Abstract
The endoplasmic reticulum (ER) is a dynamic pleiomorphic organelle containing continuous but distinct subdomains. The diversity of ER structures parallels its many functions, including secretory protein biogenesis, lipid synthesis, drug metabolism and Ca2+ signaling. Recent studies are revealing how elaborate ER structures arise in response to subtle changes in protein levels, dynamics, and interactions as well as in response to alterations in cytosolic ion concentrations. Subdomain formation appears to be governed by principles of self-organization. Once formed, ER subdomains remain malleable and can be rapidly transformed into alternative structures in response to altered conditions. The mechanisms that modulate ER structure are likely to be important for the generation of the characteristic shapes of other organelles.
Introduction
The many roles of the endoplasmic reticulum (ER) demand a high surface area and a distribution throughout the cytoplasm. The ER organizes the large amount of membrane required [1] by folding it into tubular or lamellar structures, generating a complex architecture that varies in response to functional requirements. Variations in ER organization are found not only in different cell types but also in different regions of the organelle within the same cell. Indeed, although the ER is a single, spatially continuous compartment [2,3], it is composed of structurally and functionally different subdomains. Among these, the nuclear envelope and the peripheral ER are the most obvious [4,5], but the ER contains other specialized subcompartments, such as the junctional regions between the ER and essentially every other organelle, and the exit sites where COPII-coated transport vesicles are generated [6]. As revealed by live cell imaging, many of these subdomains appear to be in a constant state of flux [3], suggesting that the ER has tremendous flexibility to alter structural organization, as necessary, to adapt to constantly changing cellular requirements. Indeed, physiological and developmental processes may rely on rapid ER restructuring, like that occurring in egg cells upon fertilization, in response to changes in cytosolic Ca2+ concentration [7].
Perhaps the first subdivision of the ER to be recognized was that between rough (ribosome-covered) and smooth (ribosome-free) domains (RER and SER) [8]. In many cells, RER and SER do not occupy spatially segregated regions, and small ribosome-free areas are interspersed with ribosome-covered regions. This type of ER is generally organized in sheets (cisternae) or in a branching tubular network typically seen in many cultured mammalian cells. Such networks are characterized by fairly straight tubules, which branch at tripartite junctions to generate a polygonal meshwork [5,9]. Only in some cells (e.g. hepatocytes, steroid-synthesizing cells and neurons) do the smooth and rough portions of the ER occupy different regions of the cytoplasm. In these cases, the SER differs from the RER on the basis not only of its ribosome-free surface but also of its distinctive spatial organization.
In this review, we will discuss recent work on the factors that determine the formation of the ER branching tubular network, as well as on the mechanisms underlying the diverse architectural arrangements of the SER and its segregation from RER.
Homotypic fusion
As is true for all membrane-bounded organelles, ER elements are capable of fusing with each other and this activity is necessary both for the dynamic restructuring of the network that constantly occurs in vivo [2,3,10•], and for the in vitro reconstitution of networks from ER-derived vesicles ([11••] and references therein). The bulk of existing evidence suggests that more than one fusion mechanism is involved.
Using a biochemical assay, Latterich et al. [12] first showed that S. cerevisiae Cdc48, an AAA ATPase with homology to the well-characterized fusion protein N-ethylmaleimide sensitive factor (NSF or Sec18), is required for ER-derived vesicle fusion. At the same time, the mammalian orthologue of Cdc48, p97, was shown to be involved in the reconstitution of Golgi cisternae from mitotic Golgi fragments [13]. Later, evidence was presented that Cdc48 works in conjunction with the ER-localized target-soluble NSF attachment protein (SNAP) receptor (t-SNARE) Ufe1p [14]. Subsequent work has confirmed the role of p97 in the dynamics of ER architecture [15,16], but also indicates the participation of additional mechanisms. Thus, antibodies against p97 or its cofactors only partially disrupt ER organization in living cells [17] or in cell-free reconstituted systems [18]. Moreover, S. cerevisiae Cdc48 mutants appear to have normal ER organization [19].
At least one p97-independent fusion activity appears to be intrinsic to the ER membrane itself. ER-derived microsomes presumably depleted of all cytosolic proteins (such as NSF or p97) show GTP-dependent (but ATP-independent) fusion activity [11••,20–22], suggesting the involvement of a cytosol-independent fusion machine tightly associated with the ER membrane. Although this fusion activity was described many years ago, its molecular components have not yet been identified.
Formation of tubular network
Phospholipid bilayers do not spontaneously arrange into highly curved structures; therefore, there is much interest in the factors that force organelle membranes to form tubules [23,24••]. All the compartments of the exo-endocytic pathway are capable of forming tubules, and this process is fundamental for membrane traffic. However, in the case of the ER, tubules represent a basic anatomical feature. Much attention has been focused on the role of the cytoskeleton in organizing ER tubular networks [3,10•,25], but recent work has demonstrated that proteins intrinsic to the ER underlie tubule formation independently of cytoskeletal elements.
Proteins mediating ER-microtubule interactions
The role of the cytoskeleton in regulating the spatial distribution of the ER is well known. In animal cells, microtubules are the main players, whereas in fungal and plant cells actin cables play a more prominent role (for reviews, see [2,26]). The interaction of the ER with microtubules is mediated both by motor proteins, which permit extension of tubules along stationary microtubules, and by non-motor proteins that mediate movement of ER tubules attached to motile or polymerizing microtubules [10•]. More than one ER protein is involved in the latter type of interaction. The first one to be identified was CLIMP-63, an integral ER membrane protein excluded from the nuclear envelope [27], whose cytosolic domain is reported to directly interact with microtubules [28]. Overexpression of CLIMP-63 causes the ER to reorganize, forming circular strands that align with micro-tubule bundles [28]. A subsequent study [29•] has shown that the interaction of CLIMP-63 with microtubules is regulated by mitotic phosphorylation, suggesting a mechanism for disruption of ER–microtubule interaction during mitosis. Interestingly, the authors found that overexpression of a mitotic phospho-mimetic form of CLIMP-63 leads to a collapse of the ER around the nucleus, suggesting that the mutant CLIMP-63 competes with the endogenous protein for factor(s) that mediate or facilitate the interaction with microtubules. Consistent with this idea, Farah et al. [30] have recently reported that CLIMP-63 interacts with the microtubule-associated protein MAP-2, and that this association explains the specific localization of RER in neuronal dendrites and its exclusion from the axon.
Novel players in the ER-microtubule interaction are the VAP-B/Nir3 couple [31•] and p22 [32••]. p22 is a myristoylated, EF-Hand-domain-containing protein that binds microtubules and Ca2+. It associates with ER membranes in a Ca2+-dependent manner, and thus provides a Ca2+-regulated link between the ER and the microtubule cytoskeleton. Concerning the VAP-B/Nir 3 couple, VAPs (vesicle-associated membrane protein interacting proteins) are C-tail-anchored proteins [33] of the ER that function as adaptors for a variety of peripheral proteins carrying a particular sequence, the FFAT (two phenylalanines in an acidic tract) motif [34,35,36••]. The majority of known FFAT-containing proteins possess lipid binding domains and are implicated in interorganellar lipid transport. Also, Nir3, a member of the highly conserved family of Nir/rdgB proteins, has a phosphatidylinositol transfer domain [37]; however, when overexpressed together with VAP-B, it causes rearrangement of the ER along circular microtubule bundles [31•], similar to the structures seen after overexpression of CLIMP-63 [28]. Interestingly, a point mutation in the VAP-B protein causes familial amyotrophic lateral sclerosis in eight Brazilian families with common Portuguese ancestry [38,39], illustrating once more the importance of cell biological studies for the understanding of disease mechanisms.
In vitro reconstitution of tubular networks
A molecular dissection of factors required for ER tubule formation requires the development of suitable cell-free systems. Tubule network formation from cell extracts or isolated microsomes has been followed by differential interference contrast by darkfield illumination microscopy, or, more frequently, by fluorescence microscopy of appropriately labeled microsomal membranes [3]. Several studies in which network formation was observed on a glass surface demonstrated a direct role for microtubules and microtubule motors in tubule formation (reviewed in [2]). Recently, two groups [40,41] have analyzed tubule generation from giant unilamellar lipid vesicles tethered to microtubules via microtubule motors. Interestingly, in the presence of energy and in the absence of any other additional proteins, branching tubules were readily created in these in vitro systems.
Different results have been obtained by the Rapoport group, who developed an assay that follows tubule formation in bulk solution [11••,22]. Using this assay, Dreier and Rapoport [22] showed that tubule generation from Xenopus oocyte-derived light microsomes could occur in the absence of microtubules. Under appropriate salt conditions, Voeltz et al. [11••] could observe tubule formation even in the absence of any cytosolic protein, prompting the authors to search for ER membrane proteins involved in the process. Taking advantage of the exquisite sensitivity of the tubulation reaction to sulfhydryl blocking reagents, the authors were able to identify the integral membrane protein Reticulon (Rtn)4a as the sulfhydryl-group-containing polypeptide implicated in tubule formation. Rtn4a is a member of a family of highly expressed ER-localized membrane proteins that have been associated with a variety of roles [42]. Consistent with the proposed role of Rtn 4a in ER tubulation, Voeltz et al. [11••] found that in animal cells Rtn4a was enriched on ER tubules but depleted from sheets and from the nuclear envelope. A similar distribution was found for the Rtn homologues in yeast. Moreover, knockout of the two yeast Rtn homologues together with an interacting partner, DP1/Yop1p, disrupted the tubular organization the yeast cortical ER. Finally, the authors present evidence that reticulons have a hairpin topology in the cytoplasmic leaflet of the ER membrane. They suggest that this unusual topology could produce a wedge-shaped intra-membrane domain that would cause bending of the phospholipid bilayer, similar to the hypothesized action of caveolin at the cell surface [43].
Voeltz et al. [11••] bring a new and unexpected ingredient to the field of ER morphogenesis, and their results will undoubtedly lead to future work aimed at testing the predictions of their model. For instance, cells should be able to modulate the extent of tubular networks by changes in reticulon levels. It will be interesting to investigate the function of reticulons in microsomes from sources other than Xenopus oocytes and to investigate whether reticulon levels relative to total ER decrease in differentiating cells such as developing plasma cells or pancreatic acinar cells that accumulate stacked rough ER cisternae. In this respect, the model of Voeltz et al. [11••] may involve additional complexities. For example, Hetzer et al. [18] combined Xenopus microsomes with chromatin to form smooth nuclear-envelope-like structures around the chromatin. Nuclear envelope assembly involved the formation of a tubular network intermediate before progressing to the final sheet structures. In this system, reticulons would be expected to remain in the sheet structure, and the tubular transition state thus suggests that membrane curvature can be regulated without changes in membrane protein composition. An intriguing possibility is that reticulon function is regulated by post-translational modification(s).
Although necessary for cortical tubular ER formation in yeast, Rtns and DP1/Ypo1p are not the only proteins required for network formation, since deletion of another ER protein (Ice2p) also causes disruption of the yeast cortical meshwork [44•]. In addition, there is evidence that the ribosomes themselves may contribute to ER geometry [19]. Thus, multiple factors, some acting autonomously on the ER membrane, others acting in concert with cytosolic elements, define the overall structure.
Smooth ER
SER is generated when the ER surface area required to house a subset of resident membrane proteins exceeds that needed for ER-associated protein synthesis. In some tissues and in some cultured cells, SER segregates from RER and assumes distinctive architectural organizations. The most common form of SER is a tubular network with different characteristics from the polygonal meshwork typically observed in cultured cells. The SER tubules are generally more convoluted than those of RER, and the branch points are more frequent, so that a sponge-like structure is generated ([5], illustrated in [45]). We refer to this SER architecture as ‘random tubular’. In some tissues, and more commonly in tissue culture cells that overexpress ER membrane proteins, SER forms parallel arrays of stacked cisternae. Such stacked cisternae may be lined up against the nuclear envelope (karmellae), or be distributed elsewhere in the cytoplasm (lamellae); in addition, they may form concentric whorls or regular sinusoidal arrays with cubic symmetry, often referred to as ‘crystalloid’ ER. Finally, the high expression of the enzyme hydroxy-methylglutaryl (HMG)-CoA reductase induces tubules that align into bundles with hexagonal symmetry ([46••] and references therein). Snapp and coworkers [46••] introduced the term OSER (organized smooth ER) to describe all of these different types of stacked smooth ER (illustrated in Figure 1). A fascinating observation is that the structure of sinusoidal ER with cubic symmetry (cubic membranes) corresponds to mathematically described minimal periodic surfaces [47], built of repetitions of saddle-shaped elements. Although such surfaces are highly curved, they in fact have a mean mathematical curvature of zero, because at every point convexity and concavity exactly compensate for each other [48]. A similar compensation between convexity and concavity is also present at branching points in polygonal meshwork ER. Thus, branching could represent a thermo-dynamically favorable arrangement for ER tubules.
Figure 1.
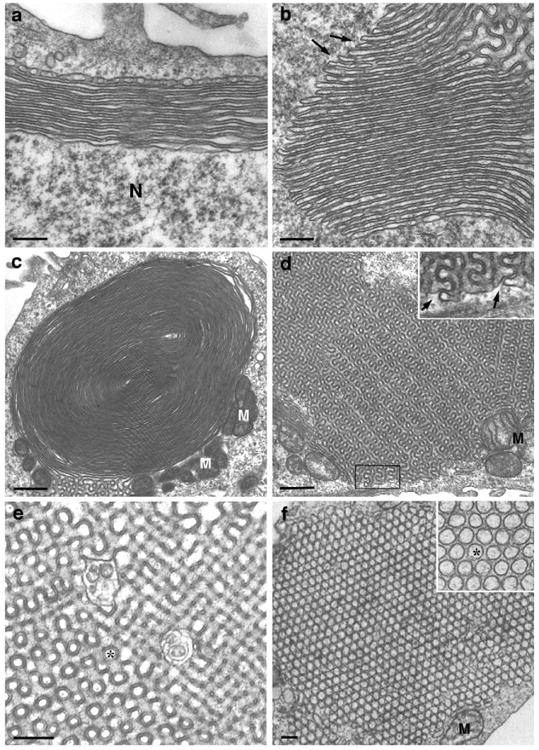
Electron micrographs illustrating different forms of OSER. Shown are: (a) a karmella, (b) a lamella, (c) a whorl, (d,e) sinusoidal arrays with cubic symmetry, and (f) a bundle of packed tubules with hexagonal symmetry. In (a), N indicates the nucleus; at the distal side of the karmella is a forming cisterna that appears to be generated by the fusion of tubules. In (b) and (d), the arrows indicate continuity with the cytosolic space; note that the electron-dense constant width space present in all the OSER structures corresponds to the cytosolic space bridging the cisternae. In (e) and (f), the asterisk indicates the lumenal space. Note, in (b) and (c), the continuity with sinusoidal ER of the lamella and whorl, respectively. Note also, in (c) and (d), the close association of mitochondria (M) with OSER structures. Structures shown in (a–e) were induced in cells transfected with a GFP–cytochrome b5 fusion protein [44•]. ‘Crystalloid’ ER with hexagonal symmetry (f) is expressed in compactin-resistant UT-1 cells expressing high levels of HMG-CoA reductase [63]. Scale bars: (a,b,c,f) 200 nm; (c,d) 600 nm. (f) is reproduced with permission from the Company of Biologists from [59].
A number of studies have implicated head-to-head dimerization of the cytosolic domains of overexpressed ER membrane proteins in cisternal stacking [49–53]. In the study of Snapp et al., this hypothesis was directly tested. Snapp and co-workers compared the ability of different GFP fusion proteins — all with the GFP moiety exposed to the cytosol — to induce OSER in mammalian cells [46••]. The commonly used EGFP dimerizes with low affinity (Kd = 0.11 mM) [54], and EGFP fusions (or spectral variants thereof) to the membrane anchors of ER-localized proteins generated prominent OSER structures. The space between the cytosolic faces of the stacked membranes was too small to lodge ribosomes but was compatible with the presence of GFP-GFP bridges (see Figure 1). When similar fusion proteins — made with GFP mutants in which the dimerization affinity is reduced nearly 100 fold — were tested, OSER induction was not observed. Instead, abundant random tubular SER was induced, suggesting that membrane proliferation had occurred without concomitant cisternal stacking. Importantly, fluorescence recovery after photobleaching (FRAP) experiments showed high diffusional mobility of all the GFP fusion proteins, including those that induced OSER: the transfected proteins diffused rapidly also within tightly packed cisternal structures and exchanged freely with the remaining ER, organized as a polygonal meshwork. In other words, ER cisternae that appear to be ‘zipped’ together and to be arranged into ‘crystalloid’ structures in fact allow free diffusion of the dimerizing proteins responsible for the packing. Thus, low-affinity transient interactions can cause dramatic restructuring of the ER into self-organized domains. The simple properties of protein–protein interactions, lipid organization, and levels of the interacting proteins combine to create emergent structures that were not predicted on the basis of the individual components.
The work of Snapp and coworkers [46••] has implications for the mechanisms of both physiological and pathological membrane stacking. Thus, the stacked arrangement of membranes, as observed with Golgi cisternae [55] or thylakoid membranes, could be caused by weak, transient interactions like the ones involved in OSER formation, rather than requiring a specific matrix or ‘glue’ for holding them together. Such transient interactions could also underlie junctions between the ER and other organelles and could be subject to regulation. For instance, Takei et al. [49] found that in Purkinje cells stacking of IP3-receptor-rich ER cisternae occurs rapidly in response to hypoxic conditions, suggesting post-translational regulation of OSER formation.
Regarding the implications for pathology, OSER has been observed to form in response to the expression of mutant, disease-causing proteins, like torsin A [56], ΔF-cystic fibrosis transmembrane conductance regulator (CFTR) [57] and mutant peripheral myelin protein-22 (PMP-22) [58], which are responsible for early onset generalized torsion dystonia, cystic fibrosis and inherited peripheral neuropathies (Dejerine Sottas syndrome and Charcot-Marie-Tooth 1A), respectively. OSER formation by these mutant proteins may in some way depend on their interaction with the ER chaperone calnexin [57–59]. However, the role of calnexin in OSER induction is unclear and deserves further investigation.
An open question on OSER formation concerns which factors determine the prevalence of different OSER forms. While karmellae, lamellae and whorls are generally observed with all OSER-inducing proteins, the formation of ‘crystalloid’ ER with hexagonal or cubic symmetry appears to be protein-specific. For example, HMG-CoA reductase, which has multiple transmembrane domains, induces OSER with hexagonal symmetry, while many single spanning membrane proteins induce sinusoidal ER with cubic symmetry. Thus, integral membrane proteins modulate the architecture of SER as well as that of polygonal ER networks.
In most studies on SER architecture, segregated SER was induced by overexpression of an ER membrane protein. Overexpression of single ER membrane proteins, whether induced by drugs or by transfection, often causes proliferation of ER membranes [60]. Thus, in these studies, ER proliferation correlated with ER restructuring and the question of whether ER restructuring can occur independently of proliferation was not addressed. Recently, Sprocati et al. have developed a system to reversibly induce segregation of random tubular smooth ER from polygonal meshwork ER in the absence of membrane proliferation [61•]. The system consists of a cell line stably transfected with a GFP tail-anchored construct. The ER of these cells, although more abundant than in nontransfected cells, maintains the normal polygonal meshwork structure. However, treatment with the drug 1-Phenyl-2-decanoyl-amino-3-morpholino-1-propanol hydrochoride (PDMP) induces the segregation of random tubular SER patches, which maintain connectivity with the rest of the ER and partially exclude RER markers. Interestingly, lipid dyes partition differently into the patches: the dye DiI-C16, which at the cell surface prefers more rigid lipid domains, concentrates more in the SER patches than does the dye FAST DiI, which has a preference for fluid domains [62], suggesting that the segregated SER domains differ from the remaining polygonal meshwork in lipid composition. Most importantly, these patches are formed without detachment of ribosomes from the ER and are reabsorbed into the rest of the ER within minutes of withdrawal of the PDMP. Thus, this system provides a useful model to investigate rapid structural rearrangements of the ER structure.
Conclusions
Biochemistry and genomics have helped define the distinct molecular constituents of each organelle and the mechanisms by which these molecules are targeted to the correct intracellular compartment. However, it is equally important to understand how these molecules modulate organelle structure. Research of the past few years suggests that many mechanisms compete and cooperate to determine ER architecture and that elaborate structures may be formed through processes of self-organization. Perturbations in the steady state levels of the structural proteins rapidly shift ER organization and permit the ER to adapt to changing requirements of the cell. The complex and plastic ER represents a paradigm for understanding the factors that shape membrane-bounded organelles. Unraveling the different factors and how they interact represents a major challenge in the field of organelle biogenesis.
Acknowledgments
We thank Francesca Lombardo for collaboration in the EM studies (panels a-e of Figure 1). Work in the laboratory of N. Borgese was supported by grants from Ministero della Istruzione, Università e Ricerca (MIUR), Ministero della Sanità (ALS grant 2002) and CNR grant ME-P02-IN-C2-M001 to the Institute of Neuroscience. Erik Snapp is an Ellison Medical Foundation New Scholar in Aging.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Weibel ER, Staubli W, Gnagi HR, Hess FA. Correlated morphometric and biochemical studies on the liver cell. I Morphometric model, stereologic methods, and normal morphometric data for rat liver. J Cell Biol. 1969;42:68–91. doi: 10.1083/jcb.42.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 3.Snapp EL. Endoplasmic reticulum biogenesis: proliferation and differentiation. In: Mullins C, editor. The biogenesis of cellular organelles Molecular Biology Intelligence Unit. C. Landes Bioscence, Georgetown TX and Kluwer Academic/Plenum Publishers; New York: 2004. pp. 63–95. [Google Scholar]
- 4.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kepes F, Rambourg A, Satiat-Jeunemaitre B. Morphodynamics of the secretory pathway. Int Rev Cytol. 2005;242:55–120. doi: 10.1016/S0074-7696(04)42002-6. [DOI] [PubMed] [Google Scholar]
- 6.Levine T, Rabouille C. Endoplasmic reticulum: one continuous network compartmentalized by extrinsic cues. Curr Opin Cell Biol. 2005;17:362–368. doi: 10.1016/j.ceb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Stricker SA. Structural reorganizations of the endoplasmic reticulum during egg maturation and fertilization. Semin Cell Dev Biol. 2006 doi: 10.1016/j.semcdb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Palade GE. The endoplasmic reticulum. J Biophys Biochem Cytology. 1956;2:85–97. doi: 10.1083/jcb.2.4.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terasaki M, Song J, Wong JR, Weiss MJ, Chen LB. Localization of endoplasmic reticulum in living and glutaraldehyde-fixed cells with fluorescent dyes. Cell. 1984;38:101–108. doi: 10.1016/0092-8674(84)90530-0. [DOI] [PubMed] [Google Scholar]
- 10•.Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7:639–646. doi: 10.1111/j.1600-0854.2006.00419.x. An interesting recent review, which deals more extensively with the molecular mechanisms of ER-microtubule interaction than has been possible in the present short article. [DOI] [PubMed] [Google Scholar]
- 11••.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. This important paper shows that a family of ER integral membrane proteins is implicated in the formation of branching ER tubules, independently of cytoskeletal elements. The transmembrane domains of the proteins are proposed to induce curvature of ER membranes, similar to the manner in which caveolin bends the membranes of caveolae. [DOI] [PubMed] [Google Scholar]
- 12.Latterich M, Frohlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- 13.Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 14.Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- 15.Potaryaev D, Squirrel JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell. 2005;16:2139–2153. doi: 10.1091/mbc.E04-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kano F, Kondo H, Yamamoto A, Tanaka AR, Hosokawa N, Nagata K, Murata M. The maintenance of the endoplasmic reticulum network is regulated by p47, a cofactor of p97, through phosphorylation of cdc2 kinase. Genes to Cells. 2005;10:334–344. doi: 10.1111/j.1365-2443.2005.00837.x. [DOI] [PubMed] [Google Scholar]
- 17.Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol. 2001;3:1086–1091. doi: 10.1038/ncb1201-1086. [DOI] [PubMed] [Google Scholar]
- 19.Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paiement J, Beaufay H, Godelaine D. Coalescence of microsomal vesicles from rat liver: a phenomenon occurring in parallel with enhancement of the glycosylation activity during incubation of stripped rough microsomes with GTP. J Cell Biol. 1980;86:29–37. doi: 10.1083/jcb.86.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sokoloff AV, Whalley T, Zimmerberg J. Characterization of N-ethylmaleimide-sensitive thiol groups required for the GRP-dependent fusion of endplasmic reticulum membranes. Biochem J. 1995;312:23–30. doi: 10.1042/bj3120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreier L, Rapoport TA. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J Cell Biol. 2000;148:883–898. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farsad K, De Camilli P. Mechanisms of membrane deformation. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 24••.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. This review, together with [23], summarizes current understanding of the induction of membrane curvature. Five different membrane-bending mechanisms are discussed. The second one (influence of integral membrane proteins) has been implicated in ER tubule formation [11••] [DOI] [PubMed] [Google Scholar]
- 25.Dabora SL, Sheetz MP. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cell extracts. Cell. 1988;54:27–35. doi: 10.1016/0092-8674(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 26.Terasaki M. Recent progress on structural interactions of the endoplasmic reticulum. Cell Motil Cytoskeleton. 1990;15:71–75. doi: 10.1002/cm.970150203. [DOI] [PubMed] [Google Scholar]
- 27.Klopfenstein DR, Klumperman J, Lustig A, Kammerer RA, Oorschot V, Hauri HP. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal α-helical segment. J Cell Biol. 2001;153:1287–1300. doi: 10.1083/jcb.153.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. Embo J. 1998;17:6168–6177. doi: 10.1093/emboj/17.21.6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Vedrenne C, Klopfenstein DR, Hauri HP. Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol Biol Cell. 2005;16:1928–1937. doi: 10.1091/mbc.E04-07-0554. Together with [31•], this article illustrates regulatory mechanisms governing the interaction between the ER and the microtubule cytoskeleton. An integral ER membrane protein's ability to associate with microtubules is shown here to be regulated by phosphorylation and to be cell-cycle-dependent. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farah CA, Liazoghli D, Perreault S, Desjardins M, Guimont A, Anton A, Lauzon M, Kreibich G, Paiement J, Leclerc N. Interaction of microtubule-associated protein-2 and p63: a new link between microtubules and rough endoplasmic reticulum membranes in neurons. J Biol Chem. 2005;280:9439–9449. doi: 10.1074/jbc.M412304200. [DOI] [PubMed] [Google Scholar]
- 31•.Amarilio R, Ramachandran S, Sabanay H, Lev S. Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J Biol Chem. 2005;280:5934–5944. doi: 10.1074/jbc.M409566200. VAPs are integral ER membrane proteins that recruit peripheral proteins, many of which have lipid binding domains. This study reports that, in addition to mediating lipid exchange between the ER and other organelles, VAPs and their partners may have a role in determining ER structure (see also [36••] [DOI] [PubMed] [Google Scholar]
- 32••.Andrade J, Zhao H, Titus B, Timm Pearce S, Barroso M. The EF-hand Ca2+-binding protein p22 plays a role in microtubule and endoplasmic reticulum organization and dynamics with distinct Ca2+-binding requirements. Mol Biol Cell. 2004;15:481–496. doi: 10.1091/mbc.E03-07-0500. Together with [29•], this article illustrates regulatory mechanisms governing the interaction between the ER and the microtubule cytoskeleton. Here, a peripheral membrane protein that modulates interactions between the ER and microtubules is identified. Changes in the calcium-modulated activity of this protein change both microtubule organization and the density of branching ER networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three familes of lipid binding proteins and in Opi1p binds VAP. Embo J. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loewen CJ, Levine TP. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem. 2005;280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- 36••.Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. VAPs bind peripheral proteins carrying a FFAT motif (two phenylalanines in an acidic tract) [35] that are implicated in lipid exchange between the ER and other organelles. In this study, the crystal structure of rat VAP-A MSP homology domain, alone and in complex with a FFAT motif, is solved. On the basis of the structure, the authors produce a mutant defective in FFAT binding and investigate its effects on ER structure. [DOI] [PubMed] [Google Scholar]
- 37.Lev S. The role of the Nir/rdgB protein family in membrane trafficking and cytoskeleton remodeling. Exp Cell Res. 2004;297:1–10. doi: 10.1016/j.yexcr.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 38.Nishimura AL, Mitne-Neto M, Silva HCA, Richieri-Costa A, Middleton S, Cascio D, Kok F, Oliveira JRM, Gillingwater T, Webb J, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura AL, Al-Chalabi A, Zatz M. A common founder for amyotrophic lateral sclerosis type 8 (ALS8) in the Brazilian population. Hum Genet. 2005;118:499–500. doi: 10.1007/s00439-005-0031-y. [DOI] [PubMed] [Google Scholar]
- 40.Koster G, VanDuijn M, Hofs B, Dogterom M. Membrane tube formation from giant vesicles by dynamic association of motor proteins. Proc Natl Acad Sci USA. 2003;100:15583–15588. doi: 10.1073/pnas.2531786100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leduc C, Campas O, Zeldovich KB, Roux A, Jolimaitre P, Bourel-Bonnet L, Goud B, Joanny JF, Bassereau P, Prost J. Cooperative extraction of membrane nanotubes by molecular motors. Proc Natl Acad Sci USA. 2004;101:17096–17101. doi: 10.1073/pnas.0406598101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oertle T, Schwab ME. Nogo and its paRTNers. Trends Cell Biol. 2003;13:187–194. doi: 10.1016/s0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 43.Parton RG, Richards AA. Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic. 2003;4:724–738. doi: 10.1034/j.1600-0854.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- 44•.Estrada de Martin P, Du Y, Novick P, Ferro-Novick S. Ice2p is important for the distribution and structure of the cortical ER network in Saccharomyces cerevisiae. J Cell Sci. 2004;118:65–77. doi: 10.1242/jcs.01583. By random mutagenesis of S. cerevisiae and visual screening, the authors identify a novel protein, predicted to span the membrane seven times, that is required to maintain the normal structure of the cortical tubular ER network and for its distribution in mother and daughter cells. [DOI] [PubMed] [Google Scholar]
- 45.Fawcett DW. The Cell. 2nd. Philadelphia: W.B. Saunders Company; 1981. [Google Scholar]
- 46••.Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. Several distinct OSER structures are induced by a variety of different resident ER membrane proteins that share a single common feature, a cytoplasmic domain capable of low affinity homotypic interactions. The OSER-inducing proteins retain full diffusional mobility within the membrane of the tightly packed cisternae. This mechanism is potentially relevant to the formation of other stacked membrane organelles [55] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Landh T. From entangled membranes to eclectic morphologies: cubic membranes as subcellular space organizers. FEBS Lett. 1995;369:13–17. doi: 10.1016/0014-5793(95)00660-2. [DOI] [PubMed] [Google Scholar]
- 48.Hyde S, Andersson S, Larsson K, Blum Z, Landh T, Lidin S, Ninham BW. The Language of Shape - The role of curvature in condensed matter: physics, chemistry and biology. Amsterdam: Elsevier; 1997. [Google Scholar]
- 49.Takei K, Mignery GA, Mugnaini E, Sudhof TC, De Camilli P. Inositol 1,4,5-trisphosphate receptor causes formation of ER cisternal stacks in transfected fibroblasts and in cerebellar Purkinje cells. Neuron. 1994;12:327–342. doi: 10.1016/0896-6273(94)90275-5. [DOI] [PubMed] [Google Scholar]
- 50.Gong FC, Giddings TH, Meehl JB, Staehelin LA, Galbraith DW. Z-membranes: artificial organelles for overexpressing recombinant integral membrane proteins. Proc Natl Acad Sci U S A. 1996;93:2219–2223. doi: 10.1073/pnas.93.5.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto A, Masaki R, Tashiro Y. Formation of crystalloid endoplasmic reticulum in COS cells upon overexpression of microsomal aldehyde dehydrogenase by cDNA transfection. J Cell Sci. 1996;109:1727–1738. doi: 10.1242/jcs.109.7.1727. [DOI] [PubMed] [Google Scholar]
- 52.Profant DA, Roberts CJ, Koning AJ, Wright RL. The role of the 3-hydroxy 3-methylglutaryl coenzyme A reductase cytosolic domain in karmellae biogenesis. Mol Biol Cell. 1999;10:3409–3423. doi: 10.1091/mbc.10.10.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukuda M, Yamamoto A, Mikoshiba K. Formation of crystalloid endoplasmic reticulum induced by expression of synaptotagmin lacking the conserved WHXL motif in the C terminus. J Biol Chem. 2001;276:41112–41119. doi: 10.1074/jbc.M106209200. [DOI] [PubMed] [Google Scholar]
- 54.Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;269:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 55.Wang Y, Seemann J, Pypaert M, Shorter J, Warren G. A direct role for GRASP65 as a mitotically regulated Golgi stacking factor. Embo J. 2003;22:3279–3290. doi: 10.1093/emboj/cdg317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kock N, Naismith TV, Boston HE, Ozelius LJ, Corey DP, Breakefield XO, Hanson PI. Effects of genetic variations in the dystonia protein torsinA: identification of polymorphism at residue 216 as protein modifier. Hum Mol Genet. 2006;15:1355–1364. doi: 10.1093/hmg/ddl055. [DOI] [PubMed] [Google Scholar]
- 57.Okiyoneda T, Harada K, Takeya M, Yamahira K, Wada I, Shuto T, Suico MA, Hashimoto Y, Kai H. Delta F508 CFTR pool in the endoplasmic reticulum is increased by calnexin overexpression. Mol Biol Cell. 2004;15:563–574. doi: 10.1091/mbc.E03-06-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dickson KM, Bergeron JJM, Shames I, Colby J, Nguyen DT, Chevet E, Thomas DY, Snipes GJ. Association of calnexin with mutant peripheral myelin protein-22 ex vivo: a basis for ‘gain-of-function’ ER diseases. Proc Natl Acad Sci USA. 2002;99:9852–9857. doi: 10.1073/pnas.152621799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fontanini A, Chies R, Snapp EL, Ferrarini M, Fabrizi GM, Brancolini C. Glycan-independent role of calnexin in the intracellular retention of Charcot-Marie-Tooth 1A Gas3/PMP22 mutants. J Biol Chem. 2005;280:2378–2387. doi: 10.1074/jbc.M405104200. [DOI] [PubMed] [Google Scholar]
- 60.Federovitch CM, Ron D, Hampton RY. The dynamic ER: experimental approaches and current questions. Curr Opin Cell Biol. 2005;17:409–414. doi: 10.1016/j.ceb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 61•.Sprocati T, Ronchi P, Raimondi A, Francolini M, Borgese N. Dynamic and reversible restructuring of the endoplasmic reticulum induced by PDMP in cultured cells. J Cell Sci. 2006 doi: 10.1242/jcs.03058. in press. doi:03058. The authors show that the drug 1-Phenyl-2-decanoyl-amino-3-morpho-lino-1-propanol hydrochloride (PDMP) can rapidly and reversibly induce the formation of SER patches in cultured cells. The study provides an interesting model system in which to study ER restructuring events in the absence of membrane proliferation. [DOI] [PubMed] [Google Scholar]
- 62.Pierini L, Holowka D, Baird B. FcεRI-mediated association of 6-micron beads with RBL-2H3 mast cells results in exclusion of signaling proteins from the forming phagosome and abrogation of normal downstream signaling. J Cell Biol. 1996;134:1427–1439. doi: 10.1083/jcb.134.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson RG, Orci L, Brown MS, Garcia-Segura LM, Goldstein JL. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J Cell Sci. 1983;63:1–20. doi: 10.1242/jcs.63.1.1. [DOI] [PubMed] [Google Scholar]


