malaysia. Gut Pathog 4, 20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. P. et al. Insights from the Genome Sequence of a Salmonella enterica Serovar Typhi Strain Associated with a Sporadic Case of Typhoid Fever in Malaysia. J Bacteriol 194, 5124–5125 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddam R. et al. Whole-Genome Sequences and Comparative Genomics of Salmonella enterica Serovar Typhi Isolates from Patients with Fatal and Nonfatal Typhoid Fever in Papua New Guinea. J Bacteriol 194, 5122–5123 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J. et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413, 848–852 (2001). [DOI] [PubMed] [Google Scholar]
- Ong S. Y. et al. Complete Genome Sequence of Salmonella enterica subsp enterica Serovar Typhi P-stx-12. J Bacteriol 194, 2115–2116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thong K. L. et al. Molecular analysis of isolates of Salmonella typhi obtained from patients with fatal and nonfatal typhoid fever. J Clin Microbiol 34, 1029–1033 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza S., Kariuki S., Mamun K. Z., Beeching N. J. & Hart C. A. Analysis of plasmid and chromosomal DNA of multidrug-resistant Salmonella enterica serovar typhi from Asia. J Clin Microbiol 38, 1449–1452 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidgell C. et al. Salmonella typhi, the causative agent of typhoid fever, is approximately 50,000 years old. Infect Genet Evol 2, 39–45 (2002). [DOI] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L. & Beatson S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC genomics 12, 402 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Riley D., Cattuto C. & Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11, 472–477 (2008). [DOI] [PubMed] [Google Scholar]
- McClelland M. et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet 36, 1268–1274 (2004). [DOI] [PubMed] [Google Scholar]
- Nuccio S. P. & Baumler A. J. Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. MBio 5, e00929–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuenne C. et al. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC genomics 14, 47 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T. et al. Massive gene decay in the leprosy bacillus. Nature 409, 1007–1011 (2001). [DOI] [PubMed] [Google Scholar]
- Olson M. V. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet 64, 18–23 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer L., Hocquet D. & Miller S. I. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol 19, 341–348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurelli A. T., Fernandez R. E., Bloch C. A., Rode C. K. & Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci U S A 95, 3943–3948 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M. et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS genet 5, e1000344 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis R. S. & Horn F. Enteropathogenic Escherichia coli, Samonella, Shigella and Yersinia: cellular aspects of host-bacteria interactions in enteric diseases. Gut Pathog 2, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I. et al. Tablet-next generation sequence assembly visualization. Bioinformatics 26, 401–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich C., Buechlein A., Podicheti R., Revanna K. V. & Dong Q. F. An Ergatis-based prokaryotic genome annotation web server. Bioinformatics 26, 1122–1124 (2010). [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Harmon D., Kasif S., White O. & Salzberg S. L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res 27, 4636–4641 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P., Brooks A. N. & Lowe T. M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33, W686–W689 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K. et al. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res 35, 3100–3108 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B. & Perna N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PloS one 5, e11147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts D. E., Brinkac L., Beck E., Inman J. & Sutton G. PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res 40, e172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agren J., Sundstrom A., Hafstrom T. & Segerman B. Gegenees: Fragmented Alignment of Multiple Genomes for Determining Phylogenomic Distances and Genetic Signatures Unique for Specified Target Groups. PloS one 7, e39107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D. H. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73 (1998). [DOI] [PubMed] [Google Scholar]
- Akhter S., Aziz R. K. & Edwards R. A. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40, e126 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Liang Y., Lynch K. H., Dennis J. J. & Wishart D. S. “PHAST: A Fast Phage Search Tool”. Nucleic Acids Res 39, W347–352 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P., Perochon J., Lestrade L., Mahillon J. & Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34, D32–D36 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille M. G. & Brinkman F. S. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics 25, 664–665 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. & Roos D. S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W. et al. Pan-genomic analysis provides insights into the genomic variation and evolution of Salmonella Paratyphi A. PLoS One 7, e45346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Table S2
Table S3