Abstract
EPAS1 involves in the hypoxic response and is suggested to be responsible for the genetic adaptation of high-altitude hypoxia in Tibetans. However, the detailed molecular mechanism remains unknown. In this study, a single nucleotide polymorphism rs56721780:G>C and an insertion/deletion (indel) polymorphism −742 indel in the promoter region showed divergence between Tibetans and non-Tibetan lowlanders. rs56721780:G>C regulated the transcription of EPAS1 by IKAROS family zinc finger 1 (IKZF1), which was identified as a new transcriptional repressor for EPAS1 gene. It demonstrated that the C allele of rs56721780:G>C decreased the binding of IKZF1, leading to the attenuated transcriptional repression of EPAS1 gene. The insertion at −742 indel provided a new binding site for Sp1 and was related to the activation of EPAS1 promoter. Further functional analysis revealed that lysyl oxidase (LOX) gene, which was reported to be responsible for extracellular matrix protein cross-linking of amnion previously, was a direct target of EPAS1. The CC genotype at rs56721780:G>C and the insertion genotype at −742 indel were found associated with higher EPAS1 and LOX expression levels in amnion, as well as higher birth weight of Tibetan newborns, suggesting that EPAS1 gene might play important roles in the development of amnion, fetus growth and high-altitude adaptation of Tibetans.
Lack of oxygen occurs in a number of processes, e.g., embryonic development1, postnatal maturation2, and tumorigenesis3. Under hypoxia condition, hypoxia-inducible factors (HIFs) are activated and function as transcriptional regulators of genes involved in the hypoxic response4,5,6. HIFs are heterodimers consisting of an oxygen-labile α subunit (HIFα) and a stable β subunit (HIFβ). HIFα in mammals includes three isoforms, of which HIF1α and HIF2α are the best characterized7. In environments with sufficient oxygen, HIFα subunits are hydroxylated at conserved proline residues by prolyl hydroxylases (PHDs) and rapidly degraded by ubiquitin-proteosome system. While in the hypoxic environment, activity of PHDs is diminished, and as a consequence, HIFα subunits become stabilized. The resulting stabilized HIFα proteins dimerize with HIFβ, and activate the transcription of the target genes8. HIF1α is expressed ubiquitously, while the expression of HIF2α is more restricted, with highest expression levels in heart, lung and placenta9.
HIF2α is encoded by endothelial PAS domain protein 1 (EPAS1) gene. Studies on EPAS1 inactivation revealed that mice with loss of EPAS1 developed a syndrome of multiple-organ pathology10 and may die at mid-gestation11. Thus it is hypothesized that certain EPAS1 expression levels are important for its proper functions in the course of embryo development. Genetic variation in the regulatory region of EPAS1 gene may influence the transcription levels of EPAS1 and may further contribute to the changes of the biological functions. Recently, genome-wide studies on high-altitude adaptation have suggested several SNPs in the regulatory region (intron and 3′ downstream) of EPAS1 gene responsible for the genetic adaptation of high-altitude hypoxia in Tibetans12,13,14. However, whether there are variants in the promoter region of EPAS1 gene related to high-altitude adaptation awaits further studies. Functional studies focusing on the molecular mechanism need to investigate the direction and the magnitude of gene expression changes associated with the variants and figure out what tissues, developmental time points, and what the downstream target genes involved.
Thus, we had three specific aims in this study. Firstly, we were interested to identify the variants in the promoter region with divergence between Tibetans and Han Chinese. Secondly, we wanted to explore the direction and the magnitude of EPAS1 gene expression changes associated with the variants. Thirdly, we intended to investigate the candidate targets that are regulated by EPAS1.
Results
Eleven variants were identified in the promoter region of EPAS1 gene
As compared with the 1000 Genome Data, we found that Tibetans have several specific variants in the promoter region of EPAS1 gene (Table 1), among which four variants have minor allele frequency (MAF) larger than 0.05. A 40-bp insertion was identified at −742 bp relative to the transcriptional start site of EPAS1 gene (NM_001430.4, Supplementary Fig. S1) in Tibetans but was absent in Chinese Han individuals (Supplementary Fig. S2). Another three variants newly identified in Tibetans lay at −706 bp, −138 bp and −129 bp, respectively, and two of them (−138 bp and −129 bp) were in strong LD with the −742 indel polymorphism (R2 = 1, Supplementary Fig. S3). Moreover, rs56721780:G>C and rs13428739:C>T lying at −886 bp and −607 bp, respectively, had much higher MAF in Tibetans than that of Han Chinese (0.372 vs. 0.010 and 0.39 vs. 0.010, allele frequency in Han Chinese was from 1000 Genomes Project) and were also in strong LD with the −742 indel polymorphism (R2 = 0.8 and 0.95, respectively). The frequency of the haplotypes constructed by the three variants (rs56721780:G>C, −742 indel and rs13428739:C>T) in strong LD was listed in Table 2.
Table 1. Variants in the promoter region of EPAS1 gene identified in Tibetans.
| Number | SNP IDa | Chromosomal position (bp) | Position relative to the TSSb | Allele Variantsc | MAF (Tibetan) | MAF (Han) | Variantd type |
|---|---|---|---|---|---|---|---|
| 1 | NA | 46522745 | −1796 | T/C | 0.036 | 0.000 | SNP |
| 2 | rs59862329 | 46523463 | −1078 | T/C | 0.140 | 0.005 | SNP |
| 3 | rs56721780 | 46523655 | −886 | G/C | 0.372 | 0.010 | SNP |
| 4 | NA | 46523799 | −742 | DEL/IN | 0.395 | 0.000 | INDEL |
| 5 | NA | 46523835 | −706 | C/T | 0.256 | 0.000 | SNP |
| 6 | rs13391305 | 46523921 | −620 | T/C | 0.134 | 0.005 | SNP |
| 7 | rs13428739 | 46523934 | −607 | C/T | 0.384 | 0.013 | SNP |
| 8 | NA | 46524015 | −526 | A/C | 0.012 | 0.000 | SNP |
| 9 | NA | 46524250 | −291 | C/T | 0.012 | 0.000 | SNP |
| 10 | NA | 46524403 | −138 | G/C | 0.395 | 0.000 | SNP |
| 11 | NA | 46524412 | −129 | G/A | 0.395 | 0.000 | SNP |
Note: aNA, not available; bTSS, transcriptional start site; cthe second allele is minor allele; dINDEL, insertion and deletion.
Table 2. Information of the haplotypes defined by rs56721780:G>C, −742 indel and rs13428739:C>T.
| Number | Haplotype | Estimated frequency | Stand Error |
|---|---|---|---|
| 1 | G/deletion/C | 0.591 | 0.0046 |
| 2 | C/insertion/T | 0.346 | 0.0054 |
| 3 | G/insertion/T | 0.037 | 0.0046 |
| 4 | C/deletion/C | 0.013 | 0.0048 |
| 5 | C/insertion/C | 0.012 | 0.0028 |
C allele of rs56721780:G>C decreased the binding affinity of IKZF1 to the cis-acting element in EPSA1 promoter
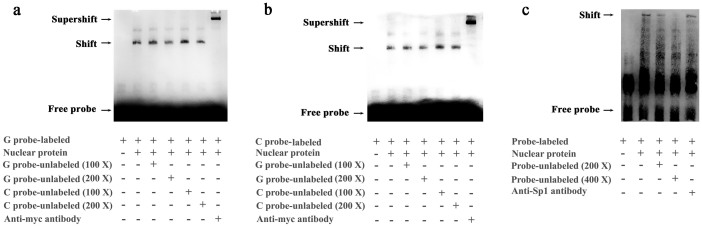
To further analyze the function of the three variants used to construct haplotypes, function prediction was performed by TFSEARCH software. The Prediction results demonstrated that transcription factor IKZF1 isoform 3 and 5 (also known as Ik-2 an Ik-3, respectively15) bind to the cis-acting element with allele G at rs56721780:G>C site, while the binding effect is missing for allele C (Supplementary Fig. S1). In this study, EMSA experiment was designed to confirm the binding effect of IKZF1 to the target site of EPAS1 promoter. Biotin-labeled G allele or C allele probe was incubated with nuclear extract from HEK293 cells transfected with pcDNA3.1-IKZF1-myc plasmid expressing IKZF1 isoform 3, and probe-protein complexes were observed for both of the two probes (Figure 1a and 1b). To verify the binding specificity, excess unlabeled G or C probe competitor was added to the EMSA reaction, and the probe-protein complex binding revealed a different affinity for the two alleles. Higher concentration of unlabeled C competitor was required to compete for binding to both of the G probe and C probe, as compared to unlabeled G competitor. The addition of anti-myc antibody targeting IKZF1 resulted in a supershift of the upper complex (Figure 1a and 1b). These results suggest IKZF1 binds more avidly to the G allele, compared to the C allele.
Figure 1. G allele of rs56721780:G>C increased the transcription binding affinity of IKZF1 and the 40-bp insertion provided the binding site for Sp1.
Electrophoretic mobility shift assay was performed with biotin-labeled (a) G probe or (b) C probe and (c) 40-bp insertion probe. These experiments were repeated more than three times independently with similar results.
The 40-bp insertion fragment in EPSA1 promoter provided the binding site for Sp1
The 40-bp insertion fragment at −742 position was predicted to have the binding site for transcription factor Sp1 (Supplementary Fig. S1). EMSA experiment observed a shift band for the probe-protein complex (Figure 1c). The intensity of the shift band decreased upon pre-incubation with excess unlabeled 40-bp competitor or anti-Sp1 antibody, confirming the binding effect of Sp1 to the 40-bp insertion fragment. As to the −607 site of rs13428739:C>T, no transcription factor binding site was found in the EPAS1 promoter region by the prediction software.
C allele of rs56721780:G>C and the 40-bp insertion increased the promoter activity of EPAS1 gene
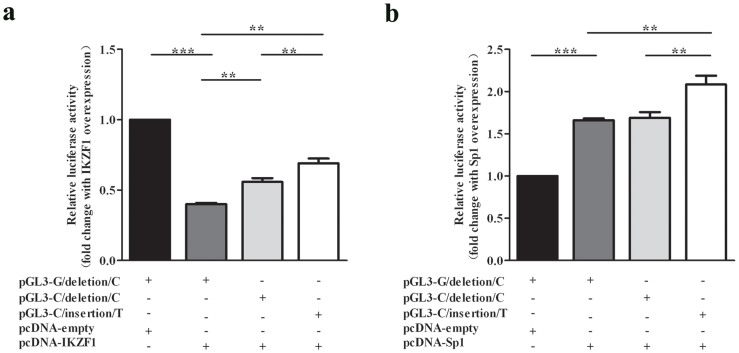
To examine the potential function of IKZF1 and Sp1 in regulating EPAS1 gene, pcDNA3.1-IKZF1-myc or pcDNA3.1-Sp1-myc and pGL3-EPAS1 promoter-luciferase plasmid (pGL3-G/deletion/C or pGL3-C/deletion/C or pGL3-C/insertion/T) were co-transfected into HEK293 cells. The results showed that IKZF1 could decrease the transcriptional activity of EPAS1-G/deletion/C promoter, EPAS1-C/deletion/C promoter and EPAS1-C/insertion/T promoter by 60%, 43%, 31%, respectively (Figure 2a), suggesting IKZF1 was a potential transcriptional repressor for EPAS1 gene and had stronger inhibition effects on EPAS1-G/deletion/C promoter than those on EPAS1-C/deletion/C and EPAS1-C/insertion/T promoters. Our results also demonstrated that Sp1 could increase the transcription activity of EPAS1-G/deletion/C, EPAS1-C/deletion/C and EPAS1-C/insertion/T promoters to 1.66 times, 1.69 times, 2.09 times, respectively (Figure 2b), indicating Sp1 was a potential transcriptional activator for EPAS1 gene and had stronger activation effects on EPAS1-C/insertion/T promoter, as compared to those on EPAS1-G/deletion/C and EPAS1-C/deletion/C promoters.
Figure 2. IKZF1 and Sp1 were a potential repressor and activator for EPAS1 gene, respectively.
Luciferase activity was measured after HEK293 cells were co-transfected with pGL3-G/deletion/C or pGL3-C/deletion/C or pGL3-C/insertion/T luciferase reporter construct, and (a) pcDNA3.1-IKZF1 plasmid or (b) pcDNA3.1-Sp1 plasmid. pcDNA3.1-empty plasmid was used as negative controls. The relative luciferase activity was calculated as the ratio of Firefly luciferase activity vs. Renilla luciferase activity. Fold changes were calculated as the ratio of relative luciferase activity with IKZF1 or Sp1 over-expression vs. relative luciferase activity without IKZF1 or Sp1 over-expression. Data represent as mean ± S.E. from 3 independent experiments with three replicates in each experiment. Statistical analysis was performed using t test. **P < 0.01.
LOX was identified as a direct target of EPAS1
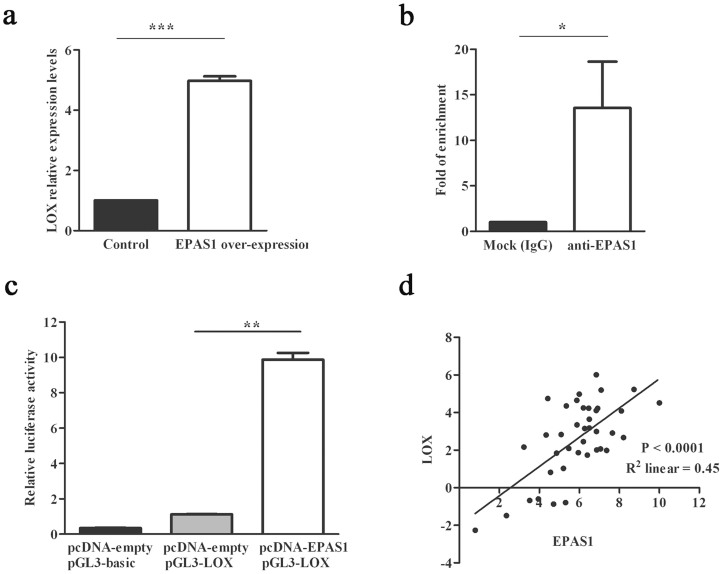
To investigate changes in gene expression associated with EPAS1, global gene expression profiling analysis of Flp-In-293 cells with/without stably expressed EPAS1 was carried out on microarrays. In total, 25 significantly up- or down-regulated (P < 9 × 10−7) genes were identified (Supplementary Table S1). The increased expression of lysyl oxidase (LOX) gene was particularly intriguing as (i) Both of the two probes of LOX gene were significantly up regulated with the most significant P values, 2.78 × 10−32 and 1.35 × 10−31, respectively; (ii) LOX gene encodes an extracellular copper enzyme that catalyzes formation of aldehydes from lysine residues in collagen and elastin precursors, resulting in cross-linking of collagen and elastin16; (iii) LOX gene was highly expressed in amnion tissues17, which contained a large percentage of collagen and were easier to collect compared with other tissues of human. Subsequent qRT-PCR analysis confirmed the results found in global gene expression profiling analysis (Figure 3a). ChIP experiment further verified that two hypoxia inducing elements (HREs), which contain the core recognition sequence 5′-RCGTG-3′, located upstream 183 to 230 bp relative to the transcriptional start site of LOX might be the binding sites for EPAS1 (Figure 3b). Luciferase expression derived by LOX promoter could be induced about 10-fold of increase by EPAS1 over-expression (Figure 3c) in HEK293 cells. Due to LOX involving in extracellular matrix protein cross-linking of amnion18, we further analyzed LOX and EPAS1 mRNA expression levels in amnion from Tibetan newborns. Our results showed that LOX mRNA levels were highly positively correlated with EPAS1 mRNA levels in amnion (Pearson correlation coefficient R2 = 0.45, P value < 0.0001, Figure 3d).
Figure 3. LOX was a direct target of EPAS1.
(a) LOX was up-regulated when EPAS1 was over-expressed in Flp-In 293 cell line by qRT-PCR analysis. (b) LOX was enriched with anti-EPAS1 antibody as compared to normal IgG by ChIP-qPCR analysis. (c) EPAS1 over-expression could induce human LOX promoter luciferase expression up to 10-fold. (d) mRNA expression levels of EPAS1 and LOX in amnion were highly correlated by Pearson correlation analysis. Both of x axis and y axis were plotted in ΔCt manner, with GAPDH as the endogenous control. Data were represented as mean ± S.E. from three independent experiments. *P < 0.05, **P < 0.01, **P < 0.001.
Three variants were associated with EPAS1 and LOX expression in the amnion from Tibetan newborns
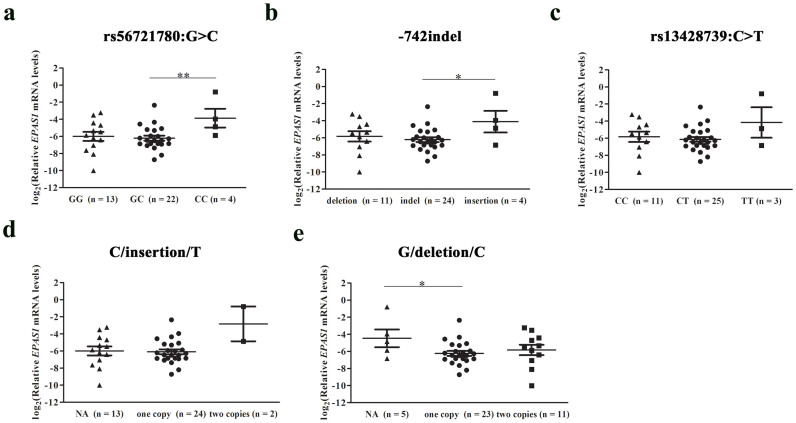
Although we have showed that rs56721780:G>C and −742 indel regulated EPAS1 expression in human cells, whether the regulation effect works in human tissues was not clear. To further understand the role of rs56721780:G>C and −742 indel in transcriptional regulation, we compared the mRNA levels of EPAS1 and LOX between different genotype groups in amnions from Tibetan newborns. The results showed that the individuals with GG or GC genotypes had much lower EPAS1 and LOX mRNA levels compared with the individuals with CC genotypes at rs56721780:G>C (Figure 4a and Supplementary Fig. S4). Similarly, the individuals with deletion allele at −742 indel (Figure 4b and Supplementary Fig. S4) or C allele at rs13428739:C>T (Figure 4c and Supplementary Fig. S4) had much lower EPAS1 and LOX mRNA levels compared with the individuals without the corresponding alleles. As to the effects of different haplotypes on the expression of EPAS1 and LOX genes, it was found G/deletion/C haplotype (Figure 4e and Supplementary Fig. S4) was related to decreased EPAS1 and LOX mRNA levels and individuals with two copies of C/insertion/T haplotype had much higher EPAS1 and LOX mRNA levels (Figure 4d and Supplementary Fig. S4). Western blot analysis demonstrated that IKZF1 and Sp1 were present in amnion (Supplementary Fig. S5), suggesting that rs56721780:G>C and −742 indel may also regulate EPAS1 expression by means of IKZF1 and Sp1 binding, respectively, in human amnion.
Figure 4. Three variants in the promoter region of EPAS1 gene were correlated with EPAS1 gene expression levels in amnion.
Individuals possessing wild-type alleles at the positions of (a) rs56721780:G>C, (b) −742 indel and (c) rs13428739 had lower EPAS1 expression levels in amnion. Individuals with two copies of (d) C/insertion/T haplotype had much higher EPAS1 expression levels in amnion, whereas the (e) G/deletion/C haplotype decreased EPAS1 expression levels. Relative expression levels were log2 transformed for plotting, with GAPDH as the endogenous control. Data were presented as mean ± S.E. Statistical analysis was performed using the t test or t test with Welch's correction implemented in the software of Graphpad Prism. *P < 0.05, **P < 0.01. The numbers in parentheses indicated the counts of individuals with the corresponding genotypes. NA, not available or without the haplotype indicated.
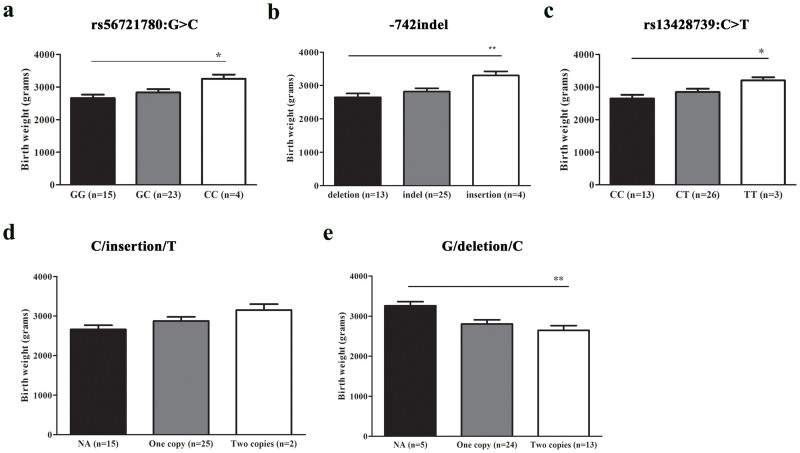
Three variants were associated with birth weight of Tibetan newborns
LOX gene encodes lysyl oxidase, which is responsible for normal extracellular matrix protein cross-linking. The reduced activity of lysyl oxidase is thought to be associated with abnormal development of the fetus, such as bone abnormalities, decreased integrity of vessels19, and preterm births17 with low birth weight fetus. Thus in this study, we were interested to test whether there was relationship between the expression levels of LOX and birth weight, and we found that individuals with the genotypes or haplotype associated with lower mean LOX expression levels also had lower mean birth weight (Figure 5). The results demonstrated that the individuals with G allele at rs56721780:G>C (Figure 5a), deletion allele at −742 indel (Figure 5b) or C allele at rs13428739:C>T (Figure 5c) had much lower birth weight compared with the individuals without the corresponding alleles. The haplotype G/deletion/C (Figure 5e) was found to be related to decreased birth weight and individuals with two copies of C/insertion/T haplotype correlated with higher birth weight (Figure 5d).
Figure 5. Three variants in the promoter region of EPAS1 gene were associated with birth weight of Tibetan newborns.
Individuals possessing wild-type alleles at the positions of (a) rs56721780:G>C, (b) −742 indel and (c) rs13428739 had lower birth weight. Individuals with (d) C/insertion/T haplotype had higher birth weight, whereas the (e) G/deletion/C haplotype decreased birth weight. Data were presented as mean ± S.E. Statistical analysis was performed using the t test or t test with Welch's correction implemented in the software of Graphpad Prism. *P < 0.05, **P < 0.01. The numbers in parentheses indicated the counts of individuals with the corresponding genotypes. NA, not available or without the haplotype indicated.
Discussion
Mutations in coding sequence of EPAS1 gene have been found to be associated with several kinds of human disorders, such as polycythemia20,21, paragangliomas or somatostatinomas22. Such mutations affect the stability of HIF2α proteins and thus lead to dis-regulation of HIF2α. In addition to mutations, a common SNP rs17039192:C>T in the exon 1 of EPAS1 gene (+18 bp relative to the transcription start site) was reported to be associated with knee osteoarthritis in Japanese and the susceptibility allele C showed higher EPAS1 promoter activity in chondrogenic cells23. However, little is known about the underlying mechanisms of common SNPs in promoter region regulating EPAS1 transcription. In this study, we found a SNP variation rs56721780:G>C and an indel variation −742 indel in EPAS1 promoter region regulated EPAS1 expression by means of binding with two transcription factors, IKZF1 and Sp1, respectively. Both of the two minor alleles enriched in Tibetans, C allele at rs56721780:G>C and insertion allele at −742 indel, conferred the elevated transcription of EPAS1 gene. IKZF1 encodes a transcription factor that belongs to the family of zinc-finger DNA binding proteins, and has been shown to regulate expression of its target genes in activation or repression manner via chromatin remodeling24, playing a key role in a variety of cellular functions, such as cell proliferation25, cell apoptosis26, and Notch cell signal transduction pathway27,28. IKZF1 is initially characterized as a hemo-lymphopoietic system-restricted transcriptional factor to regulate lymphocyte differentiation and subsequent studies also demonstrated that IKZF1 expresses in placenta and transactivates the expression of leucine aminopepetidse/insulin-regulated aminopeptidase gene29. IKZF1 gene possesses at least 13 spliced transcript variants encoding different isoforms (isoform 1–13). All isoforms share C-terminal domain containing two zinc finger motifs that required for hetero- or homo-dimerization, and for interactions with other proteins, but differ in the number of N-terminal zinc finger motifs containing nuclear localization signal and requiring for DNA binding30. Both of the two isoforms (isoform 3 and 5) predicted as transcription regulators for EPAS1 gene contain requisite three N-terminal zinc fingers, but only isoform 3 has normal nuclear localization30, and thus confers high affinity binding to a specific core DNA sequence element in the promoters of target genes. Thus for in vitro experiment, we only studied IKZF1 isoform 3. The prediction results (Supplementary Fig. S1) showed G allele of rs56721780:G>C lies in the sequence of GGTTTCCCAGGA (the letter in bold), which is specific for IKZF1 binding to EPAS1 promoter. When G is changed to C, the binding of IKZF1 to EPAS1 promoter might be eliminated. In fact, our EMSA results revealed that the C allele only decreased the binding of IKZF1 to EPAS1 promoter. The inconsistency of the EMSA results with the prediction on the G>C change might due to the location of the G>C allele in the binding site of IKZF1 to EPAS1 promoter, i.e. it was not included in the core sequence (TTCCCA) of the high affinity binding element of IKZF131. IKZF1 was identified as a new transcriptional repressor for EPAS1 gene in this study. The results demonstrated that G allele of rs56721780:G>C increased the binding of IKZF1 to the cis-acting element, leading to the enhanced transcription repression of EPAS1 gene. The decreased inhibition of IKZF1 on EPAS1-C/insertion/T promoter as compared with that on EPAS1-C/deletion/C promoter (Figure 2A) might be caused by the steric hindrance of other transcriptional factors binding to the 40-bp insertion fragment. Sp1 is a zinc finger transcription factor that binds to GC-rich motifs of many promoters acting as an activator32 or a repressor33, which is involved in many cellular processes, including cell differentiation, cell growth, apoptosis34, immune responses35 and so on. Sp1 contains three spliced transcript variants encoding different isoforms (isoform 1–3). We focused on the longest isoform (isoform 1) in this study. We found Sp1 could bind to the 40-bp insertion fragment at −742 indel site and activate the transcription of EPAS1 gene. The results were not unexpected given a previous study demonstrating that Sp1 and the sequence −478/−445 in the promoter region of EPAS1 gene were responsible for the activation of EPAS1 expression during adipogenesis in 3T3-L1 cell36.
Another highlight in this study is that LOX gene was found to be a direct target of EPAS1. Numerous gene array studies37,38,39 including ours searching for novel HIF-regulated genes have drawn LOX as one of the highest regulated genes. Although functional studies have been successfully identified the binding site in the LOX gene for HIF140, whether there is direct binding effect of HIF2 to LOX gene remains unknown. In this study, ChIP experiment successfully determined the binding effect of HIF2α/EPAS1 to the promoter of LOX gene, and the binding effect was further confirmed to be functional by luciferase report assay.
LOX gene encodes an extracellular copper enzyme, which has been well studied in human fetal membranes because of its importance in cross-linking both of collagens and elastin, thus increasing the tensile strength of these proteins17. Such strength is particularly required in the third trimester of pregnancy when the growth of the fetal membranes fails to keep pace with the growth of the fetus41. If the membranes fail to distend in the third trimester of pregnancy, they may rupture prematurely almost always resulting in preterm delivery17,18 with low-birth-weight newborns, who are likely to experience physical health problems or die during the early age of life as compared to the infants with normal weight18. The human fetal membranes constitute of amnion and chorion, and resistance to tearing and rupture is provided primarily by the amnion18. This is the main reason that amnion tissues were used in this study. All the amnion tissues in our study were collected from laboring vaginal deliveries of healthy pregnant women from Shigatse (~3,800 meters in altitude), Tibet. Although a recent report demonstrated that placentas subjected to labor are oxidatively stressed compared with non-labored placentas delivered by cesarean section42, laboring placentas at high altitude (3,100 m) have little or no oxidative stress at the time of delivery irrespective of whether they are labored or delivered by cesarean section43. Thus, it is inferred that labor would be not an inducer of oxidative stress of the amnion tissues collected at ~3,800 m in our study. Our results showed that LOX mRNA levels were highly correlated with EPAS1 mRNA levels, and Tibetans with the genotypes or haplotype associated with lower mean LOX expression levels also had lower mean birth weight. The relationship between LOX and birth weight has also been observed in a previous study investigating the effect of copper depletion throughout gestation on neonates44. The results showed the offspring born to female rabbits fed marginal copper diets (1.5 parts per million copper) had lower birth weight than that of the group fed control diets (10 parts per million copper). And the lungs of pups from the marginal copper diet group also had lower LOX activities. The findings indicating that lower LOX activities in lung or other organs might be related to the lower birth weight of the neonates. However, clear reports about the regulation of birth weight by LOX were limited. More efforts should be made to verify the regulation of birth weight by LOX in future studies.
It is possible that the decreased LOX expression regulated by EPAS1 in amnion might cause inadequate cross-linking of interstitial collagen and reduced tensile strength, leading to premature rupture of the membranes or intrauterine growth restriction of the Tibetan fetus, and producing babies with lower birth weight. Reduced birth weight likely compromises survival during the early postnatal period. Thus, it is presumed that the genotypes and haplotype which were enriched in Tibetans and associated with higher LOX expression levels in amnion might be helpful for fetal growth, and provide Tibetans with high infant survival rate that tightly correlates with heavier birth weight, and may contribute to high-altitude adaptation of Tibetans45.
A previous genome-wide study13 on high-altitude adaptation of Tibetans showed that H1 haplotype of EPAS1 gene was overrepresented in Tibetans (with a frequency of 89.1%) but was significantly less prevalent in other populations (e.g., 3.4% in East Asian populations), indicating the roles of the H1 haplotype in high-altitude adaptation of Tibetans. In this study, we constructed a C/insertion/T haplotype with a frequency of 34.6% in Tibetans but absent in Chinese Han individuals. All the Tibetans with C/insertion/T haplotype also possessed the EPAS1-H1 haplotype, suggesting the C/insertion/T haplotype was full concordance with EPAS1-H1 haplotype.
In conclusion, we showed that EPAS1 gene was regulated by IKZF1 and Sp1 at the sites of rs56721780:G>C and −742 indel, respectively, and might play important roles in the development of amnion, fetus growth and high-altitude adaptation of Tibetans by regulating LOX expression. Further studies will increase our understanding of the important roles of EPAS1 during development.
Methods
Amnion and umbilical cord blood
Fresh placental tissues and umbilical cord blood samples were obtained from 43 newborns delivered at the People's Hospital of Shigatse District, Tibet. All the deliveries were vaginal deliveries of healthy pregnant women from Shigatse (~3,800 meters in altitude). Amnion was manually separated from choriodecidua and immediately washed extensively with sterile physiological salt solution and cut into segments before storing in RNAlater (Ambion, Austin, TX, USA) at −20°C following the manufacture's instructions. Birth weight of the newborns was measured immediately after delivery using a regularly calibrated weighing scale. This study also randomly selected 25 Chinese Han DNA samples from our database to serve as controls13. This study was conducted with an approval from the Internal Review Board of the School of Life Sciences at Fudan University in Shanghai, China. The methods were carried out in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all the subjects included in the study or their parents.
Gene cloning and plasmid construction
Since IKZF1 is mainly expressed in the hemo-lymphopoietic system, Jurkat cells which are an immortalized line of T lymphocyte cells were used for IKZF1 isoform 3 (NCBI accession no. NM_001220766.1) cloning. Approximately 1 × 106 cultured Jurkat cells were subjected to total RNA extraction and cDNA synthesis. The coding region of human IKZF1 isoform 3 was amplified using primers pcDNA3.1-IKZF1-F and pcDNA3.1-IKZF1-R (Table S2) at 98°C, 5 min; 98°C, 30 s; 68°C, 2.5 min for 35 cycles; then 72°C, 10 min and was cloned into the EcoRI and HindIII restriction sites of pcDNA3.1/myc-His(-)A vector. The generated plasmid was named pcDNA3.1-IKZF1-myc.
The coding sequence clone of human EPAS1 gene (NCBI accession no. NM_001430.4) was purchased from Genecopoeia (Guangzhou, China). The coding sequence was subcloned into the 3× flag-pEGFP-N1 vector which was provided by Dr. Jing Zheng, East China University of Science and Technology. The fragment containing coding sequence and 3× flag tag was then subcloned to pcDNA5/FRT/TO expression vector (Invitrogen, Carlsbad, CA, USA). The coding sequence of human Sp1 gene was subcloned into the pcDNA3.1 vector using the pcDNA3.0-Sp1 expression plasmid as a template which was kindly provided by Dr. Xingzhong Wu from Fudan University46. A 2,088 bp of DNA fragment (from −1988 bp to +100 bp relative to the transcription start site) of the human EPAS1 gene (Figure S1) and the promoter sequence from −268 bp to +300 bp relative to the transcriptional start site47 of lysyl oxidase (LOX) gene (NCBI accession no. NM_002317.5) were cloned by PCR utilizing the genomic DNA as the template. The amplified products were inserted into pGL3-basic vector (Promega, Madison, WI, USA) to generate pGL3-EPAS1 (pGL3-G/deletion/C) plasmid and pGL3-LOX plasmid, respectively. pGL3-G/deletion/C plasmid was further used as a template to generate pGL3-C/deletion/C plasmid by PCR-based site-directed mutagenesis at the position of SNP rs56721780:G>C (−886 bp relative to the transcription start site). pGL3-C/insertion/T plasmid was constructed using the genomic DNA of the individual with C/insertion/T haplotye as the template. The PCR primers used for DNA amplification were listed in Supplementary Table S2. All plasmids were sequence-verified before use.
Cell culture and stable cell line establishment
Human embryonic kidney (HEK) 293 cell and Jurkat cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences, and were cultured under recommended conditions. Flp-In-293 cells (Invitrogen) were maintained in DMEM supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2 mM L-glutamine, 100 μg/mL of Zeocin, 15 μg/mL of blasticidin at 37°C with 5% (v/v) CO2. Stably transfected Flp-In-293 cell lines were generated by integrating the pcDNA5/FRT/TO vector containing EPAS1 coding sequence into a single defined genomic locus via Flp recombinase.
RNA extraction, microarray assay and quantitative real-time PCR (qRT-PCR)
Total RNA from Flp-In-293 cells and amnion was extracted with TRIzol Reagent (Sigma-Aldrich, St. Louis, MO, USA). Microarray assays were performed to figure out the differentially expressed genes between Flp-In-293 cells stably expressed EPAS1 gene and the control Flp-In-293 cells according to the manufacturer's protocol (Affymetrix, Santa Clara, CA, USA). Briefly, total RNA was used to produce double-stranded cDNA, which was then used to synthesize biotin-labeled antisense RNA (aRNA) by in vitro transcription. aRNA was cleaned, fragmented, and hybridized to Affymetrix HG U133 Plus 2.0 GeneChips followed by washing and scanning. Standardized Fold Change method designed in house (Wang Y, et al.) was used for microarray gene expression difference analysis. The gene that displayed the most significant change between the samples was selected for further investigation. qRT-PCR was performed on an Applied Biosystems 7900 HT analyzer using SYBR Green dyes (Takara, Otsu, Shiga, Japan) according to the manufacturer's protocol. First-strand cDNA for qRT-PCR was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) with total RNA as template. Relative quantification was performed by means of the 2−ΔΔCt method, with GAPHD as the endogenous control. qRT-PCR primers used were listed in Supplementary Table S2.
Cell transient transfection and dual-luciferase reporter gene assay
Prior to transfection, 4 × 104 HEK293 cells were split into 24-well plates and incubated in DMEM. Next day, pcDNA3.1-IKZF1 plasmid or pcDNA3.1-Sp1 plasmid or pcDNA3.1-empty plasmid was co-transfected with pGL3-G/deletion/C or pGL3-C/deletion/C plasmid or pGL3-C/insertion/T plasmid at a ratio of 300 ng:180 ng into the HEK293 cells using Lipofectamine 2000 (Invitrogen). pcDNA5-EPAS1 plasmid or pcDNA5-empty plasmid was co-transfected with pGL3-LOX plasmid at a ratio of 240 ng:240 ng into the HEK293 cells to investigate the regulation of EPAS1 on LOX promoter. In addition, 20 ng of pRL-SV40 plasmid was used as an internal control in each transfection. Luciferase activity was measured with Dual-luciferase reporter assay system (Promega). The relative luciferase activity was calculated as the ratio of Firefly luciferase activity vs. Renilla luciferase activity for each sample.
Electrophoretic mobility shift assay (EMSA)
Non-radioactive EMSAs were performed with IKZF1 isoform 3 proteins and biotin-labeled double stranded probes, which were selected on the basis of predicted IKZF1 binding sites in the promoter of the EPAS1 gene (Supplementary Fig. S1). Non-radioactive EMSAs were also performed with Sp1 proteins and biotin-labeled double stranded probes which were made by the 40-bp insertion fragment. EMSAs were performed using a LightShift chemiluminescent electrophoretic mobility shift assay kit (Thermo Scientific) according to the manufacturer's instructions.
Chromatin immunoprecipitation (ChIP) analysis
ChIP experiment was carried out using ChIP Assay Kit (Millipore) according to the manufacturer's instructions. Briefly, Flp-In-293 cells stably expressed EPAS1 were treated with 1 μg/mL tetracycline (Invitrogen) for 24 h and treated with chemical hypoxia-mimetic 2,2′-dipyridyl during the last 16 h before harvest. Cells were then crosslinked with 1% formaldehyde for 10 min at 37°C. Nuclear lysate was harvested and sonicated to obtain 250 bp to 1,000 bp DNA fragments. The chromatin was immunoprecipitated with rabbit polyclonal EPAS1 antibodies (Novus Biologicals, Littleton, CO, USA) or normal rabbit IgG (Santa cruz, Dallas, TX, USA). The precipitated genomic DNA crosslinks were reversed by heating at 65°C overnight with NaCl, recovered by phenol/chloroform extraction, and analyzed by quantitative PCR (qPCR). Primers used for ChIP-qPCR were listed in Supplementary Table S2.
DNA extraction and sequencing
Genomic DNA was extracted from umbilical cord blood using AxyPrep Multisource Genomic DNA Minprep kit (Axygen Scientific, Union City, CA, USA) following the procedure detailed in the kit. A 2,088 bp DNA fragment (from −1988 bp to +100 bp relative to the transcription start site) of the human EPAS1 gene (NCBI accession no. NM_001430.4) was amplified by PCR and the variants included this region were determined by direct dye terminator sequencing of the PCR products with the Applied Biosystems Prism BigDye system according to the manufacturer's instructions. The products for sequencing were run on an Applied Biosystems 3730 automated sequencer and analyzed by SeqMan software. Insertion and deletion (indel) variants were further verified by PCR method. Primers used for PCR and sequencing were presented in Supplementary Table S2.
Statistical analysis
Linkage disequilibrium (LD) and haplotype analyses for the variants identified in the promoter of EPAS1 gene were performed in the 43 unrelated individuals. Pair-wise R2 for six variants with MAF larger than 0.25 were calculated and LD structures were plotted by the program haploview48. The Hardy–Weinberg equilibrium for each variant was also tested by haploview program. Population haplotypes for the variants in strong LD (0.8 ≤ R2 < 1) were inferred using PHASE v2.1.1 software49. Potential transcription factor binding sites were predicted using TFSEARCH (version 1.3, threshold score 85.0, classification: vertebrate)50.
The data were presented as mean ± S.E. unless otherwise indicated. The data were tested for normal distribution and homogeneity of variances by Ryan-Joiner test implemented in Minitab software and by Levene's test implemented in SPSS software, respectively. If the data were normal distributed and had similar variances, Student's t test was performed to compare means between measured groups. Otherwise, data transformation was conducted before t test or Mann-Whitney test or t test with Welch's correction was performed using GraphPad Prism software. The correlation between mRNA levels of the interested genes was presented by Pearson coefficient using GraphPad Prism software. P values less than 0.05 were considered significant.
Author Contributions
X.X.H. and H.X.W. designed the study, carried out the molecular biological experiments and data analysis, and drafted the manuscript. Q.L., L.Y.N., W.Y., L.C., M.Y.Y. and S.K. helped in data collection and data analysis. L.Q.M. and Q.F. helped in performing molecular biological experiments. J.L. and W.J.C. participated in the design of the study, conceived of the study and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary information
Acknowledgments
We thank Dr. Jing Zheng and Dr. Xingzhong Wu for providing the plasmids. This research was supported by grants from the Key Program of National Natural Science Foundation of China (31330038), the Science and Technology Committee of Shanghai Municipality (11DJ1400102), National Basic Research Program (2012CB944600), and National High-Tech Research and Development Program (2012AA021802).
References
- Patel J., Landers K., Mortimer R. H. & Richard K. Regulation of hypoxia inducible factors (HIF) in hypoxia and normoxia during placental development. Placenta 31, 951–957 (2010). [DOI] [PubMed] [Google Scholar]
- Bedo G., Vargas M., Ferreiro M. J., Chalar C. & Agrati D. Characterization of hypoxia induced gene 1: expression during rat central nervous system maturation and evidence of antisense RNA expression. Int J Dev Biol 49, 431–436 (2005). [DOI] [PubMed] [Google Scholar]
- Criscimanna A. et al. PanIN-Specific Regulation of Wnt Signaling by HIF2alpha during Early Pancreatic Tumorigenesis. Cancer Res 73, 4781–4790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccia A. J., Simon M. C. & Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev 18, 2183–2194 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol 13, 167–171 (2001). [DOI] [PubMed] [Google Scholar]
- Schofield C. J. & Ratcliffe P. J. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol 5, 343–354 (2004). [DOI] [PubMed] [Google Scholar]
- Majmundar A. J., Wong W. J. & Simon M. C. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40, 294–309 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loboda A., Jozkowicz A. & Dulak J. HIF-1 versus HIF-2–is one more important than the other? Vascul Pharmacol 56, 245–251 (2012). [DOI] [PubMed] [Google Scholar]
- Tian H., McKnight S. L. & Russell D. W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev 11, 72–82 (1997). [DOI] [PubMed] [Google Scholar]
- Scortegagna M., Morris M. A., Oktay Y., Bennett M. & Garcia J. A. The HIF family member EPAS1/HIF-2alpha is required for normal hematopoiesis in mice. Blood 102, 1634–1640 (2003). [DOI] [PubMed] [Google Scholar]
- Tian H., Hammer R. E., Matsumoto A. M., Russell D. W. & McKnight S. L. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev 12, 3320–3324 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol 28, 1075–1081 (2011). [DOI] [PubMed] [Google Scholar]
- Xu S. et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol Biol Evol 28, 1003–1011 (2011). [DOI] [PubMed] [Google Scholar]
- Yi X. et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzat S., Yu S. & Asa S. L. Ikaros isoforms in human pituitary tumors: distinct localization, histone acetylation, and activation of the 5′ fibroblast growth factor receptor-4 promoter. Am J Pathol 163, 1177–1184 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszar K. Lysyl oxidases: a novel multifunctional amine oxidase family. Prog Nucleic Acid Res Mol Biol 70, 1–32 (2001). [DOI] [PubMed] [Google Scholar]
- Hein S. et al. Lysyl oxidases: expression in the fetal membranes and placenta. Placenta 22, 49–57 (2001). [DOI] [PubMed] [Google Scholar]
- Casey M. L. & MacDonald P. C. Lysyl oxidase (ras recision gene) expression in human amnion: ontogeny and cellular localization. J Clin Endocrinol Metab 82, 167–172 (1997). [DOI] [PubMed] [Google Scholar]
- Uriu-Adams J. Y., Scherr R. E., Lanoue L. & Keen C. L. Influence of copper on early development: prenatal and postnatal considerations. Biofactors 36, 136–152 (2010). [DOI] [PubMed] [Google Scholar]
- Percy M. J. et al. A gain-of-function mutation in the HIF2A gene in familial erythrocytosis. N Engl J Med 358, 162–168 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z. et al. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med 367, 922–930 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. et al. Novel HIF2A mutations disrupt oxygen sensing, leading to polycythemia, paragangliomas, and somatostatinomas. Blood 121, 2563–2566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. et al. Transcriptional regulation of endochondral ossification by HIF-2alpha during skeletal growth and osteoarthritis development. Nat Med 16, 678–686 (2010). [DOI] [PubMed] [Google Scholar]
- Payne K. J. & Dovat S. Ikaros and tumor suppression in acute lymphoblastic leukemia. Crit Rev Oncog 16, 3–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L. C. et al. Ikaros inhibits proliferation and, through upregulation of Slug, increases metastatic ability of ovarian serous adenocarcinoma cells. Oncol Rep 28, 1399–1405 (2012). [DOI] [PubMed] [Google Scholar]
- Yagi T. et al. High frequency of Ikaros isoform 6 expression in acute myelomonocytic and monocytic leukemias: implications for up-regulation of the antiapoptotic protein Bcl-XL in leukemogenesis. Blood 99, 1350–1355 (2002). [DOI] [PubMed] [Google Scholar]
- Dumortier A. et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol 26, 209–220 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng A. P. et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 306, 269–271 (2004). [DOI] [PubMed] [Google Scholar]
- Yamamoto E. et al. Ikaros is expressed in human extravillous trophoblasts and involved in their migration and invasion. Mol Hum Reprod 11, 825–831 (2005). [DOI] [PubMed] [Google Scholar]
- Sun L. et al. Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 96, 680–685 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap W. H., Yeoh E., Tay A., Brenner S. & Venkatesh B. STAT4 is a target of the hematopoietic zinc-finger transcription factor Ikaros in T cells. Febs Lett 579, 4470–4478 (2005). [DOI] [PubMed] [Google Scholar]
- Hekmatnejad B., Gauthier C. & St-Arnaud R. Control of Fiat (factor inhibiting ATF4-mediated transcription) expression by Sp family transcription factors in osteoblasts. J Cell Biochem 114, 1863–1870 (2013). [DOI] [PubMed] [Google Scholar]
- Law A. Y., Yeung B. H., Ching L. Y. & Wong C. K. Sp1 is a transcription repressor to stanniocalcin-1 expression in TSA-treated human colon cancer cells, HT29. J Cell Biochem 112, 2089–2096 (2011). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. Inhibition of the transcription factor Sp1 suppresses colon cancer stem cell growth and induces apoptosis in vitro and in nude mouse xenografts. Oncol Rep 30, 1782–1792 (2013). [DOI] [PubMed] [Google Scholar]
- Menon R. et al. Gender-based blood transcriptomes and interactomes in multiple sclerosis: involvement of SP1 dependent gene transcription. J Autoimmun 38, J144–155 (2012). [DOI] [PubMed] [Google Scholar]
- Wada T. [Transcription factor EPAS1 regulates insulin signaling pathway]. Yakugaku Zasshi 127, 143–151 (2007). [DOI] [PubMed] [Google Scholar]
- Elvidge G. P. et al. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J Biol Chem 281, 15215–15226 (2006). [DOI] [PubMed] [Google Scholar]
- Wang V., Davis D. A., Haque M., Huang L. E. & Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res 65, 3299–3306 (2005). [DOI] [PubMed] [Google Scholar]
- Warnecke C. et al. The specific contribution of hypoxia-inducible factor-2alpha to hypoxic gene expression in vitro is limited and modulated by cell type-specific and exogenous factors. Exp Cell Res 314, 2016–2027 (2008). [DOI] [PubMed] [Google Scholar]
- Erler J. T. et al. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature 440, 1222–1226 (2006). [DOI] [PubMed] [Google Scholar]
- Millar L. K., Stollberg J., DeBuque L. & Bryant-Greenwood G. Fetal membrane distention: determination of the intrauterine surface area and distention of the fetal membranes preterm and at term. Am J Obstet Gynecol 182, 128–134 (2000). [DOI] [PubMed] [Google Scholar]
- Cindrova-Davies T. et al. Oxidative stress, gene expression, and protein changes induced in the human placenta during labor. Am J Pathol 171, 1168–1179 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot van Patot M. C. et al. Human placental metabolic adaptation to chronic hypoxia, high altitude: hypoxic preconditioning. Am J Physiol Regul Integr Comp Physiol 298, R166–172 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Mageed A. B., Welti R., Oehme F. W. & Pickrell J. A. Perinatal hypocuprosis affects synthesis and composition of neonatal lung collagen, elastin, and surfactant. Am J Physiol 267, L679–685 (1994). [DOI] [PubMed] [Google Scholar]
- Zhang Y. B., Li X., Zhang F., Wang D. M. & Yu J. A preliminary study of copy number variation in Tibetans. PLoS One 7, e41768 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W. et al. Regulation of integrin alphaV subunit expression by sulfatide in hepatocellular carcinoma cells. J Lipid Res 54, 936–952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietke R. et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: insights into cellular transformation processes mediated by HIF-1. J Biol Chem 285, 6658–6669 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- Stephens M. & Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet 76, 449–462 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T. et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res 26, 362–367 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information