Summary
Many cell therapies currently being tested are based on mesenchymal stromal cells (MSCs). However, MSCs start to enter the senescent state upon long-term expansion. The role of retinoblastoma (RB) protein in regulating MSC properties is not well studied. Here, we show that RB levels are higher in early-passage MSCs compared with late-passage MSCs. RB knockdown induces premature senescence and reduced differentiation potentials in early-passage MSCs. RB overexpression inhibits senescence and increases differentiation potentials in late-passage MSCs. Expression of DNMT1, but not DNMT3A or DNMT3B, is also higher in early-passage MSCs than in late-passage MSCs. Furthermore, DNMT1 knockdown in early-passage MSCs induces senescence and reduces differentiation potentials, whereas DNMT1 overexpression in late-passage MSCs has the opposite effect. These results demonstrate that RB expressed in early-passage MSCs upregulates DNMT1 expression and inhibits senescence in MSCs. Therefore, genetic modification of RB could be a way to improve the efficiency of MSCs in clinical use.
Graphical Abstract
Highlights
-
•
RB levels are higher in early-passage MSCs than in late-passage MSCs
-
•
RB knockdown induces a premature senescence and reduces differentiation potentials
-
•
RB overexpression reverses senescence and increases differentiation potentials
-
•
RB upregulates DNMT1 to inhibit senescence and promote differentiation potentials
In this article, Hung, Kao, and colleagues show that retinoblastoma (RB) levels are higher in early-passage mesenchymal stromal cells (MSCs) than in late-passage MSCs, which serves to regulate senescence and differentiation potentials. Expression of DNMT1 is also higher in early-passage MSCs than in late-passage MSCs. This study demonstrates that RB expressed in early-passage MSCs upregulates DNMT1 expression and inhibits senescence in MSCs.
Introduction
Mesenchymal stromal cells (MSCs) with self-renewal and multilineage differentiation properties can give rise to mesodermal derivatives such as osteoblasts, chondrocytes, and adipocytes. Adult MSCs can be isolated from various tissues, such as bone marrow and dental pulp, and expanded in culture to a very high number. Because of the lack of adverse effects in clinical application, many cell therapies currently being tested are based on MSCs (Caplan, 2007). After a few passages of expansion in culture, MSCs enter the senescent state. Since a large amount of cells are required for therapy and MSCs have a limited lifespan in vitro, the quality of cell therapies using MSCs might decline. Therefore, it is critical to understand the mechanism that controls the replicative senescence of MSCs (Bork et al., 2010).
P21 and p16 are cyclin-dependent kinase inhibitors that play important roles in cell-cycle regulation and tumor suppression (Poole et al., 2004; Shapiro et al., 1998), and both are highly expressed upon senescence in MSCs. Knockdown of p21 in late-passage MSCs increases stem cell properties, including the proliferation rate, differential potentials, and expression of stemness markers, and also enhances the capacity for in vivo bone repair (Yew et al., 2011). Knockdown of p16 has also been shown to reverse the senescent state and promote the proliferation ability (Shibata et al., 2007).
RB encodes the retinoblastoma (RB) protein, which controls cell-cycle progression from G1 into S phase by binding to E2F and inhibiting its activity (Li et al., 1994). It is also involved in the control of stem cell properties (Galderisi et al., 2006). The role of RB in the cellular senescence of MSCs in vitro has been studied in expanded human MSCs (Alessio et al., 2013). Moreover, the fate choice of MSCs in vivo has been studied using an oncogenic transformation mouse model that induces liposarcoma through RB deletion (Calo et al., 2010). However, the mechanism that RB mediates to inhibit replicative senescence and promote the differentiation potential of human MSCs has not been studied.
In this study, we demonstrate that RB, unlike other cell-cycle regulators that play negative roles in maintaining stem cell properties, plays a positive role in regulating MSC properties by affecting DNA methylation via upregulation of DNA methyltransferase 1 (DNMT1).
Results
RB Expression Is High in Early-Passage MSCs and Is Reduced after Expansion
Self-renewal and multipotent differentiation represent stem cell properties that are repressed in expanded MSCs (Shibata et al., 2007; Yew et al., 2011). Expansion of MSCs at a low density (∼50–100 cells/cm2) accelerates senescence and loss of MSC properties (Yew et al., 2011). We found that early-passage (P2 or P3 at low density) MSCs had a significantly increased growth rate compared with late-passage MSCs (P6 or later at low density) (Figure 1A). Consistently, early-passage MSCs decreased in cell size (Figure 1B) and β-galactosidase (β-gal; Figure 1C) expression compared with late-passage MSCs, which showed an enlarged and flat morphology corresponding with the typical morphology of senescent MSCs (Sekiya et al., 2002; Sethe et al., 2006). Moreover, we compared fluorescence dilution by flow-cytometric analysis after staining the cells with carboxyfluorescein succinimidyl ester (CFSE). These results showed that early-passage MSCs replicated as a more homogeneous population compared with late-passage MSCs. Almost all cells at early passage replicated rapidly upon reseeding, whereas only a portion of cells at late passage replicated at a slower speed than early-passage MSCs and some cells stopped replicating (Figure S1 available online). These data suggest that early-passage MSCs are a relatively more homogeneous population than late-passage MSCs. Induction of MSCs in osteogenic induction medium (OIM) followed by alizarin red S staining showed that early-passage MSCs had an increased mineralization capacity at 2 weeks of osteogenic induction (Figure 1D). Similarly, early-passage MSCs showed increased mRNA levels of osteogenesis genes, such as RUNX2 and BSP, at 1 week of osteogenic induction (Figure 1E). Likewise, induction of MSCs in adipogenic induction medium (AIM) followed by oil red O staining revealed that early-passage MSCs increased in differentiation into mature adipocytes at 2 weeks of adipogenic induction (Figure 1F). Similarly, early-passage MSCs showed increased expression of PPAR-γ2 and LPL at 1 week of adipogenic induction (Figure 1G). These data suggest that early-passage MSCs have better proliferation and differentiation potentials than late-passage MSCs.
Figure 1.
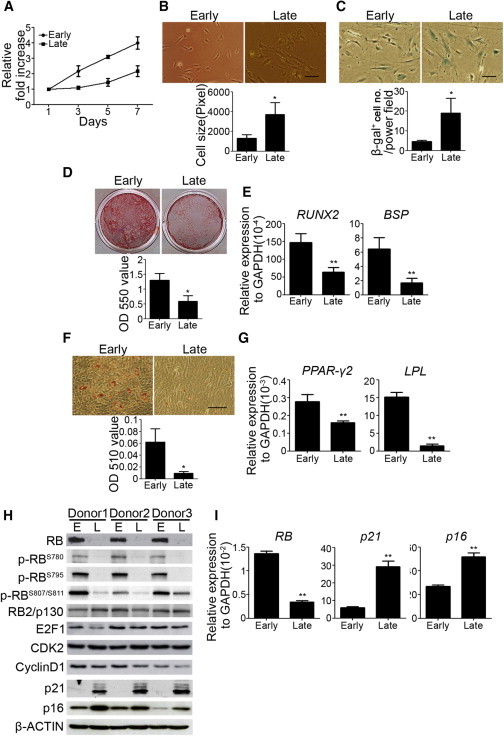
Late-Passage MSCs Show Decreased Growth, Increased Cell Size, and Loss of In Vitro Osteogenic and Adipogenic Differentiation Ability Compared with Early-Passage MSCs
(A) Early- and late-passage MSCs were seeded at 4.5 × 103 /cm2 and cultured for 7 days. MTT assay was performed during the indicated period and data are shown as the relative fold increase.
(B) Upper panel: morphology of early- and late-passage MSCs. Lower panel: cell size measurements.
(C) Early- and late-passage MSCs were stained with β-gal.
(D and E) Early- and late-passage MSCs were treated with OIM followed by alizarin red S staining. (D) Upper: representative pictures of alizarin red S staining at 2 weeks. Lower: optical density measurements of extracted alizarin red S staining.
(E) Quantitative RT-PCR showed the mRNA levels at 1 week.
(F and G) Early- and late-passage MSCs were treated with AIM. (F) Upper: representative picture of oil red O staining at 2 weeks. Lower: optical density measurements of extracted oil red O staining. (G) Quantitative RT-PCR showed the mRNA levels at 1 week.
(H and I) Western blot analysis (H) and quantitative RT-PCR (I) of early- and late-passage MSCs from three individuals.
The results are expressed as mean ± SD of three independent experiments. Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01). Scale bar, 50 μm. See also Figure S1.
To understand the roles of cell-cycle regulators in controlling the proliferation and differentiation potentials of MSCs, we checked the protein levels of several cell-cycle regulators and cyclin-dependent kinases in MSCs. Similar to what was observed in previous reports (Shibata et al., 2007; Yew et al., 2011), early-passage MSCs showed decreased expression of p21 and p16. In addition, we also found that early-passage MSCs had increased levels of RB compared with late-passage MSCs (Figure 1H). There were no significant differences in the levels of CDK2, cyclin D1, and E2F1 between early-passage and late-passage MSCs. Notably, the RB molecules of early-passage MSCs were hyperphosphorylated at the most important phosphorylation sites that are required for RB function in cell-cycle arrest (Figure 1H), including S780 (Kitagawa et al., 1996), S795 (Connell-Crowley et al., 1997), and S807/811 (Knudsen and Wang, 1996). However, we did not observe any changes in the levels of RB2/p130 between early- and late-passage MSCs (Figure 1H). Similar results were found in MSCs derived from three individuals (Figure 1H). Similarly, quantitative RT-PCR also revealed that early-passage MSCs had increased RB mRNA levels and decreased p21 and p16 mRNA levels (Figure 1I).
RB Knockdown Induces Loss of Proliferation Rate and Differentiation Potentials
To examine the roles of RB in maintaining MSC properties, we first compared the proliferation rate and population doubling time between early-passage MSCs transduced with control or one of two RB-specific small hairpin RNAs (shRNAs) throughout long-term culture. Since it was difficult to expand MSCs with RB knockdown at low density, proliferation assays were performed at high density (1,000 cells/cm2). Compared with transduction with control shRNAs, transduction of RB shRNAs reduced RB expression at both the protein and mRNA levels (Figure 2A), which increased the proliferation rate and decreased the population doubling time in early subcultures, but reduced the proliferation rate and increased the population doubling time in late subcultures (Figure 2A). Consistent with the decrease in proliferation rate in late subcultures after RB knockdown, cells with RB knockdown had a much larger size than MSCs expressing control shRNAs after the sixth subculture (Figure 2B). Similarly, knockdown of RB reduced the bromodeoxyuridine (BrdU) incorporation rate (Figure S2A). To investigate the effects of RB knockdown on the differentiation potentials of MSCs, cells were induced in OIM or AIM for up to 2 weeks. Alizarin red S staining at 2 weeks of osteogenic induction revealed that MSCs with RB knockdown had decreased osteogenic differentiation potentials compared with MSCs expressing control shRNAs (Figure 2C). Consistently, ALP activity also demonstrated that cells with RB knockdown had less active bone formation (Figure S2B). Likewise, quantitative RT-PCR revealed that RB knockdown decreased the mRNA levels of RUNX2 and BSP at 1 week of osteogenic induction (Figure 2D). Oil red O staining at 2 weeks of adipogenic induction revealed that MSCs with RB knockdown had decreased numbers of lipid-accumulated cells compared with MSCs expressing control shRNAs (Figure 2E). Similarly, MSCs with RB knockdown had lower mRNA levels of PPAR-γ2 and LPL than MSCs transduced with control shRNAs at 1 week of adipogenic induction (Figure 2F). Taken together, these results indicate that knockdown of RB in early-passage MSCs induced loss of proliferation rate and differentiation potentials.
Figure 2.
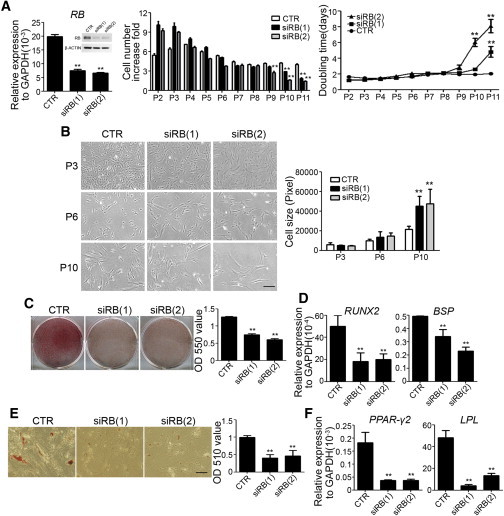
Knockdown of RB in Early-Passage MSCs Increased the Cell Size and Induced Loss of the Proliferation Rate and In Vitro Osteogenic and Adipogenic Differentiation Potentials
Early-passage MSCs were infected with lentivirus carrying control (CTR) or RB-specific shRNAs, siRB(1) or siRB(2).
(A) Left: quantitative RT-PCR and western blot analysis of RB knockdown efficiency. Cells were seeded at 103 cells/cm2 and a subculture was performed at 1-week intervals. Middle: cell number fold increase. Right: population doubling time.
(B) Left: morphology of MSCs without or with RB knockdown. Right: cell size measurements.
(C and D) MSCs without or with RB knockdown were treated with OIM. (C) Left: representative pictures of alizarin red S staining at 2 weeks. Right: optical density measurements of extracted alizarin red S staining. (D) Quantitative RT-PCR showed the mRNA levels at 1 week.
(E and F) MSCs without or with RB knockdown were treated with AIM. (E) Left: representative pictures of oil red O staining at 2 weeks. Right: optical density measurements of extracted oil red O staining. (F) Quantitative RT-PCR showed the mRNA levels at 1 week.
The results are expressed as mean ± SD of three independent experiments. Asterisks indicate significant differences (∗∗p < 0.01). Scale bar, 50 μm. See also Figure S2.
RB Knockdown Induces Premature Senescence
APO-1, p21, and p16 are all senescence markers that are expressed in late-passage MSCs (Shibata et al., 2007; Tsai et al., 2011; Yew et al., 2011). Western blot analysis showed that RB knockdown increased the protein levels of p21 and p16 (Figure S2C). The increase of these senescence-associated markers by RB knockdown was more dramatic in early-passage MSCs than in late-passage MSCs, consistent with the increased expression of RB in early-passage MSCs (Figure S2C). Quantitative RT-PCR also revealed that knockdown of RB in early-passage MSCs increased APO-1 mRNA levels compared with MSCs transduced with control shRNAs (Figure S2D). Moreover, β-gal staining revealed that knockdown of RB in early-passage MSCs increased the number of cells stained by β-gal (Figure S2E). Taken together, these results suggest that knockdown of RB in early-passage MSCs induces premature senescence.
Overexpression of RB Prevents Loss of Proliferation and Differentiation Potentials and Inhibits Senescence
To further confirm the roles of RB in maintaining stem cell properties and preventing senescence, we overexpressed RB in late-passage MSCs and evaluated its effects on proliferation, differentiation potentials, and senescence marker expression. Overexpression of RB in late-passage MSCs increased the proliferation rate (Figure 3A) and differentiation potentials (Figures 3B and 3C); decreased the expression of senescence-associated markers such as p21, p16 (Figure 3D), and APO-1 (Figure 3E); and reduced β-gal staining (Figure 3F). Similarly, RB overexpression via retroviral transduction in late-passage MSCs also increased the proliferation rate (Figure S3A), decreased the expression of p21 and p16 (Figure S3B), and reduced β-gal staining (Figure S3C). However, overexpression of RB in IMR90 fibroblasts decreased the proliferation rate (Figure S3D) and increased β-gal staining (Figure S3E), consistent with a previous report that showed RB’s role in replicative senescence of normal fibroblasts (Li et al., 1994). Together, these data suggest that RB plays an essential role in maintaining stem cell properties and prevents senescence in MSCs, but induces replicative senescence in fibroblasts.
Figure 3.
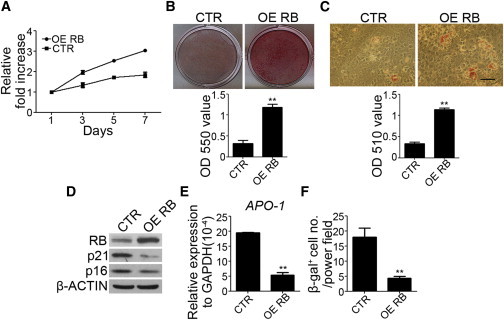
Overexpression of RB in Late-Passage MSCs Increased the Proliferation Rate and In Vitro Osteogenic and Adipogenic Differentiation Potential, and Reduced Senescence Markers
Late-passage MSCs were transfected with control (CTR) or RB overexpression vectors (OE RB).
(A) MSCs without or with RB overexpression were seeded at 4.5 × 103 /cm2 and cultured for 7 days. MTT assay was performed during the indicated period, and data are shown as the relative fold increase.
(B) MSCs without or with RB overexpression were treated with OIM. Upper: representative pictures of alizarin red S staining at 2 weeks. Lower: optical density measurements of extracted alizarin red S staining.
(C) MSCs without or with RB overexpression were treated with AIM. Upper panel: representative pictures of oil red O staining at 2 weeks. Lower panel: optical density measurements of extracted oil red O staining.
(D–F) Western blot analysis (D), quantitative RT-PCR (E), and β-gal staining (F) of MSCs without or with RB overexpression.
The results are expressed as mean ± SD of three independent experiments. Asterisks indicate significant differences (∗∗p < 0.01). Scale bar, 50 μm. See also Figure S3.
RB Maintains MSC Properties through Upregulation of DNMT1
Decreased DNMT1 activity in cellular senescence has been demonstrated in aging fibroblasts, and RB has been reported to be involved in the transcriptional regulation of DNMT1 (Lopatina et al., 2002; Slack et al., 2001). It has also been demonstrated that DNA methylation regulated by DNMT1 involves the expression of p21 and p16 (Tsai et al., 2012). Therefore, we compared the expression of DNMTs in MSCs under different conditions. Interestingly, the protein levels of DNMT1 and DNMT3b, but not DNMT3a, were higher in early-passage MSCs than in late-passage MSCs from three individuals (Figure 4A). Similarly, early-passage MSCs had increased mRNA levels of DNMT1 (Figure 4B). Moreover, MSCs transduced with RB shRNAs had decreased protein levels of DNMT1, but not DNMT3b, and the effect was more obvious in early-passage MSCs compared with late-passage MSCs (Figure 4C). As expected, overexpression of RB in late-passage MSCs increased the protein levels of DNMT1 (Figure 4D). Moreover, loss-of-function mutations in each of the phosphorylation sites were associated with a decrease in the upregulation of DNMT1 by RB (Figure S4A). These data suggest that RB upregulates DNMT1 expression in early-passage MSCs, which is dependent on RB phosphorylation.
Figure 4.
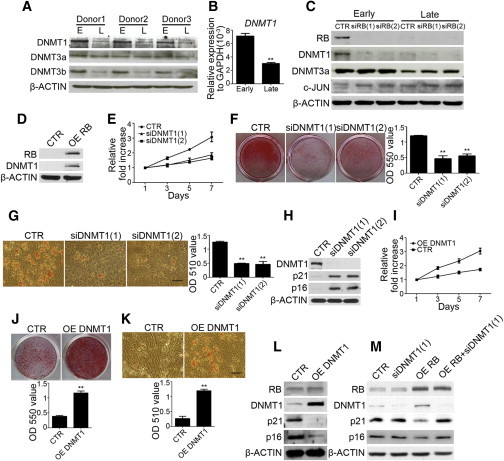
RB Maintains the Proliferation Rate and Differentiation Potential through Upregulation of DNMT1 in MSCs
(A and B) Western blot analysis (A) and quantitative RT-PCR (B) of early- and late-passage MSCs from three individuals.
(C) MSCs at passage 3 and passage 6 were infected with lentivirus carrying control (CTR) or RB-specific shRNAs, siRB(1) or siRB(2) followed by western blot analysis.
(D) Late-passage MSCs were transfected with control (CTR) or RB overexpression vectors (OE RB) followed by western blot analysis.
(E) MSCs without or with DNMT1 knockdown were seeded at 4.5 × 103 /cm2 and cultured for 7 days. MTT assay was performed during the indicated period, and data are shown as the relative fold increase.
(F) MSCs without or with DNMT1 knockdown were treated with OIM. Left: representative pictures of alizarin red S staining at 2 weeks. Right: optical density measurement of extracted alizarin red S staining.
(G) MSCs without or with DNMT1 knockdown were treated with AIM. Left: representative pictures of oil red O staining at 2 weeks. Right: optical density measurement of extracted oil red O staining.
(H) Western blot analysis of MSCs without or with DNMT1 knockdown.
(I) Late-passage MSCs were transfected with control (CTR) or DNMT1 overexpression vectors (OE DNMT1). MSCs without or with DNMT1 knockdown were seeded at 4.5 × 103 /cm2 and cultured for 7 days. MTT assay was performed during the indicated period and data are shown as the relative fold increase.
(J) MSCs without or with DNMT1 overexpression were treated with OIM. Upper: representative pictures of alizarin red S staining at 2 weeks. Lower: optical density measurements of extracted alizarin red S staining.
(K) MSCs without or with DNMT1 overexpression were treated with AIM. Upper: representative pictures of oil red O staining at 2 weeks. Lower: optical density measurements of extracted oil red O staining.
(L) Western blot analysis of MSCs without or with DNMT1 overexpression.
(M) Western blot analysis of combinational knockdown of DNMT1 upon overexpression of RB in late-passage MSCs.
The results are expressed as mean ± SD of three independent experiments. Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01). Scale bar, 50 μm. See also Figure S4.
To demonstrate the link between DNMT1 expression and function of DNMT1 in early-passage MSCs, we applied genetic modification of DNMT1 to both early- and late-passage MSCs. Knockdown of DNMT1 in early-passage MSCs was associated with a decreased proliferation rate (Figure 4E), reduced osteogenic and adipogenic differentiation potentials (Figures 4F and 4G), and premature expression of senescence-associated markers such as p21 and p16 (Figure 4H). Overexpression of DNMT1 in late-passage MSCs also increased the proliferation rate (Figure 4I), enhanced osteogenic and adipogenic differentiation potentials (Figures 4J and 4K), and suppressed the expression of p21 and p16 (Figure 4L). However, DNMT1 enzyme lacking the catalytic domain (ΔCat) when overexpressed in late-passage MSCs did not have the same effects as wild-type DNMT1 had on the proliferation rate (Figure S4B), β-gal staining (Figure S4C), expression of p21 and p16 (Figure S4D), and differentiation potentials (Figures S4E and S4F). We also demonstrated that combinational knockdown of DNMT1 upon overexpression of RB in late-passage MSCs blocked the effects of exogenous RB on suppressing senescent genes such as p16 and p21 (Figure 4M), suggesting that DNMT1 is an effector of RB in the regulation of MSC senescence. Together, these data suggest that RB upregulates DNMT1 and thereby maintains MSC properties and prevents senescence.
RB Induces DNMT1 Expression through Cooperation with c-JUN
A previous report demonstrated that RB modulates DNMT1 expression either by binding with c-JUN to a noncanonical AP-1 site in the DNMT1 promoter and thereby activating DNMT1 promoter activity, or by binding with E2F1 and thereby blocking E2F1-induced DNMT1 promoter activity (Slack et al., 2001). To demonstrate that RB upregulates DNMT1 in early-passage MSCs through the above-mentioned mechanism, we first showed that RB bound with c-JUN rather than E2F1 in early-passage MSCs (Figures 5A and 5B). Consistently, chromatin immunoprecipitation (ChIP) analysis revealed that the AP-1 site of the DNMT1 promoter was bound by RB and c-JUN, whereas the E2F binding site of the DNMT1 promoter was only bound by E2F1, and not by RB (Figure 5C), suggesting that RB plays a coactivator role in upregulation of DNMT1 by binding c-JUN in the AP-1 site in early-passage MSCs. We also performed ChIP assays in non-MSCs. We found the E2F1 binding site, but not the AP-1 site, of the DNMT1 promoter was bound by RB in a control somatic cell line, H1299 cells (Figure S5), suggesting that RB has a differentiation role in regulating DNMT1 between early-passage MSCs and late-passage MSCs or other somatic cells. Moreover, we found that the c-JUN level was not changed between early- and late-passage MSCs or before and after RB knockdown (Figure 5D), suggesting that the increase of DNMT1 expression in early-passage MSCs was not due to upregulation of c-JUN or through RB-mediated c-JUN upregulation in early-passage MSCs. To further demonstrate that RB upregulation of DNMT1 in early-passage MSCs requires c-JUN binding, we transfected MSCs with c-JUN shRNAs. We found that c-JUN knockdown reduced DNMT1 expression in early-passage MSCs, but not late-passage MSCs (Figure 5E), although the expression of RB in early-passage MSCs was not suppressed (Figure 5E). Together, these data suggest that RB binds with c-JUN to activate DNMT1 expression in early-passage MSCs.
Figure 5.
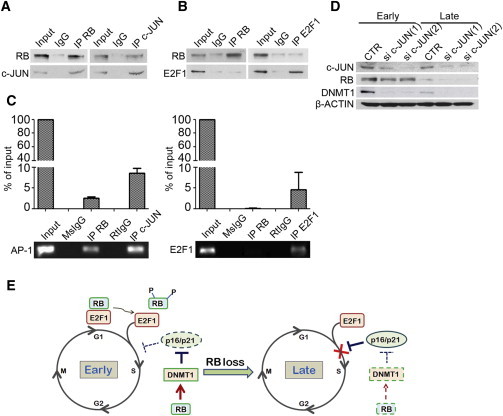
RB Induces DNMT1 Expression through Cooperation with c-JUN
(A and B) Equal aliquots of lysates from early-passage MSCs were coimmunoprecipitated by using isotype IgG or antibodies against RB, c-JUN, and E2F1, followed by western blot analysis. Input: 10% of precleared lysates (20 μg total protein).
(C) ChIP assay of early-passage MSCs. The chromatin was incubated with antibodies against RB, c-JUN, or E2F1, or their isotype IgG antibodies. Fragments of AP-1 (left) and E2F1 site (right) on the DNMT1 promoter were amplified by PCR (lower) and quantitative PCR (upper). Results are shown as the mean ± SD. MsIgG, mouse IgG2a is the isotype of anti-RB antibody. RtIgG, rabbit IgG is the isotype of anti-c-JUN and anti-E2F1 antibody. The results are expressed as mean ± SD of three independent experiments.
(D) Early- and late-passage MSCs were infected with lentivirus carrying control (CTR) or c-JUN-specific shRNAs, sic-JUN(1), or sic-JUN(2), followed by western blot analysis.
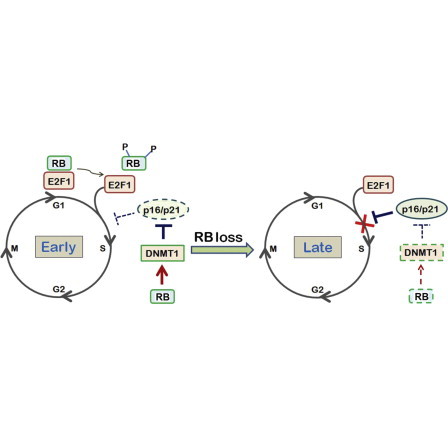
(E) Scheme of the signaling pathway involving RB in regulating the proliferation of MSCs. In early-passage MSCs, RB either binds to E2F1 and suppresses E2F1 targets or downregulates p21 and p16 by upregulating DNMT1, thereby keeping MSCs in a state of mitotic quiescence. In late-passage MSCs, loss of RB upregulates p21 and p16 and induces premature senescence.
See also Figure S5.
Similar Phenomena Are Observed in Bone-Marrow-Derived MSCs and MSCs from Other Tissues
To demonstrate that the phenomena and related signaling pathway demonstrated here are also applicable to MSCs from other tissues, we examined MSCs derived from placenta tissues (p-MSCs) of two individuals. Western blotting showed that early-passage p-MSCs (P7) had increased levels of total and phospho-RB and DNMT1 compared with late-passage p-MSCs (P23), but decreased levels of p16 and p21 (Figure 6). These data suggest that the phenomena and related signaling pathway are observed not only in bone-marrow-derived MSCs but also in MSCs derived from other tissues.
Figure 6.

Bone-Marrow-Derived MSCs and MSCs from Placenta Show Similar Phenomena
Western blot analysis of placenta MSCs from two individuals. The results show that early-passage p-MSCs (P7) had increased levels of total and phospho-RB and DNMT1 compared with late-passage p-MSCs (P23), but decreased levels of p16 and p21.
Early-Passage MSCs Delivered in Ceramic Cubes Increase the Ability to Form Bone upon Transplantation into Immunodeficient Mice
To demonstrate that early-passage MSCs have increased abilities for application in tissue engineering, we investigated the potential of MSCs delivered in ceramic cubes to form bone in vivo. At 4 weeks after implantation into the skin of immunodeficient mice, micro-computed tomography (micro-CT) analysis (Figure 7A) and Mallory’s trichrome staining (Figure 7B) revealed that the ceramic cubes delivered with early-passage MSCs had higher bone mineral density (BMD) and better collagen deposition than those delivered with late-passage MSCs. Moreover, the specimen delivered with early-passage MSCs with RB knockdown had decreased BMD (Figure 7C) and collagen deposition (Figure 7D) compared with early-passage MSCs transduced with control shRNAs. On the other hand, the specimen delivered with late-passage MSCs with RB overexpression showed increased BMD (Figure 7E) and collagen deposition (Figure 7F) compared with that delivered with late-passage MSCs transduced with control constructs. These data suggest that the current findings regarding the RB-regulated differentiation potential of early- and late-passage MSCs can be applied to tissue engineering.
Figure 7.
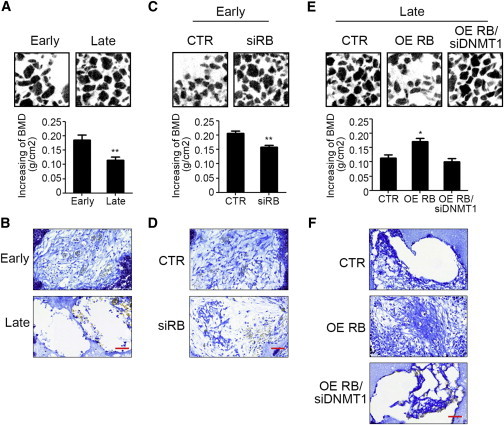
Early-Passage MSCs Delivered in Ceramic Cubes Increase the Ability to Form Bone upon Transplantation into Immunodeficient Mice
(A–F) Aliquots of 106 early- and late-passage MSCs without or with indicated genetic modification were delivered in ceramic cubes and induced in OIM for 1 week, followed by transplantation into the skin of immunodeficient mice (n = 6 for each group). At 4 weeks after implantation, micro-CT analysis (A, C, and E) and Mallory’s trichrome staining (B, D, and F) were applied to analyze BMD and collagen deposition of transplants. Results are presented as mean ± SD. Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01). Scale bar, 50 μm.
Discussion
MSCs have a wide range of clinical uses, including tissue engineering, cell therapy, immunomodulatory therapy, and anticancer cell therapy (Gómez-Barrena et al., 2011; Krampera et al., 2007). Since all of the above clinical applications require large-scale production of MSCs, a number of culture expansion techniques have been developed. However, with these techniques, MSCs have been shown to enter the senescent state and simultaneously lose their proliferation and differentiation abilities after a few passages of expansion in culture (Shibata et al., 2007; Yew et al., 2011). We demonstrated that RB maintains MSC properties by upregulating DNMT1, and thereby represses the expression of senescence markers such as p21 and p16 by increasing the DNA methylation levels of their promoters (Tsai et al., 2012). The current findings might be used to modify the properties of expanded MSCs for use in future clinical therapies.
Traditionally, RB has been considered to negatively regulate cell-cycle progression by binding to E2F1 and arresting cells in G1 phase. RB binds to E2F1 and thereby inhibits E2F1 transcriptional activity (Galderisi et al., 2006; Weinberg, 1995). While being phosphorylated, RB would liberate E2F1 and stop inhibiting the expression of E2F1 target genes, thereby letting a cell enter into S phase (Galderisi et al., 2006). Our data show that RB knockdown increased the proliferation of MSCs at the beginning, suggesting that RB inhibited cell-cycle progression. However, after a few passages of expansion, the growth curve of MSCs with RB knockdown dropped abruptly and then the proliferation rate of the MSCs with RB knockdown became slower than that of MSCs expressing control shRNAs. This dramatic change in growth might be explained by the role of RB in maintaining stem cell properties. Bone marrow stem cells, like other primitive stem cells, possess the property of mitotic quiescence, which means that a cell can temporarily exit from the cell cycle and divide slowly or rarely (Galderisi et al., 2006; Morrison et al., 1997). In this work, early-passage MSCs transduced without or with control shRNAs (representing the cells containing RB) were able to stay in the quiescent state and exhibited a growth curve that dropped comparatively slowly. In contrast, late-passage MSCs or MSCs with RB knockdown could leave the quiescent state, quickly enter cell-cycle progression, and then become exhausted in the senescent state after a few passages of subculture. Together, these data suggest that RB might maintain MSCs in the quiescent state and prevent them from undergoing premature senescence.
It was previously reported that the DNMT1 promoter could be repressed by the RB tumor suppressor in the mouse embryonal carcinoma cell line P-19 (Bigey et al., 2000). However, the same group of authors also demonstrated that RB and c-JUN could form an AP-1 complex by binding to the noncanonical AP-1 site and activating the promoter of DNMT1 (Slack et al., 2001). RB might have the opposite roles in regulating DNMT1 promoter activity. Our data showed that DNMT1 was expressed in parallel with RB expression in MSCs, and accordingly decreased and increased with RB knockdown and overexpression, respectively. Similarly to previous findings (Bigey et al., 2000; Slack et al., 2001), we demonstrated that RB binds with c-JUN, rather than E2F1, to increase the expression of DNMT1. As expected, knockdown c-JUN in early-passage MSCs blocked RB-dependent expression of DNMT1. Interestingly, we also demonstrated that RB-dependent upregulation of MSC properties was mediated through upregulation of DNMT1. Together, these data suggest that RB plays a positive role in regulating MSC properties through a pathway that involves E2F1, c-JUN, DNMT1, p21, and p16, and maintains early-passage MSCs in a relatively mitotic quiescent state, thereby protecting the cells against rapid exhaustion and premature senescence (Figure 5E).
In the future, clinical therapies may make use of MSCs for tissue engineering and cell therapy. This study describes a pathway of RB-DNMT1 that early-passage MSCs mediate to maintain MSC properties and prevent cells from undergoing replicative senescence. These data show that DNMT1 knockdown in RB-overexpressing MSCs restores the expression of 21 and p16. However, a more comprehensive approach is needed to determine whether DNMT1 is necessary or sufficient to mediate RB-dependent maintenance of pluripotency or stemness. To elucidate this further, molecular approaches and functional assays are required. Nevertheless, the molecular pathway for RB-mediated maintenance of MSC properties described here should aid in the development of new strategies for applying MSCs in clinical therapies.
Experimental Procedures
Cell Culture
These studies were approved by the Institutional Review Board of Taipei Veterans General Hospital, and informed consent was obtained from the donors who provided bone marrow aspirates. Briefly, mononuclear cells were isolated from heparinized bone marrow by density gradient centrifugation using Ficoll-Hypaque at a density of 1.077 g/l, followed by seeding into a six-well plate with complete culture medium (α-minimal essential medium [α-MEM]; GIBCO-BRL) supplemented with 16.6% fetal bovine serum [FBS], 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine) at 37°C under 5% CO2 atmosphere. At ∼10 days after seeding, cells were recovered for subculture. For low-density culture, MSCs were reseeded at a density of 50–100 cells/cm2 and the culture medium was changed twice per week. The cells were recovered 10–12 days later and reseeded at 50–100 cells/cm2. For high-density culture, cells were reseeded at a density of 103 cells/cm2 and the culture medium was changed twice per week. Subculture was performed at a density of 103 cells/cm2 every week. Primary human fetal fibroblasts (IMR90, CCL186) were obtained from the American Tissue Culture Collection and grown in Eagle’s MEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. The culture medium was changed every 3–4 days and cells were subcultured at a 1:5 ratio at 80%–90% confluence. Primary human placenta MSCs were obtained from Dr. M.L. Yen (College of Medicine, National Taiwan University) and grown in DMEM-LG medium supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were subcultured at a 1:2 ratio and the culture medium was changed every 3 days.
Cell Proliferative Assay
To compare the proliferation rates of MSCs from different conditions, cells were plated in triplicate in 96-well tissue culture plates at a density of 4.5 × 103 cells/cm2 in complete culture medium. To assay cell proliferation at 1–7 days, the cells were washed twice with PBS and the cell number was estimated using the 3-(4,5-dimethyllthiazol 2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Then 0.5 ml of 1 mg/ml MTT reagent in serum-free α-MEM was added to each well, and plates were incubated for 2 hr at 37°C. We then aspirated the reagent and added 0.5 ml of solubilizing solution (DMSO) to solubilize the formazan crystals. Absorbance was measured at 570 nm using a 630 nm differential filter.
CFSE Proliferation Assay
Aliquots of 105 MSCs were seeded in 10 cm dishes and incubated with 1 μM CFSE solution (Invitrogen) for 15 min at 37°C. The solution was then replaced with fresh prewarmed medium. Detection of CFSE by flow cytometry was performed on the indicated days after culture.
Cell Differentiation Assay
MSCs were induced with OIM (α-MEM supplemented with 16.6% FBS, 50 μg/ml ascorbate-2 phosphate, 10−8 M dexamethasone, and 10 mM β-glycerophosphate) and AIM (α-MEM supplemented with 16.6% FBS, 50 μg/ml ascorbate-2 phosphate, 10−7 M dexamethasone, 50 μM indomethacin, 0.45 mM 3-isobutyl-1-methyl-xanthine, and 10 μg/ml insulin) to differentiate into osteoblasts and adipocytes, respectively. After the appearance of morphologic features of differentiation, cells treated with OIM were assayed for alkaline phosphatase activity and stained with alizarin red S, and cells treated with AIM were stained with oil red O. To analyze alkaline phosphatase activity, aliquots of cell extracts (20 mg) were incubated with r-nitrophenol phosphate (Sigma-Aldrich) at 37°C and absorbance was read at 405 nm. Samples were run in triplicate.
β-Gal Staining
Cells were washed with PBS and fixed with 2% formaldehyde/0.2% glutaraldehyde for 10 min at room temperature. After washing, the cells were incubated at 37°C for an appropriate time with fresh β-Gal chromogenic substrate solution (1 mg/ml 5-bromo-4-chloro-3-indolyl-β-galactoside [X-Gal; Cell Signaling Technology], 40 mM citric acid [pH 6.0], 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2). The experiment was repeated three times and the number of cells expressing β-gal was calculated.
Quantitative RT-PCR
TRIzol reagent (Invitrogen) was used to extract total RNA according to the manufacturer’s specifications. RNA was reversely transcribed in 20 μl using 0.5 μg of oligo dT and 200 U Superscript III RT (Invitrogen) for 30 min at 50°C, followed by 2 min at 94°C to inactivate the reverse transcriptase. Quantitative real-time PCR was performed using cDNA as the template in a 20 μl reaction mixture containing Fast SYBR Green Master Mix (Invitrogen) and a specific primer pair of each cDNA according to the published sequences (Table S1). Real-time PCR reactions were prepared in duplicate and heated to 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 1 min, and extension at 72°C for 20 s. Standard curves (cycle threshold values versus template concentration) were prepared for each target gene and for the endogenous reference (GAPDH) in each sample. Quantification of the unknown samples was performed using the StepOne and StepOnePlus Software v2.3 supplied with the ABI Step One Real-Time PCR System machine.
Western Blot Analysis
Cells were lysed and protein was extracted using M-PER (Pierce) plus protease inhibitor cocktail (Pierce), and protein concentrations were determined using the BCA assay (Pierce). Aliquots of protein lysates were separated on SDS-10% polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membrane, which was blocked with 5% blotting-grade milk (Bio-Rad) in TBST (20 mM Tris–HCl [pH 7.6], 137 mM NaCl, and 1% Tween 20). The membrane was then hybridized with the indicated primary antibodies followed by the corresponding secondary antibodies, and then detected using a chemiluminescence assay (Millipore). Membranes were exposed to X-ray film to visualize the bands (Amersham Pharmacia Biotech). Antibodies against RB (C-2), DNMT1 (H-12), p21 (N-20), p16 (C-20), c-JUN (H-79), CDK2 (D-12), and Cyclin D1 (M-20) were purchased from Santa Cruz Biotechnology. Antibodies against E2F1 (GTX112912), DNMT3a (GTX104955), and DNMT3b (GTX62171) were purchased from GeneTex. Antibody against β-ACTIN (AC-15) was purchased from Novus Biologicals.
Immunoprecipitation
Cell extracts were incubated with antibodies against RB (Santa Cruz), c-JUN (Santa Cruz), or E2F1 (GeneTex) overnight at 4°C with gentle rotation. The immune complexes were collected by adding 5 μl protein G beads (Millipore) and incubated for 1 hr at 4°C with gentle rotation followed by centrifugation. Precipitates were washed two times with ice-cold PBS, one time with 0.05% Tween-20 in PBS, and finally one time with ice-cold PBS. The precipitates were suspended in 20 μl of 2× SDS sample buffer and analyzed by SDS-10% polyacrylamide gel. Immunoblotting was performed by antibodies against RB, c-JUN, or E2F1.
BrdU Incorporation Assay
The BrdU incorporation assay was performed using the In Situ Cell Proliferation Kit, FLUOS (Roche Diagnostics) according to the manufacturer’s instructions. In brief, 2 × 105 MSCs were plated in 10 cm dishes for 48 hr and then thymidine analog BrdU was added into each dish and incubated for 18 hr. Cells were then washed twice with PBS and fixed with an ethanol and glycine mixture (7:3) for 30 min. DNA was denatured in 2N HCl and anti-BrdU antibody was added to detect positive cells. Cells were counterstained with 1 mg/ml propidium iodide and analyzed with a FACScan (Becton Dickinson).
Plasmid Reconstruction
The pSLX-CMV-RB plasmid for RB overexpression was a gift from Dr. M.F. Chang (Institute of Biochemistry and Molecular Biology, College of Medicine, National Taiwan University). The pSLX-CMV empty plasmid was generated by cutting the RB fragment from the BamH1 site of the pSLX-CMV-RB plasmid. The full-length cDNA (wild-type; +1/+4851) of human DNMT1 and a truncated form (ΔCat; +1/+3494) of this cDNA lacking the catalytic domain were gifts from Dr. Jesús Espada (Instituto de Investigaciones Biomédicas “Alberto Sols,” CSIC-UAM; Espada et al., 2011) and were cloned in a pcDNA3.1 backbone. The open reading frames (ORFs) of RB and RB mutations (S780A, S795A, and S807/811A) in pReceiver expression vector containing the N-terminal Flag epitope were purchased from Genecopeia. The ORFs of the RB and RB mutations were recloned in pLAS2W lentiviral expression vector (National RNAi Core Facility Platform, Academia Sinica) for lentivirus production.
Transfection
Nucleofector technology (AMAXA Biosystems) was used to perform transfection, which was based on electroporation with each nucleofection sample containing 2–4 μg of DNA, 4 × 105 cells, and 100 μl of Human MSC Nucleofector Solution. We used the program C-17 of the Nucleofector device to carry out the transfection as recommended by the manufacturer.
Lentiviral-Mediated RNAi
The expression plasmids and the bacteria clones for RB shRNA (TRCN0000010418 and TRCN0000040167), DNMT1 shRNA (TRCN0000021891 and TRCN0000021893), and c-JUN-shRNA (TRCN0000338221 and TRCN0000338225) were obtained from the National RNAi Core Facility Platform. Subconfluent cells were infected with lentivirus in the presence of 8 μg/ml polybrene (Sigma-Aldrich). At 24 hr postinfection, the medium was removed and replaced with fresh growth medium containing puromycin (3 μg/ml) and selected for infected cells for 48 hr.
ChIP Assay
MSCs and H1299 cells were crosslinked in 1% formaldehyde for 10 min at room temperature and then halted with 125 mM glycine for 5 min. Cells were washed in cold PBS and scratched in cold PBS containing protease inhibitors. The cell pellets were resuspended in SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1]) containing protease inhibitors and incubated on ice for 10 min. The samples were sonicated with a Bioruptor Plus sonicator (Diagenode) to an average length of ∼200–1,000 bp and then microcentrifuged. The supernatant was diluted in 9 vol of ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 10 mM Tris-HCl [pH 8.1], 150 mM NaCl) and precleaned with 50 μl of protein G Mag Sepharose Xtra (GE) for 30 min at 4°C. The precleaned chromatin was incubated with 2 μg of immunoprecipitation antibodies with rotation at 4°C for 12–16 hr. The antibodies were against RB (c-15, sc-50; Santa Cruz), c-JUN (39309; Active Motif), and E2F1 (c-20, sc-193; Santa Cruz). The chromatin was immunoprecipitated, washed, and eluted with buffer containing 1% SDS and 0.1 M NaHCO3. The DNA-protein crosslinking was reversed by adding NaCl to a final concentration of 200 mM at 65°C for 4 hr. The DNA was then purified with a ChIP DNA Clean & Concentrator Kit (Zymo Research). The DNA was amplified with specific primer (Table S1) for AP-1 and E2F1 sites on the DNMT1 promoter by real-time PCR. Data were expressed as the percentage of input DNA.
In Vivo Osteogenic Differentiation
Aliquots of 106 cells were delivered in 5 × 5 × 5 mm ceramic cubes (Zimmer) followed by induction with OIM. One week after induction, the cell-containing cubes were transplanted subcutaneously into immunodeficient mice. The specimens were harvested for BMD analysis by micro-CT, and collagen deposition was assessed by Mallory trichrome staining 4 weeks later.
Statistical Analysis
All values were expressed as mean ± SD. Comparisons between two groups were analyzed by Student’s t test. Comparisons within three groups were analyzed by ANOVA. A value of p < 0.05 was considered statistically significant.
Author Contributions
F.-Y.C., S.-Y.K, and S.-C.H. conceived and designed the study. S.-P.L., F.-Y.C., Y.W., and M.-L.Y. collected and/or assembled the data. S.-P.L., F.-Y.C., Y.W., S.-Y.K., and S.-C.H. analyzed and interpreted the data. S.-P.L., S.-Y.K., and S.-C.H. wrote the manuscript. All authors gave final approval of the manuscript.
Acknowledgments
We thank the staff of the Medical Science and Technology Building of Taipei Veterans General Hospital for providing experimental space and facilities. This study was supported by grants from the National Science Council (NSC 102-2321-B-010-011 and 101-2314-B-010-028-MY3), Taipei Veterans General Hospital (V100E1-011), and National Yang-Ming University.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Shou-Yen Kao, Email: sykao@vghtpe.gov.tw.
Shih-Chieh Hung, Email: hungsc@vghtpe.gov.tw.
Supplemental Information
References
- Alessio N., Bohn W., Rauchberger V., Rizzolio F., Cipollaro M., Rosemann M., Irmler M., Beckers J., Giordano A., Galderisi U. Silencing of RB1 but not of RB2/P130 induces cellular senescence and impairs the differentiation potential of human mesenchymal stem cells. Cell. Mol. Life Sci. 2013;70:1637–1651. doi: 10.1007/s00018-012-1224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigey P., Ramchandani S., Theberge J., Araujo F.D., Szyf M. Transcriptional regulation of the human DNA Methyltransferase (dnmt1) gene. Gene. 2000;242:407–418. doi: 10.1016/s0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- Bork S., Pfister S., Witt H., Horn P., Korn B., Ho A.D., Wagner W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2010;9:54–63. doi: 10.1111/j.1474-9726.2009.00535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J.A., Danielian P.S., Nedelcu S., Berman S.D., Lees J.A. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A.I. Adult mesenchymal stem cells for tissue engineering versus regenerative medicine. J. Cell. Physiol. 2007;213:341–347. doi: 10.1002/jcp.21200. [DOI] [PubMed] [Google Scholar]
- Connell-Crowley L., Harper J.W., Goodrich D.W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espada J., Peinado H., Lopez-Serra L., Setién F., Lopez-Serra P., Portela A., Renart J., Carrasco E., Calvo M., Juarranz A. Regulation of SNAIL1 and E-cadherin function by DNMT1 in a DNA methylation-independent context. Nucleic Acids Res. 2011;39:9194–9205. doi: 10.1093/nar/gkr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galderisi U., Cipollaro M., Giordano A. The retinoblastoma gene is involved in multiple aspects of stem cell biology. Oncogene. 2006;25:5250–5256. doi: 10.1038/sj.onc.1209736. [DOI] [PubMed] [Google Scholar]
- Gómez-Barrena E., Rosset P., Müller I., Giordano R., Bunu C., Layrolle P., Konttinen Y.T., Luyten F.P. Bone regeneration: stem cell therapies and clinical studies in orthopaedics and traumatology. J. Cell. Mol. Med. 2011;15:1266–1286. doi: 10.1111/j.1582-4934.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Higashi H., Jung H.K., Suzuki-Takahashi I., Ikeda M., Tamai K., Kato J., Segawa K., Yoshida E., Nishimura S., Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Knudsen E.S., Wang J.Y. Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J. Biol. Chem. 1996;271:8313–8320. doi: 10.1074/jbc.271.14.8313. [DOI] [PubMed] [Google Scholar]
- Krampera M., Franchini M., Pizzolo G., Aprili G. Mesenchymal stem cells: from biology to clinical use. Blood Transfus. 2007;5:120–129. doi: 10.2450/2007.0029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Nichols M.A., Shay J.W., Xiong Y. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product pRb. Cancer Res. 1994;54:6078–6082. [PubMed] [Google Scholar]
- Lopatina N., Haskell J.F., Andrews L.G., Poole J.C., Saldanha S., Tollefsbol T. Differential maintenance and de novo methylating activity by three DNA methyltransferases in aging and immortalized fibroblasts. J. Cell. Biochem. 2002;84:324–334. doi: 10.1002/jcb.10015. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Shah N.M., Anderson D.J. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. doi: 10.1016/s0092-8674(00)81867-x. [DOI] [PubMed] [Google Scholar]
- Poole A.J., Heap D., Carroll R.E., Tyner A.L. Tumor suppressor functions for the Cdk inhibitor p21 in the mouse colon. Oncogene. 2004;23:8128–8134. doi: 10.1038/sj.onc.1207994. [DOI] [PubMed] [Google Scholar]
- Sekiya I., Larson B.L., Smith J.R., Pochampally R., Cui J.G., Prockop D.J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530–541. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- Sethe S., Scutt A., Stolzing A. Aging of mesenchymal stem cells. Ageing Res. Rev. 2006;5:91–116. doi: 10.1016/j.arr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Shapiro G.I., Edwards C.D., Ewen M.E., Rollins B.J. p16INK4A participates in a G1 arrest checkpoint in response to DNA damage. Mol. Cell. Biol. 1998;18:378–387. doi: 10.1128/mcb.18.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata K.R., Aoyama T., Shima Y., Fukiage K., Otsuka S., Furu M., Kohno Y., Ito K., Fujibayashi S., Neo M. Expression of the p16INK4A gene is associated closely with senescence of human mesenchymal stem cells and is potentially silenced by DNA methylation during in vitro expansion. Stem Cells. 2007;25:2371–2382. doi: 10.1634/stemcells.2007-0225. [DOI] [PubMed] [Google Scholar]
- Slack A., Pinard M., Araujo F.D., Szyf M. A novel regulatory element in the dnmt1 gene that responds to co-activation by Rb and c-Jun. Gene. 2001;268:87–96. doi: 10.1016/s0378-1119(01)00427-9. [DOI] [PubMed] [Google Scholar]
- Tsai C.C., Chen Y.J., Yew T.L., Chen L.L., Wang J.Y., Chiu C.H., Hung S.C. Hypoxia inhibits senescence and maintains mesenchymal stem cell properties through down-regulation of E2A-p21 by HIF-TWIST. Blood. 2011;117:459–469. doi: 10.1182/blood-2010-05-287508. [DOI] [PubMed] [Google Scholar]
- Tsai C.C., Su P.F., Huang Y.F., Yew T.L., Hung S.C. Oct4 and Nanog directly regulate Dnmt1 to maintain self-renewal and undifferentiated state in mesenchymal stem cells. Mol. Cell. 2012;47:169–182. doi: 10.1016/j.molcel.2012.06.020. [DOI] [PubMed] [Google Scholar]
- Weinberg R.A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- Yew T.L., Chiu F.Y., Tsai C.C., Chen H.L., Lee W.P., Chen Y.J., Chang M.C., Hung S.C. Knockdown of p21(Cip1/Waf1) enhances proliferation, the expression of stemness markers, and osteogenic potential in human mesenchymal stem cells. Aging Cell. 2011;10:349–361. doi: 10.1111/j.1474-9726.2011.00676.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


