Summary
Gene activation by the CRISPR/Cas9 system has the potential to enable new approaches to science and medicine, but the technology must be enhanced to robustly control cell behavior. We show that the fusion of two transactivation domains to Cas9 dramatically enhances gene activation to a level that is necessary to reprogram cell phenotype. Targeted activation of the endogenous Myod1 gene locus with this system led to stable and sustained reprogramming of mouse embryonic fibroblasts into skeletal myocytes. The levels of myogenic marker expression obtained by the activation of endogenous Myod1 gene were comparable to that achieved by overexpression of lentivirally delivered MYOD1 transcription factor.
Graphical Abstract
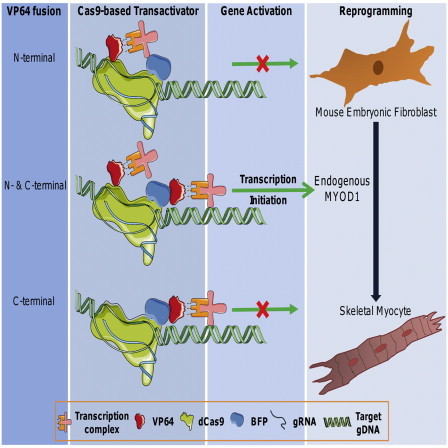
Highlights
-
•
RNA-guided VP64dCas9-BFPVP64 fusion protein robustly activates endogenous Myod1
-
•
Transactivated Myod1 can reprogram mouse embryonic fibroblasts to skeletal myocytes
-
•
VP64 fusion to both the N and C terminus of dCas9-BFP facilitates reprogramming
-
•
Myogenic gene expression is comparable to MYOD1 overexpression-based reprogramming
In this article, Leong and colleagues show skeletal reprogramming of mouse fibroblasts by RNA-guided VP64dCas9-BFPVP64-based transactivation of endogenous Myod1. Dual fusion of VP64 transactivation domains to Cas9 is critical for activating the Myod1 gene to a level necessary for the direct reprogramming. The resultant myogenic gene expression levels are comparable to that achieved by overexpression of MYOD1 transgene.
Introduction
The type II clustered regularly interspaced short palindromic repeat (CRISPR) systems and the associated Cas9 nucleases have evolved in archaea and bacteria for sequence-specific recognition of DNA targets via a single-stranded RNA intermediate (Jinek et al., 2012). In an engineered version of the CRISPR system, the Streptococcus pyogenes Cas9 nuclease is directed by guide RNAs (gRNAs) targeting 20 bp sequences adjacent to a 5′-NRG-3′ sequence motif, and the resultant cleavage has been used to edit the genome in several species (Cho et al., 2013; Cong et al., 2013; Hwang et al., 2013; Jinek et al., 2013; Mali et al., 2013b). A mutated nuclease-inactive Cas9 (dCas9) regulates gene expression by physically blocking transcription or through fusion to a transactivator (VP64, Ω subunit of RNA polymerase) or repressor domain (KRAB, SID) (Bikard et al., 2013; Cheng et al., 2013; Farzadfard et al., 2013; Gilbert et al., 2013; Kearns et al., 2014; Konermann et al., 2013; Maeder et al., 2013; Mali et al., 2013a; Perez-Pinera et al., 2013; Qi et al., 2013). While transgene overexpression has been used to achieve cellular reprogramming (Davis et al., 1987; Takahashi and Yamanaka, 2006), reprogramming via direct activation of an endogenous gene has only been recently demonstrated through the use of transcription activator-like effectors (TALEs) (Gao et al., 2013). However, difficulty in designing and codelivering multiple TALE expression constructs precludes simple screening and multiplexed gene activation that is straightforward with the dCas9-VP64 system. In this study, we used dCas9-based transactivators combined with an efficient lentivirus-based gene delivery system to induce cellular reprogramming.
Results and Discussion
We tested the efficacy of a VP64dCas9-BFPVP64 fusion protein to activate expression of the endogenous Myod1 gene locus for a sufficient duration and magnitude to ultimately induce the reprogramming of mouse embryonic fibroblasts (MEFs) to skeletal myocytes (SkMs) (Figure 1A). Although we and others have previously used the single C-terminal fusion of VP64 to dCas9 to activate gene expression (Cheng et al., 2013; Farzadfard et al., 2013; Gilbert et al., 2013; Kabadi et al., 2014; Maeder et al., 2013; Perez-Pinera et al., 2013), preliminary studies indicated that this approach did not lead to levels of expression sufficient for cell reprogramming. Therefore, we tested whether two VP64 domains flanking dCas9 (VP64dCas9-BFPVP64) would yield higher Myod1 gene expression levels (Figures 1B and 1C and Figure S1A available online). dCas9 was also fused to blue fluorescent protein (BFP) to monitor expression (Figures 1B, 1C, S1A, and S1B). Lentiviral VP64dCas9-BFPVP64 was placed under the transcriptional control of a doxycycline-inducible promoter (Figures 1C and S1B). To avoid steric hindrance that may prevent transcriptional complex recruitment, we included flexible glycine-serine linkers adjacent to the VP64 domains (Figure S1A). We also added a third nuclear localization signal (NLS), which improved the nuclear localization of VP64dCas9-BFPVP64 by ∼10-fold (Figures 1C–1F). Initially, we transfected C3H10T1/2 cells with the VP64dCas9-BFPVP64 plasmid. However, transfection was inefficient as transgene expression was detectable only in a few cells, presumably due to the large size of the plasmid (13.5 kb). We then used a lentiviral gene delivery system, allowing stable transduction of more cells as evidenced by both BFP and immunofluorescence staining of the FLAG epitope (Figure 1F). Approximately 50% of the transduced cells were found to be BFP positive as compared with 5% by transfection (Figure 1G). To ensure efficient gRNA codelivery, we developed a lentivirus-based U6 promoter-driven gRNA delivery system (Figure 1C). In separate experiments, we observed an ∼6-fold upregulation of Myod1 mRNA levels when all components were delivered by transfection (Figure S1C) compared with ∼60-fold by lentiviral transduction (Figure 1H).
Figure 1.
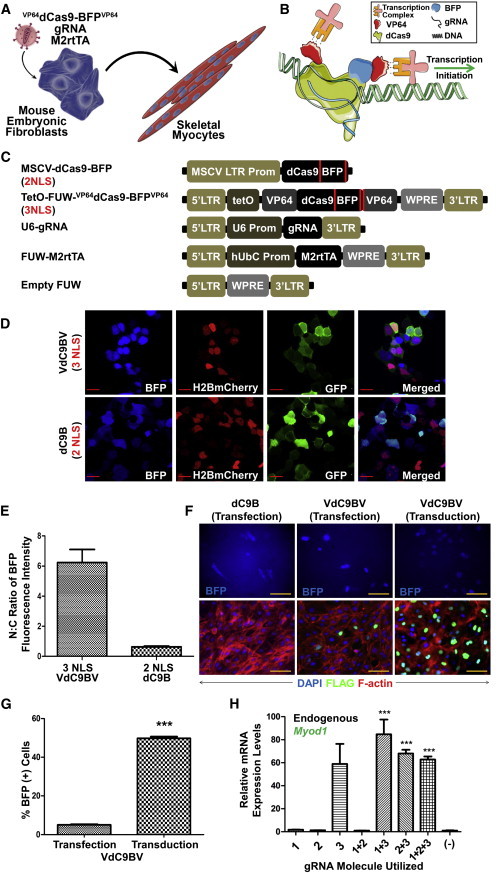
Engineering an RNA-Guided, Nuclease-Inactive VP64dCas9-BFPVP64—VdC9BV—Fusion Protein to Enable Robust Transactivation of the Endogenous Myod1 Gene
(A) Illustration depicting the reprogramming of MEFs to SkMs initiated by the lentiviral transduction of MEFs with the gRNA, reverse tetracycline transactivator (M2rtTA), and a doxycycline-inducible VdC9BV.
(B) Illustration of gRNA-guided transactivation by the VdC9BV fusion protein.
(C) The lentiviral constructs utilized to activate endogenous genes.
(D) Confocal images of live HEK293T cells transfected with VdC9BV or dC9B, H2BmCherry fluorescent protein (nuclear-localized), and GFP (predominantly cytoplasmic) showing that inclusion of an additional NLS in VdC9BV (3 NLS) enables better nuclear localization than the 2 NLS dCas9-BFP (dC9B) parent construct. The scale bar represents 20 μm.
(E) Nucleus-to-cytoplasm (N:C) ratio of BFP fluorescence intensity quantified from confocal images of live transfected HEK293T cells. The analysis shows that the 3 NLS form of dCas9 (VdC9BV) localizes better to the nucleus than the original 2 NLS dCas9 (dC9B) form (n = 20 cells, error bars represent SEM).
(F) Lentiviral vector enables robust delivery of nuclear-localized VdC9BV. dC9B (2 NLS) transfected into C3H10T1/2 cells shows lack of nuclear localization. VdC9BV (3 NLS) enables better nuclear localization. However, few cells expressed VdC9BV when transfected into the cells. Transduction of C3H10T1/2 with the lentiviral form of VdC9BV enables higher efficiency of delivery of the construct, as evidenced by BFP fluorescence imaging of live cells and immunofluorescence staining of the FLAG epitope in fixed cells. The scale bar represents 100 μm.
(G) Lentiviral delivery of VdC9BV is more efficient than delivery by transfection. By flow cytometry analysis, ∼50% of the transduced cells were found to be BFP positive compared with ∼5% by transfection (two-tailed unpaired t test, p ≤ 0.0001, n = 3 biological replicates; error bars represent SEM).
(H) qRT-PCR evaluation of relative Myod1 mRNA expression in C3H10T1/2 on day 6 postinduction of VdC9BV expression in the presence of various combinations of three gRNA molecules (p ≤ 0.001, one-way ANOVA, Dunnett’s post hoc comparing all the groups to the [−] gRNA control group [M2rtTA + VdC9BV + Empty FUW], n = 3 biological replicates, error bars represent SEM).
We initially tested this system’s capacity to induce epigenetic reprogramming toward the SkM lineage in C3H10T1/2 cells previously shown to readily undergo this transformation (Davis et al., 1987). We designed three separate gRNAs (1–3) targeted to different positions proximal to the Myod1 transcription start site and initially codelivered all three gRNAs (Figure S1D). We found that VP64 fusion and the presence of the gRNA were essential for endogenous Myod1 gene activation (Figure S1C). Activation of the endogenous Myod1 gene in C3H10T1/2 was sufficient to initiate the SkM reprogramming process, as determined by gene expression analysis, phase contrast imaging, and immunocytochemistry (Figures 2A–2C and S1B). Myotubes were visible as early as 4.5 days after transduction. Nuclear-localized blue fluorescence was observed in the myotubes during the time VP64dCas9-BFPVP64 was kept induced by doxycycline addition (Figure S1B). Importantly, the myotubes stained positive for the skeletal transcription factors (TFs) MYOD1 and MYOG, and the sarcomeric proteins actinin, desmin, myosin heavy chain, and titin (Figures 2C and S2A). The myotubes were also multinucleated, indicating cell fusion, one of the hallmarks of myogenic reprogramming (Figure 2C). When examining the effect of individual gRNA molecules in inducing Myod1 mRNA expression, we determined that gRNA3 alone was as potent as all three gRNAs combined (Figure 1H). We speculate that factors, including distance from transcription initiation site, low binding affinity, and binding site competition with endogenous TFs, are the possible causes of gRNA1 and gRNA2 inactivity. C3H10T1/2 cells fused in the 8 days that Myod1 was expressed under doxycycline-induced VP64dCas9-BFPVP64 expression, as demonstrated quantitatively by a recombination-based cell fusion assay. The fusion process continued unabated even after the withdrawal of doxycycline, thereby indicating stability of this phenotypic transformation (Figure 2D).
Figure 2.
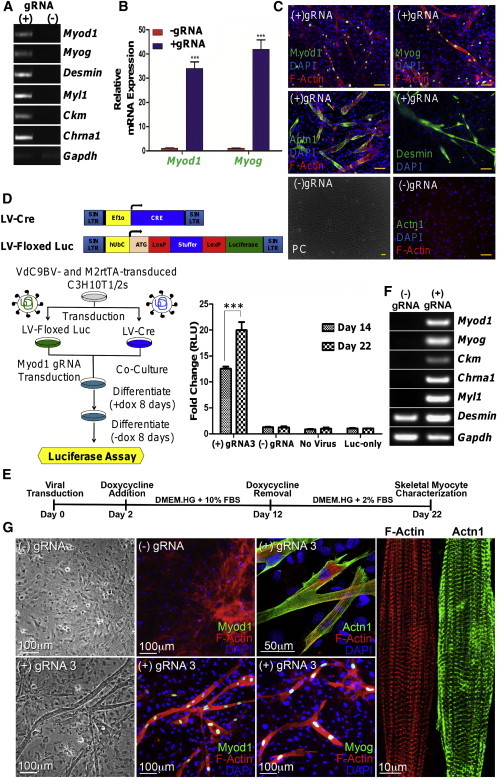
CRISPR/VP64dCas9-BFPVP64 Transactivates the Endogenous Myod1 Gene and Reprograms C3H10T1/2 Cells and MEFs into SkMs
(A) VP64dCas9-BFPVP64 (VdC9BV), in the presence of three gRNAs targeting the Myod1 locus, results in upregulation of major SkM genes in C3H10T1/2 cells detected by RT-PCR on day 18 posttransduction.
(B) Reprogramming of C3H10T1/2 cells by VdC9BV is characterized by significantly higher levels of Myod1 and Myog mRNA detected by qRT-PCR on day 18 posttransduction (two-way ANOVA, Bonferroni post tests, p ≤ 0.001 for both Myod1 and Myog, n = 3 biological replicates, error bars represent SEM). Fold change in expression is relative to the (−) gRNA control group.
(C) Immunocytochemistry (ICC) of reprogrammed C3H10T1/2 cells for SkM markers. The multinucleated myotubes stained for nuclear MYOD1 and MYOG proteins and sarcomeric proteins desmin and ACTN1 (α-actinin). Reprogrammed myotubes were absent in the control group (phase contrast [PC] imaging and ICC). The scale bar represents 100 μm.
(D) Cell fusion assay. Cell fusion in the reprogramming cells was quantified by detecting the luciferase released after mixing two lentivirally transduced cell populations: one expressing Cre recombinase (LV-Cre) and a second containing a Cre recombinase responsive Floxed-Stuffer Luciferase cassette (LV-Floxed Luc). Fold change in relative light units (RLUs) was calculated relative to a Luc-only group (VdC9BV, M2rtTA, and LV-Floxed Luc). The RLU fold change of the (+) gRNA3 group is significantly higher on day 22 than on day 14 (two-way repeated measures [RM] ANOVA, Bonferroni post hoc test, p ≤ 0.001, n = 3 biological replicates, error bars = SEM). Overall, the (+) gRNA3 group has significantly higher luciferase activity than other groups (two-way RM ANOVA, p ≤ 0.0001).
(E) Timeline denoting the main steps of the MEF reprogramming process.
(F) Gene expression analysis (RT-PCR) performed on reprogrammed and control MEFs.
(G) Imaging of the reprogrammed MEFs shows the presence of multinucleated myotubes containing SkM markers. No myogenesis was observed in the (−) control group. Confocal imaging shows cross-striated organization of ACTN1 and F-actin.
We tested whether coexpression of a single targeting gRNA molecule (gRNA3) along with VP64dCas9-BFPVP64 would be sufficient to induce reprogramming of primary MEFs (Figure 2E). Using gene expression analysis, we readily detected transcriptional activation of the mature myotube markers Myl1, Ckm, Desmin, and Chrna1 (Figure 2F). We detected formation of multinucleated myotubes expressing skeletal TFs and striated sarcomeres indicating a high degree of cytoskeletal organization and maturity (Figures 2G and S2B). Within 3 weeks of cell transduction, we observed myotubes exhibiting spontaneous intermittent twitching (Movie S1).
To test whether transient expression of VP64dCas9-BFPVP64 is sufficient to induce sustained expression of Myod1, we induced transgene expression between days 2 and 10 posttransduction. We detected an initial robust upregulation of VP64dCas9-BFPVP64 expression followed by a gradual downregulation suggesting transgene silencing in the reprogramming cells (Figure 3A). Doxycycline removal coincided with rapid VP64dCas9-BFPVP64 decline to levels measured prior to initial induction. VP64dCas9-BFPVP64 expression also coincided with an increase in Myod1 expression. Importantly, Myod1 mRNA levels remained elevated for the duration of the 18-day experiment even after doxycycline removal (day 10), suggesting that maintenance of endogenous activation of Myod1 is stable and independent of VP64dCas9-BFPVP64 activity.
Figure 3.
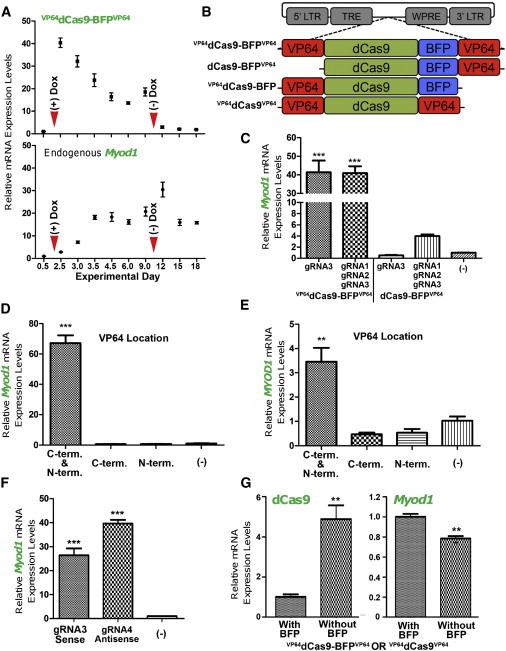
Characteristics of gRNA-Guided VP64dCas9-BFPVP64-Mediated Activation of the Endogenous Myod1 Gene Locus
(A) VdC9BV-mediated activation of the endogenous murine Myod1 gene is stable and sustained following doxycycline removal. The red arrowheads indicate the time points on which addition and removal of doxycycline are performed. dCas9 expression kinetics is significantly different from that of transactivated Myod1 (p ≤ 0.0001, two-way ANOVA). Fold change in expression is relative to levels at day 0.5 posttransduction (n = 4 biological replicates, error bars represent SEM).
(B) Illustration of dCas9-based activators.
(C) The C-terminal-only fusion of VP64 to dCas9, dCas9-BFPVP64, fails to activate the endogenous murine Myod1 gene in C3H10T1/2 cells even in the presence of multiple gRNAs. VdC9BV significantly activated transcription of the endogenous Myod1 gene locus when compared with all other groups (p ≤ 0.001, one-way ANOVA, Tukey’s post hoc test, n = 3). Control group: M2rtTA + Empty FUW. Fold change in expression is relative to the (−) control group.
(D) The N-terminal-only VP64 fusion to dCas9 (VP64dCas9-BFP) also failed to match the ability of VdC9BV to transactivate significant levels of endogenous Myod1 expression in the presence of a single gRNA (p ≤ 0.001, one-way ANOVA, Dunnett’s post hoc test, n = 3). Control: M2rtTA +Empty FUW. Fold change in expression is relative to the (−) control group.
(E) Only the N- and C-terminal VP64 fusion to dCas9 (VdC9BV) significantly activated the human MYOD1 locus of HEK293T cells in the presence of a gRNA (p ≤ 0.01, one-way ANOVA, Dunnett’s post hoc test, n = 3).
(F) A gRNA targeting the antisense strand (gRNA4) can significantly activate endogenous Myod1 expression to levels similar to that achieved by the gRNA targeting the sense strand (gRNA3) (p ≤ 0.001, one-way ANOVA, Dunnett’s post hoc comparing all the groups to the [−] gRNA group, n = 3). (−) gRNA group: M2rtTA + VdC9BV + Empty FUW virus. Fold change in expression is relative to the (−) control group.
(G) In the absence of the BFP domain, VP64dCas9VP64 activates Myod1 expression to a lesser extent than the VdC9BV group, although dCas9 is expressed at a significantly higher level using the same MOI (C3H10T1/2 cells, unpaired two-tailed t test p = 0.0051 [dCas9], p = 0.0044 [Myod1], n = 3). Fold change in expression is relative to the VdC9BV group. Measurements for (C–G) occurred at posttransduction day 6, and the data from independent biological replicates are represented as mean ± SEM.
dCas9 with VP64 fused to its C terminus (dCas9-BFPVP64) failed to activate the Myod1 locus to levels sufficient to initiate cellular reprogramming even in the presence of three gRNAs (Figures 3B, 3C, and S3A). However, in the presence of three gRNAs, the mean fold-activation level by dCas9-BFPVP64 was higher than with a single gRNA (Figure 3C), consistent with recent reports of synergistic effects of multiple gRNAs with dCas9 and a single VP64 fusion (Cheng et al., 2013; Maeder et al., 2013; Mali et al., 2013a; Perez-Pinera et al., 2013). The N-terminal-only VP64 fusion protein (VP64dCas9-BFP) also failed to match the ability of VP64dCas9-BFPVP64 to activate sufficient endogenous Myod1 for reprogramming in the presence of a single gRNA (Figures 3B, 3D, and S3A). Interestingly, VP64dCas9-BFPVP64 was able to activate the endogenous human MYOD1 locus significantly in the presence of a single gRNA, whereas the N-terminal-only VP64 fusion protein (VP64dCas9-BFP) and the C-terminal-only VP64 fusion protein (dCas9-BFPVP64) failed to do so (Figure 3E). These observations suggest that the inclusion of both VP64 activator domains fused at the two termini of dCas9-BFP significantly improves the activation capacity of this targeting platform. However, the endogenous human MYOD1 transactivation was an order of magnitude lower than that observed in the murine system. Transactivation of higher levels of MYOD1 mRNA may require extensive optimization of the gRNA target sequences in the future.
To test whether the direction of the genomic strand targeted by the gRNA molecule was a factor in its capacity to recruit dCas9 and activate expression of the endogenous locus, we designed a gRNA molecule (gRNA4) targeting the minus strand of the same region for which gRNA3 was designed (PAM separated by 14 bases) (Figure S1D). Interestingly, gRNA4 was also able to activate Myod1 expression and initiate reprogramming, indicating the system is insensitive to the target strand (Figures 3F and S3B).
We also evaluated the effect of BFP fusion on the activity of VP64dCas9-BFPVP64. Following transduction at equivalent multiplicity of infection (MOIs), the construct without BFP showed statistically higher levels of dCas9 expression, although it was less efficacious than the BFP-fused form in inducing expression of Myod1 (Figures 3B and 3G). However, both forms led to reprogramming (Figure S3C). The omission of BFP from the C-terminal-only VP64 fusion protein (dCas9VP64) failed to impart ability to activate the endogenous Myod1 locus (Figure S3D). We speculate that the positive effect of BFP on Myod1 expression may be due to increased spacing between the two VP64 domains or increased flexibility of the domains.
We also compared the VP64dCas9-BFPVP64-mediated activation of endogenous Myod1 gene to transgenic MYOD1 overexpression for its ability to reprogram cells. C3H10T1/2 cells were transduced with VP64dCas9-BFPVP64 and M2rtTA. The resultant BFP-positive cells were sorted for BFP expression and the BFP+ cells were transduced with either gRNA or doxycycline-inducible transgenic human MYOD1 at equivalent MOIs (MOI = 10) (Figures 4A and 4B). During the induction phase of the reprogramming process, endogenous Myod1 expression over the entire duration was significantly higher in the VP64dCas9-BFPVP64/gRNA-mediated activation group than the transgenic MYOD1 overexpression group. Expression kinetics of Myog and Desmin were similar in both the groups (Figure 4C).
Figure 4.
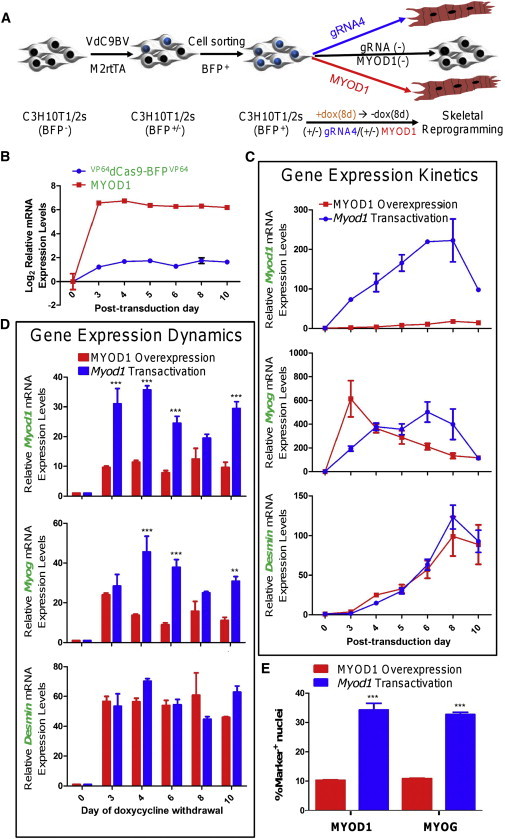
Comparison of Skeletal Reprogramming by gRNA-Guided VP64dCas9-BFPVP64-Mediated Transactivation of the Endogenous Myod1 Gene Locus and by Transgenic MYOD1 Overexpression
(A) Illustration depicting the experimental setup for the comparison of skeletal reprogramming by Myod1 transactivation and by transgenic MYOD1 overexpression.
(B) Expression kinetics of VdC9BV and MYOD1 transgenes under doxycycline induction from days 2 to 10 posttransduction.
(C) Expression kinetics of myogenic genes (Myod1, Myog, and Desmin) in C3H10T1/2 cells detected by qRT-PCR during the 8 days of doxycycline induction of both the systems. The endogenous Myod1 expression over the entire duration of induction is significantly higher in the Myod1 transactivation group than the transgenic MYOD1 overexpression group (p < 0.0001). Expression of Myog (p = 0.3740) and Desmin (p = 0.6588) is similar in both the groups (two-way ANOVA, n = 3). Fold change in expression is relative to the (−) gRNA (−) MYOD1 transgene group.
(D) The effect of duration of reprogramming system induction by doxycycline on myogenic marker expression dynamics in C3H10T1/2 cells detected by qRT-PCR on day 18 posttransduction. The endogenous Myod1 and Myog expression is significantly higher in the Myod1 transactivation group than the transgenic MYOD1 overexpression group (p < 0.0001). Expression of Desmin (p = 0.5348) is similar in both the groups (two-way ANOVA, Bonferroni post tests, n = 3). Fold change in expression is relative to the (−) gRNA (−) MYOD1 transgene control group.
(E) Percentage of 4',6-diamidino-2-phenylindole-stained nuclei that also express MYOD1/ MYOG protein in C3H10T1/2 cells on day 18 posttransduction is higher in the Myod1 transactivation group (mean = ∼34% MYOD1+, 33% MYOG+) than the MYOD1 overexpression group (mean = ∼10% MYOD1+, 11% MYOG+) (p = 0.0004 [MYOD1], <0.0001 [MYOG], two-tailed unpaired t test, n = 3 biological replicates). Doxycycline induction of VdC9BV and MYOD1 transgenes was carried out for 8 days till day 10 posttransduction.
Data from independent biological replicates for (B)–(E) are represented as mean ± SEM.
The effects of limiting the activation of the VP64dCas9-BFPVP64/gRNA system (by varying the duration of doxycycline exposure) on myogenic gene expression were compared with the MYOD1 overexpression system. Results showed that even a single day of doxycycline exposure (from day 2 to day 3 posttransduction) was adequate to activate the downstream myogenic genes (Myog and Desmin) to similar levels in both the groups and similar to levels achieved after 8 days of induction (Figure 4D). Almost all the markers on day 18 posttransduction after a maximum 8 days of induction had similar expression levels in both the groups (Figures 4D and S4A). However, both Myod1 expression and the percentage of MYOD1 TF+ nuclei were approximately 3-fold higher in the Myod1 transactivation group than the MYOD1 overexpression group (Figures 4D, 4E, and S4B). It indicates that the higher expression of Myod1 on quantitative RT-PCR (qRT-PCR) was probably a result of more MYOD1 TF+ cells rather than higher expression levels in individual cells. Myog expression in C3H10T1/2 followed a pattern similar to Myod1 (Figures 4D and 4E). Similar differential Myod1 expression was also observed in the reprogrammed MEFs. However, expression of all the other myogenic markers and the percentage of MYOG+ nuclei were similar in both groups (Figures S4C and S4D). It may be speculated that VP64dCas9-BFPVP64/gRNA action renders the endogenous Myod1 locus more receptive to MYOD1 TF-positive feedback response (Zingg et al., 1994). However, other cellular factors needed in conjunction with MYOD1 TF to initiate skeletal reprogramming may not be present in adequate quantities, thereby explaining the lack of a significant difference for other downstream late myogenic genes.
This study shows that the dual fusion of the VP64 transactivation domain to both the N and C terminus of dCas9 enables a high level of endogenous Myod1 activation for the direct conversion of primary murine fibroblasts into SkMs. This potentiation of the transactivation process can be explained by an increased probability of the TFs homing onto two VP64 domains compared with just one VP64 domain. As a result, transcription can be initiated more frequently in the presence of VP64dCas9-BFPVP64. Moreover, a synergistic transactivation effect resulting from favorable interactions of the TF complexes assembled on both the terminus of VP64dCas9-BFPVP64 can also help in explaining its potency. Improved nuclear localization of dCas9 afforded by increasing the number of NLS sequences, efficient lentiviral transgene delivery, inclusion of a BFP spacer sequence to decrease steric hindrance, and the identification of an efficient gRNA together complemented the effect of an additional VP64 domain in VP64dCas9-BFPVP64 to efficiently transactivate the endogenous Myod1 locus. The results also demonstrate that CRISPR/Cas9-based transactivation performs comparably to the traditional MYOD1 overexpression-based skeletal reprogramming in upregulating some major myogenic genes. It augurs well for the potential use of this tool in reprogramming protocols that require complex and multiple TF activation.
In conclusion, we expect that this VP64dCas9-BFPVP64 platform, which is significantly more potent than previous versions, will encourage the adoption of CRISRP/Cas9-based TF technology to achieve multiplexed gene activation for reprogramming as well as nonreprogramming applications in basic science, biotechnology, and medicine.
Experimental Procedures
gRNA Design
The gRNAs were designed by utilizing UCSC genome browser tracks (Feng Zhang Lab, MIT). The mouse Myod1 locus-specific gRNAs 1–3 were selected to represent specific locations at a decreasing distance from the designated Myod1 transcription initiation site, respectively. gRNA4 was selected to target the genome in the immediate vicinity of gRNA3, albeit on the opposite strand (Figure S1D). The human gRNA was picked to target the genome within 200 bases upstream of the MYOD1 transcription initiation site.
VP64dCas9-BFPVP64 Plasmid Construction
pdCas9::BFP-humanized plasmid (Addgene plasmid 44247) was used as the source of the dCas9-BFP fusion cassette. gBlock DNA fragments (Integrated DNA Technologies) were utilized to generate the VP64dCas9-BFPVP64 construct from dCas9-BFP and subsequently cloned into a lentiviral vector.
Cell Culture, Transfection, and Viral Transduction
All of the cell types used in this study were initially cultured with a high serum media (10% fetal bovine serum in Dulbecco’s modified Eagle’s medium-high glucose [DMEM-HG]). The cells were seeded at a density of 25,000 per well of a 12-well plate for transfection or transduction with components of the reprogramming system. Liposome-based transfection and lentiviral transduction were done while maintaining a constant dosage among the tests and controls. Low serum medium (2% horse serum in DMEM-HG) was used to induce skeletal differentiation.
Cell Staining and qRT-PCR
Four percent paraformaldehyde fixed cells were permeabilized with 0.2% Triton X-100 and subsequently stained with primary antibodies and fluorescent-labeled secondary antibodies. Relative quantification of gene expression (delta-delta Ct method) was performed on the RT-PCR data obtained by using SYBR green chemistry.
Statistical Analysis
Statistical analysis was done by GraphPad Prism 5.0 software. The convention followed to denote significance: ∗p ≤ 0.05, ∗∗p ≤ 0.01, and ∗∗∗p ≤ 0.001.
Acknowledgments
Support from NIH (K.W.L.: EB015300, AI096305, UH2TR000505; C.A.G.: DP2OD008586, R01DA036865), National Science Foundation (CBET-1151035), American Heart Association (10SDG3060033), and the Muscular Dystrophy Association (MDA277360) is acknowledged. N.C. was supported by the Flight Attendant Medical Research Institute. We thank Pablo Perez-Pinera for helpful discussions. We also acknowledge Stanley Qi, Marius Wernig, and David Baltimore for Addgene plasmids 44247, 27152, and 14883, respectively. C.A.G. is an inventor on patent applications related to genome engineering and a scientific advisor to Editas Medicine.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., Lassar A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Farzadfard F., Perli S.D., Lu T.K. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth. Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Yang J., Tsang J.C., Ooi J., Wu D., Liu P. Reprogramming to pluripotency using designer TALE transcription factors targeting enhancers. Stem Cell Rev. 2013;1:183–197. doi: 10.1016/j.stemcr.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabadi A.M., Ousterout D.G., Hilton I.B., Gersbach C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku749. Published online August 13, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns N.A., Genga R.M., Enuameh M.S., Garber M., Wolfe S.A., Maehr R. Cas9 effector-mediated regulation of transcription and differentiation in human pluripotent stem cells. Development. 2014;141:219–223. doi: 10.1242/dev.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S., Brigham M.D., Trevino A.E., Hsu P.D., Heidenreich M., Cong L., Platt R.J., Scott D.A., Church G.M., Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P., Kocak D.D., Vockley C.M., Adler A.F., Kabadi A.M., Polstein L.R., Thakore P.I., Glass K.A., Ousterout D.G., Leong K.W. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Zingg J.M., Pedraza-Alva G., Jost J.P. MyoD1 promoter autoregulation is mediated by two proximal E-boxes. Nucleic Acids Res. 1994;22:2234–2241. doi: 10.1093/nar/22.12.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


