Summary
The increased incidence of obesity and metabolic disease underscores the importance of elucidating the biology of adipose tissue development. The recent discovery of cell surface markers for prospective identification of adipose precursor cells (APCs) in vivo will greatly facilitate these studies, yet tools for specifically targeting these cells in vivo have not been identified. Here, we survey three transgenic mouse lines, Fabp4-Cre, PdgfRα-Cre, and Prx1-Cre, precisely assessing Cre-mediated recombination in adipose stromal populations and mature tissues. Our data provide key insights into the utility of these tools to modulate gene expression in adipose tissues. In particular, Fabp4-Cre is not effective to target APCs, nor is its activity restricted to these cells. PdgfRα-Cre directs recombination in the vast majority of APCs, but also targets other populations. In contrast, adipose expression of Prx1-Cre is chiefly limited to subcutaneous inguinal APCs, which will be valuable for dissection of APC functions among adipose depots.
Highlights
-
•
Genetic tools to target adipose precursors in vivo assessed by flow cytometry
-
•
Fabp4-Cre is active only in a small subset of white adipose precursors
-
•
PdgfRα-Cre is active in nearly all white and brown adipose precursors
-
•
In adipose tissue, Prx1-Cre activity is limited to subcutaneous inguinal precursors
The discovery of cell surface markers that label adipose progenitors identifies new avenues of stem cell and metabolic research. Here, Krueger et al. use these markers to survey Cre-mediated recombination in the adipose depots of several transgenic mouse lines. These data provide key insights into the suitability of these tools to modulate gene expression in adipose tissues.
Introduction
Adipose precursor cells (APCs) are a subpopulation of lineage (Lin)−SCA1+ adipose-derived stromal cells competent to undergo adipogenesis in vivo and generate a functional adipose depot after transplantation (Rodeheffer et al., 2008; Joe et al., 2009; Schulz et al., 2011). It is widely anticipated that these cells will have an important role in regenerative medicine (Ong and Sugii, 2013). Furthermore, the recent isolation and characterization of APCs from white adipose tissue (WAT) using cell surface markers (Rodeheffer et al., 2008) open new avenues of stem cell and metabolic research. These markers can be used to quantify precursor cells within the adipose depot or to isolate cells for transplant or ex vivo assays (Rodeheffer et al., 2008; Joe et al., 2010; Schulz et al., 2011; Berry and Rodeheffer, 2013). However, it would be of great utility to manipulate gene expression in APCs in their physiologic in vivo context. Such experiments would require the identification of promoter/enhancer sequences that can direct gene expression specifically to this population. Previously, the promoter regions of several genes expressed in mesenchymal progenitors have been isolated and their activity characterized (Logan et al., 2002; Roesch et al., 2008; Berry and Rodeheffer, 2013). These regulatory elements, when used to drive expression of Cre recombinase, may be useful for genetic manipulation of APCs.
Here we analyze recombination directed by several Cre lines in stromal populations of WAT and brown adipose tissue (BAT). We find that the most commonly used line to direct adipose-specific expression, Fabp4-Cre (He et al., 2003), is not active in most WAT APCs. We confirm previous studies that PdgfRα-Cre (Roesch et al., 2008) is active in most WAT APCs (Berry and Rodeheffer, 2013) and also find activity in most BAT Lin−SCA1+ cells. However, we also detect activity in other adipose stromal cells. In contrast, a line previously reported to direct expression in uncommitted mesenchymal progenitors (Calo et al., 2010), Prx1-Cre, has activity restricted to adipose precursors of the subcutaneous inguinal fat pad, with little recombination in other cell types within the fat pads examined. Together, these data serve as a guide for the use of these tools in the manipulation of gene expression in adipose tissues in vivo.
Results
The stromal compartment of adipose depots includes lymphocytes, macrophages, endothelial cells (ECs), fibroblasts, and APCs and, when isolated from adipocytes, is collectively referred to as the stromal vascular fraction (SVF) (Sanchez-Gurmaches and Guertin, 2014). Using a combination of four markers (CD45, CD11b, CD31, and SCA1), SVF cells can be categorized into five populations (Figures 1A and 1B), including APCs (CD45−CD11b−CD31−SCA1+). In culture, these isolated Lin−SCA1+ cells are highly responsive to standard adipogenic induction cocktail (Figure 1C and Figure S1A available online). Although cells from BAT with the APC marker profile differentiate ex vivo (Sanchez-Gurmaches et al., 2012) and express markers of BAT (Schulz et al., 2011; Liu et al., 2013), these cells are less well studied than WAT APCs; therefore, we refer to these as Lin−SCA1+ cells. Cells not labeled by any of the cell surface markers in our scheme (CD45−CD31−SCA1−, hereafter referred to as SCA1−CD31− cells) are heterogeneous in size and morphology (Figures S1B and S1C). In order to thoroughly document expression in all stromal cells, we report the recombination data (as in Figure S1D) but have not further characterized this population.
Figure 1.
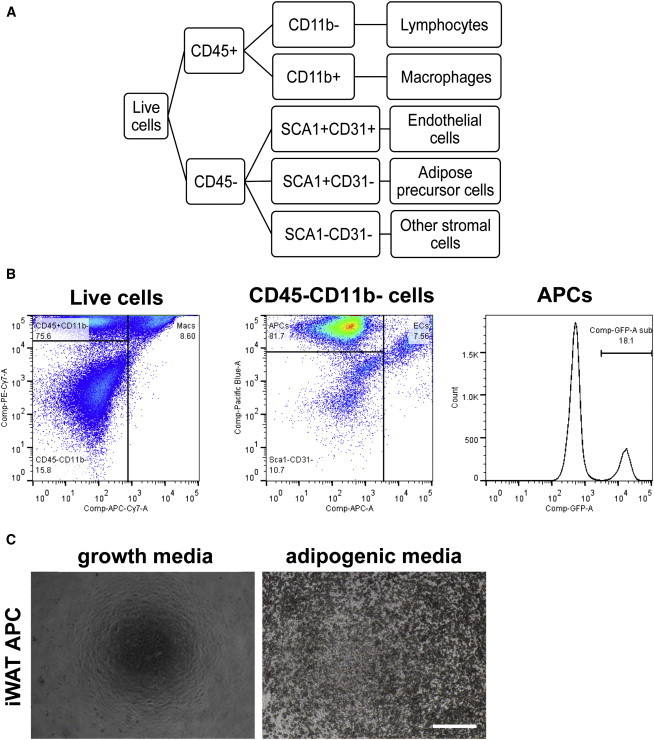
Flow Cytometry Schema to Profile Adipose Stromal Cells and Validation by Ex Vivo Differentiation
(A) Flow cytometry schema of adipose stromal cell types using combinations of cell surface markers, applied to all Cre;Rosa-EYFP adipose depots.
(B) Representative flow cytometry plots of inguinal WAT live cells. (Left panel) Plot of live cells stained with CD45 (PE-Cy7) and CD11b (APC-Cy7). (Middle panel) Plot of CD45−CD11b− cells stained with SCA1 (Pacific Blue) and CD31 (APC). (Right panel) Histogram of EYFP fluorescence within APCs (Lin−SCA1+). Label above plots indicates parent populations.
(C) iWAT APCs cultured in growth media (left) or adipogenic induction media (right), shown 6 days after induction. Scale bar represents 0.5 mm.
See also Figure S1.
To test whether transgenic Cre mouse lines are capable of specific targeting APCs in vivo, mice carrying the Cre recombinase-responsive Enhanced Yellow Fluorescent Protein (EYFP) allele in the Rosa locus (Rosa-EYFP) (Srinivas et al., 2001) were crossed to one of three Cre lines. These lines were chosen for their common usage in studies of adipose tissue (Fabp4-Cre) (Mullican et al., 2013; Berry et al., 2014) or because of previous reports of activity of these Cre lines in adipose or mesenchymal progenitors (PdgfRα-Cre, Prx1-Cre) (Calo et al., 2010; Lee et al., 2012; Berry and Rodeheffer, 2013). The flow cytometry analysis scheme (Figure 1A) was applied to the SVF of fat pads from Cre;Rosa-EYFP double transgenic mice.
Fabp4-Cre
The Fabp4 (also called aP2) promoter is a widely used tool to direct adipose-specific gene expression and, when used to direct Cre recombinase, adipose-specific gene deletion (Mullican et al., 2013; Berry et al., 2014). Multiple Fabp4-Cre lines have been generated (Barlow et al., 1997; Abel et al., 2001; He et al., 2003; Tang et al., 2008), but the line described by He et al. (2003) is most commonly used (Mullican et al., 2013). Despite confirmation of Fabp4-Cre activity in adipose tissues, prior studies did not quantitatively assess the expression of Fabp4-Cre in adipose stromal cells or specifically in APCs in vivo. We analyzed the SVF of subcutaneous inguinal and gonadal WAT (iWAT and gWAT, respectively) and of interscapular BAT from male Fabp4-Cre;Rosa-EYFP mice by flow cytometry. We found that Fabp4-Cre directs low levels of recombination in WAT APCs, ranging from 15% in iWAT to 20% in gWAT (Figure 2A). However, ∼41% of BAT Lin−SCA1+ cells are recombined (Figure 2A). In contrast, EYFP expression is observed in 23%–38% of ECs of BAT and WAT. Fapb4-Cre directs modest levels of recombination in SCA1−CD31− cells (6%–7% across depots) and macrophages (7%–13% across depots), with very few nonmacrophage white blood cells (CD45+CD11b−) expressing EYFP (Figure 2A). Similar results were obtained when female mice were examined (Figure 2B).
Figure 2.
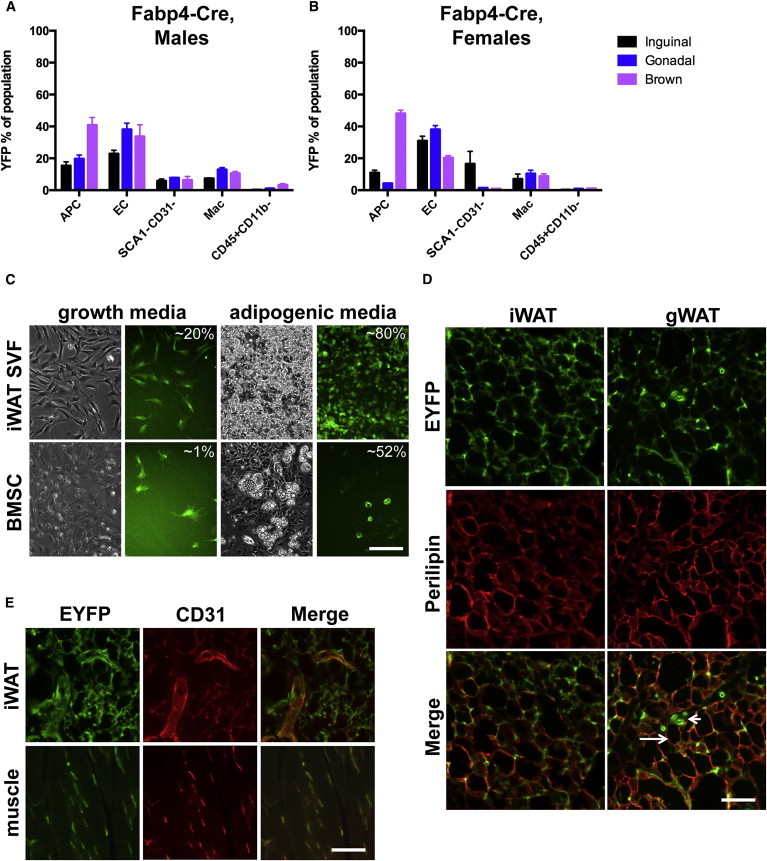
Fabp4-Cre Recombinase Activity in Adipose Stromal Populations, Culture, and Tissue Sections
(A) Flow cytometry profile of Fabp4-Cre;Rosa-EYFP male mice (n = 3), where each n is one animal. Data are represented as mean ± SEM.
(B) Flow cytometry profile of Fabp4-Cre:Rosa-EYFP female mice (n = 3), where each n is one animal. Data are represented as mean ± SEM.
(C) Expression of EYFP in cultured cells, prior to or after adipogenic induction. The estimated percentage of EYFP+ cells or adipocytes is indicated. Scale bar represents 0.2 mm.
(D) Expression of EYFP and perilipin (red), a marker of adipocytes, in sections of adipose tissue. Arrows indicate nonrecombined adipocytes. Arrowheads indicate putative endothelial structures labeled by EYFP. Scale bar represents 0.1 mm.
(E) Expression of EYFP and CD31 (red) in sections of adipose tissue or skeletal muscle. Scale bar represents 0.1 mm.
Mac, macrophage. See also Figure S2.
The low levels of recombination observed in iWAT APCs led us to wonder about the properties of those select APCs in which Fabp4-Cre is active. First, we evaluated gene expression in iWAT APCs for markers of more committed preadipocytes. Interestingly, cells in which Fabp4-Cre is active (EYFP+) express higher levels of Fabp4 and Pparγ as compared with EYFP− cells (Figure S2A). These findings are consistent with the EYFP+ cells being more differentiated than the EYFP− cells. Next, we considered whether these recombined cells are more adipogenic than nonrecombined cells. EYFP+ and EYFP− iWAT APCs were sorted, cultured, and induced to differentiate using standard induction cocktail. Both populations differentiated equally well (Figure S2B), indicating that adipogenic capability, at least ex vivo, does not correlate exclusively with Fabp4-Cre activity.
Fabp4 is expressed at low levels in undifferentiated SVF in culture, but rapidly increases its expression with adipogenesis and is highly expressed in mature adipocytes (Shan et al., 2013). We therefore hypothesized that adipogenic differentiation would increase Fabp4-Cre-mediated recombination. Both SVF and bone marrow stromal cell (BMSC) cultures contain preadipocytes and are commonly used as ex vivo models of adipogenesis. We monitored Fabp4-Cre recombinase activity in cultured iWAT SVF and BMSCs from Fabp4-Cre;Rosa-EYFP mice. In undifferentiated cells, ∼20% of SVF and ∼1% of BMSCs expressed EYFP (Figure 2C, left panels). Strikingly, following adipogenic induction, ∼80% of the SVF-derived adipocytes are EYFP+, but far fewer (∼52%) of the BMSC-derived adipocytes appear recombined (Figure 2C, right panels). Thus, Fabp4-Cre labeling of adipose precursors and adipocytes is depot dependent.
To monitor Fabp4-Cre-mediated recombination in vivo, adipose tissues were fixed, sectioned, and stained for EYFP and markers of adipocytes (perilipin) or ECs (CD31). Since there is green autofluorescence in adipose tissues, single transgenic controls were processed and stained alongside double transgenic tissues for comparison (Figure S2C). As expected, immunostaining of adipose tissue revealed many EYFP+ adipocytes in both WAT and BAT (Figure 2D), although some adipocytes appear unlabeled (Figure 2D, arrows). We also observed perilipin− structures clearly labeled with EYFP (Figure 2D, arrowheads), some of which are ECs (Figure 2E). Importantly, CD31+ structures in muscle are also colabeled with EYFP (Figure 2E). Consistent with the findings of others (Morán-Salvador et al., 2013; Mullican et al., 2013), we also detect widespread recombined cells in the liver and intestine (Figure S2D). Together, these results demonstrate that Fabp4-Cre is expressed in many but not all adipocytes, is not restricted to the adipose lineage, and is not specific to adipose tissues.
PdgfRα-Cre
PdgfRα is expressed in many cells of neuroectodermal or mesenchymal origin, including oligodendrocyte precursors (Rivers et al., 2008), neural crest cells (Soriano, 1997), the developing intestine (Karlsson et al., 2000), testes (Brennan et al., 2003), and heart (Chong et al., 2011) and may also mark bone marrow mesenchymal stem cells (Morikawa et al., 2009). A mouse in which Cre is directed by PdgfRα regulatory elements (Roesch et al., 2008) was shown to label WAT APCs when combined with other cell surface markers (Berry and Rodeheffer, 2013). Consistent with these latter findings, our analysis of PdgfRa-Cre;Rosa-EYFP mice suggests that Cre is active in nearly all (96%–99%) WAT APCs (Figure 3A). Additionally, we find that most (89%) BAT Lin−SCA1+ cells are also recombined (Figure 3A). Moreover, we observe that PdgfRα-Cre activity in iWAT is specific for APCs, as only low levels of recombination (<11%) are detected in white blood cells and ECs. However, PdgfRα-Cre induces recombination in 56% of SCA1−CD31− cells in gWAT (Figure 3A). Thus, while this Cre line effectively targets the vast majority of APCs, it is clearly not exclusive to this cell type across adipose depots. Similar results were obtained when female mice were examined (Figure 3B). In agreement with previous reports of endogenous PdgfRα expression (Morikawa et al., 2009) and in contrast to Fabp4-Cre, PdgfRα-Cre also directs expression to most BMSC preadipocytes, as evidenced by the large fraction of EYFP+ adipocytes generated following adipogenic induction (Figure 3C).
Figure 3.
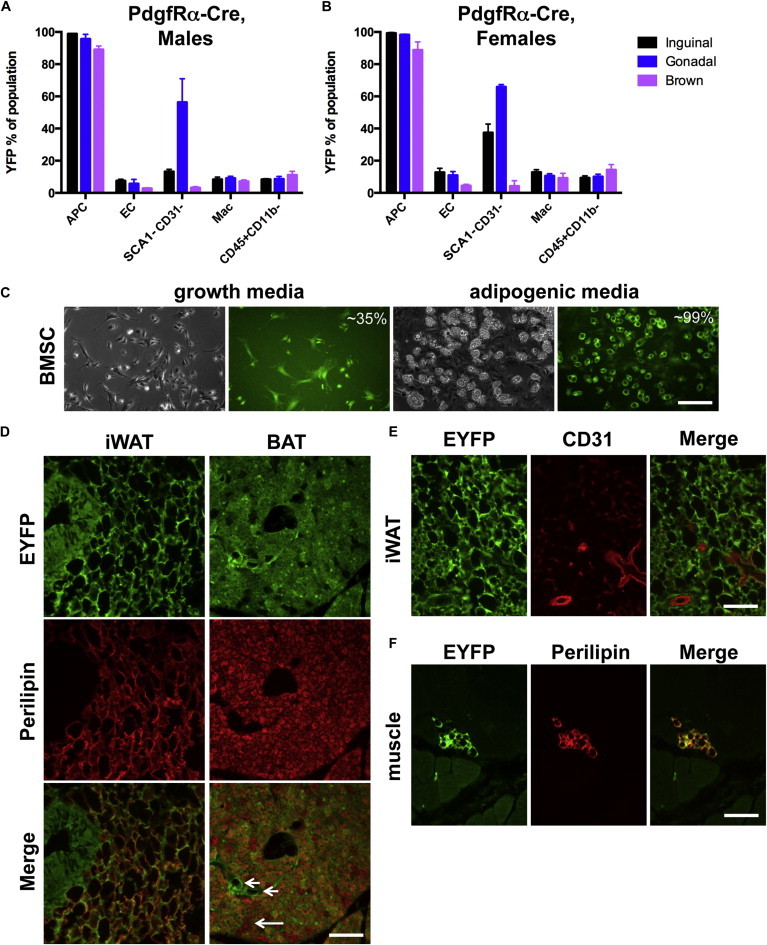
PdgfRα-Cre Recombinase Activity in Adipose Stromal Populations and in Tissue Sections
(A) Flow cytometry profile of PdgfRα-Cre;Rosa-EYFP male mice (n = 3), where each n is one animal. Data are represented as mean ± SEM.
(B) Flow cytometry profile of PdgfRα-Cre;Rosa-EYFP female mice (n = 3), where each n is one animal. Data are represented as mean ± SEM.
(C) Expression of EYFP in cultured BMSCs, prior to or after adipogenic induction. The estimated percent EYFP+ cells or adipocytes is indicated. Scale bar represents 0.2 mm.
(D) Expression of EYFP and perilipin (red) in sections of WAT and BAT. Arrows indicate nonrecombined adipocytes. Arrowheads indicate putative vascular structures labeled by EYFP. Scale bar represents 0.1 mm.
(E) Expression of EYFP and CD31 (red) in sections of adipose tissue. Scale bar represents 0.1 mm.
(F) Expression of EYFP and perilipin (red) in skeletal muscle. Scale bar represents 0.1 mm.
Mac, macrophage. See also Figure S3.
Imaging of histological sections of WAT from PdgfRα-Cre;Rosa-EYFP animals confirms the flow cytometry results, as a high level of recombination in adipocytes is observed (Figure 3D). Interestingly, not all brown adipocytes are EYFP+, perhaps reflecting the slightly lower recombination in BAT Lin−SCA1+ cells versus WAT APCs (Figures 3A and 3D, arrow). We also noted recombination in cells surrounding CD31+ ECs (Figure 3E). This finding is intriguing in light of other reports of adipose progenitor cells associated with blood vessels as part of a perivascular niche (Tang et al., 2008). In contrast to Fabp4-Cre, very few EYFP+ cells were detected in muscle sections from PdgfRα-Cre;Rosa-EYFP mice, and most of these are intramuscular adipocytes (Figure 3F). These studies show that PdgfRα-Cre is highly active in, but not limited to, APCs of adipose lineages.
Prx1-Cre
Prx1-Cre directs expression to the embryonic limb bud mesenchyme, flank mesoderm, and a subset of the cranial mesenchyme (Logan et al., 2002), as well as to uncommitted mesenchymal progenitors of several other tissues (Calo et al., 2010). Therefore, it was expected that recombination would be observed in many SVF cells of all adipose depots. However, in striking contrast to the other Cre lines examined, adipose Prx1-Cre activity is mostly restricted to iWAT APCs: we observed recombination in >82% of APCs in both males and females (Figures 4A and 4B). Intriguingly, modest recombination is observed in gWAT APCs of female mice (∼18%), as well as in SCA1−CD31− cells of either depot (14%–22%) (Figure 4B), but is absent in males (Figure 4A). Neither sex displays significant recombination in any BAT SVF cell type (Figures 4A and 4B). Thus, Prx1-Cre has unexpected specificity for APCs, and this is particularly evident in male mice. As expected, examination of cultured iWAT SVF or BMSCs revealed a strikingly high percentage of recombination, both prior to and after adipogenic induction (Figure 4C), which is consistent with previous data showing recombination in limb bud mesenchyme (Logan et al., 2002).
Figure 4.
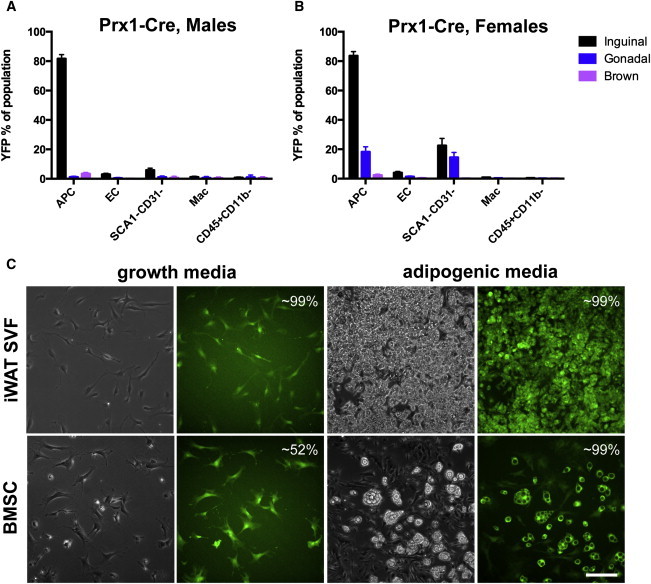
Prx1-Cre Recombinase Activity in Adipose Stromal Populations and in Cultured Cells
(A) Flow cytometry profile of Prx1-Cre;Rosa-EYFP male mice (n = 3), where each n is one animal.
(B) Flow cytometry profile of Prx1-Cre;Rosa-EYFP female mice (n = 3), where each n is one animal. Data are represented as mean ± SEM.
(C) Expression of EYFP in cultured cells, prior to or after adipogenic induction. The estimated percentage of EYFP+ cells or adipocytes is indicated. Scale bar represents 0.2 mm.
Mac, macrophage.
In sections of iWAT, nearly all perilipin+ cells also express EYFP (Figure 5A, left). Also consistent with the flow cytometry results, there are very few EYFP+ cells in BAT or gWAT sections from male mice (Figure 5A, right). However, in agreement with the Cre activity detected in APCs, there are perilipin+EYFP+ cells in female gWAT (Figure 5B). Similar to our results with PdgfRα-Cre;Rosa-EYFP mice and in contrast to our observations of Fabp4-Cre;Rosa-EYFP iWAT, some EYFP+ cells in Prx1-Cre;Rosa-EYFP iWAT surround but do not overlap with CD31+ cells (Figure 5C, upper panel).
Figure 5.
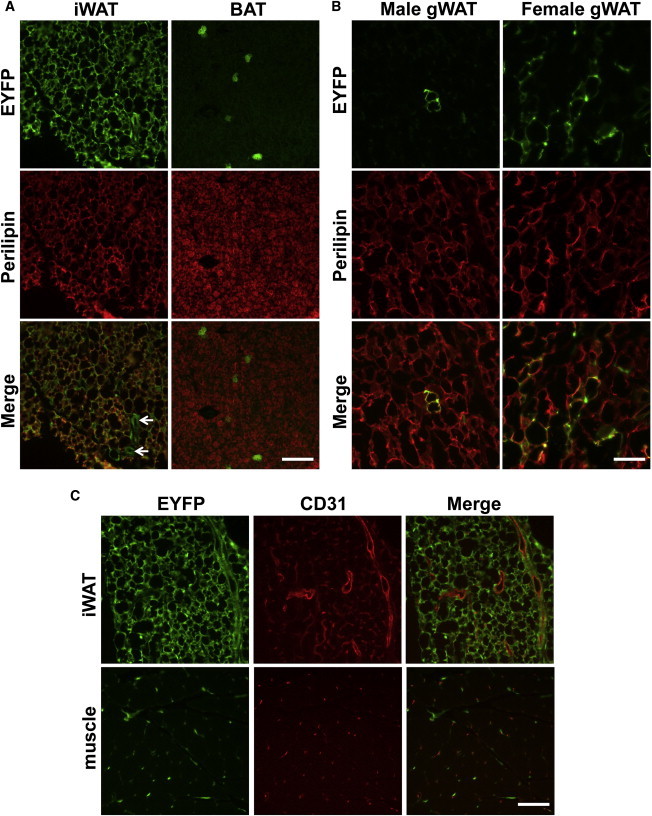
Prx1-Cre Recombinase Activity in Tissue Sections
(A) Expression of EYFP and perilipin (red) in sections of male WAT and BAT. Arrowheads indicate putative ECs labeled by EYFP. Scale bar represents 0.1 mm.
(B) Expression of EYFP and perilipin (red) in sections of gonadal tissue from male and female mice. Scale bar represents 0.1 mm.
(C) Expression of EYFP and CD31 (red) in sections of male iWAT and skeletal muscle. Scale bar represents 0.1 mm.
Mac, macrophage; gWAT, gonadal (epididymal or parametrial for male or female, respectively) WAT. See also Figure S4.
Interestingly, staining of muscle tissue reveals recombination in a number of adipocytes, as well as in scattered CD31− cells (Figure 5C, lower panel). However, EYFP expression is not detectable in the myofibers, suggesting cells expressing the Prx1-Cre transgene do not contribute to the muscle lineage, even though muscle progenitors arise from mesenchymal stem cells (Calo et al., 2010). Rare cells showing recombination are also detected in the liver and intestine of Prx1-Cre;Rosa-EYFP mice (Figure S3). In summary, our findings suggest that Prx1-Cre has a surprisingly restricted expression pattern in adipose and other tissues and a high degree of specificity for APCs in the iWAT.
Discussion
Recent studies have elucidated surface markers that can be used to distinguish and isolate APCs in adipose tissue from other stromal cell populations (Rodeheffer et al., 2008). However, continued progress in the field relies on the identification of tools for targeted manipulation of APCs in vivo. In this study, we assessed the ability of different Cre mouse lines to target APCs relative to other adipose stromal populations. While prior studies have examined recombination in selected stromal cells directed by either Fabp4-Cre (Shan et al., 2013) or PdgfRα-Cre (Berry and Rodeheffer, 2013), here we provide a quantitative report of in vivo Cre recombinase activity in all stromal populations of WAT and BAT depots.
Comparative analysis of the expression patterns directed by the various Cre lines reveals important distinctions. In particular, the most widely used line for directing adipose-specific recombination, Fabp4-Cre, is poorly suited to target APCs. A previous study of Fabp4-Cre;Rosa-TdTomato mice reported substantially higher numbers of recombined precursors (∼60% in both WAT and BAT) than reported here, using the same cell surface marker profile (Shan et al., 2013). However, Shan and colleagues evaluated cells that were cultured after sorting, whereas in our study, the EYFP reporter expression is quantified in vivo. Fabp4-Cre is neither active in the majority of APCs of any depot, nor is it specific for APCs: other stromal cells types, particularly ECs, display similar or even higher levels of recombination (Figures 2A and 2B). Since Fabp4 expression increases with adipogenic differentiation, it is plausible that the Fabp4-Cre recombination of APCs marks a more committed subset of cells (Liu et al., 2013), although additional studies are required to substantiate this hypothesis.
Despite low levels of recombination in APCs, substantial activity of Fabp4-Cre is apparent in mature adipose tissues, consistent with the initial characterization (He et al., 2003), and this should contribute to the phenotypes observed in studies that utilize this promoter. However, the lack of fidelity to the adipose lineage is clear, as recombination has been detected in muscle and liver (Figures 2E and S2D, respectively; Morán-Salvador et al., 2013; Mullican et al., 2013), embryonic limb bud and dorsal root ganglion (Urs et al., 2006), adrenal medulla and brain (Martens et al., 2010), and other tissues (Mullican et al., 2013). These other sites of expression could be a feature of this particular transgenic line, but analysis of another independently generated Fabp4-Cre line (Abel et al., 2001) also identified many nonadipose sites of recombination (Lee et al., 2013). Together, these findings suggest that Fabp4-Cre is not a suitable tool for effective and specific targeting of APCs.
In contrast to Fabp4-Cre, PdgfRα-Cre is highly active in both WAT and BAT Lin−SCA1+ cells, demonstrating the highest recombination efficiency among all examined Cre lines. However, we observed PdgfRα-Cre activity in other stromal populations (Figures 3A–3C). While further evaluation will be required, the expression of Cre in other lineages could confound the results of in vivo experiments utilizing this line to study adipose tissue biology. The PdgfRα-Cre line characterized here was generated using a large genomic fragment, from a bacterial artificial chromosome, in an effort to capture the regulatory elements required for the complex endogenous PdgfRα expression pattern (Zhang et al., 1998). However, it would be highly useful to isolate regulatory elements for particular progenitor populations. Such findings should allow more precise targeting of each cell group and may inform the lineage hierarchy among progenitors.
Surprisingly, we discovered that among adipose depots, Prx1-Cre activity is specific for iWAT APCs, particularly in male mice. These results were unexpected, given a prior study that reported widespread recombination in both WAT and BAT, purportedly due to recombination at the progenitor stage (Calo et al., 2010). However, that study utilized a different indicator line, Rosa-lacZ (Soriano, 1999), and did not quantify recombination or determine the identity of the cells in which recombination was detected. Here, we comprehensively examine stromal cells from multiple depots and quantify recombination in vivo in defined cell populations. These findings are consistent with the immunostaining results of adipose tissues: high levels of recombination in iWAT, much less in gWAT, and little to no recombination in BAT. Finally, previous work has shown the same Cre allele can direct different expression patterns depending on the mouse strain background (Hébert and McConnell, 2000). The mice utilized in our study were maintained in the C57BL/6 background, whereas the prior study utilized a mixed genetic background (Calo et al., 2010). Therefore, some differences in recombination observed between these studies may be the result of background modifiers of Prx1-Cre expression.
Our studies do not address whether this recombination pattern reflects physiological expression of Prx1, the activity of isolated Prx1 promoter elements, or results from chromosomal positional effects of this particular transgenic line. The documented expression of Prx1-Cre in the limb bud (Logan et al., 2002) and our own studies of bone marrow cultures (Figure 4C) suggest that experiments with these mice will require careful interpretation. However, for the study of genes primarily essential to adipose tissue function, our results highlight the utility of Prx1-Cre over PdgfRα-Cre, as the restricted activity of the former within adipose depots allows for the study of gene function specifically in iWAT APCs. Previous studies in mice have shown that iWAT transplanted into the visceral cavity is metabolically protective, resulting in decreased overall fat mass, improved glucose homeostasis, and insulin sensitivity, although the reasons for these benefits are not well understood (Tran et al., 2008). The specificity of Prx1-Cre activity could enable the identification of genes responsible for these and other known functional differences among fat depots, including gene expression, adipokine release, APC proliferation, and lipid homeostasis (Tchkonia et al., 2013). Interestingly, another recently characterized Cre line is active in various visceral adipose depots, including gWAT, but not in iWAT or BAT (Chau et al., 2014), and thus will be a useful complementary tool to analyze alongside Prx1-Cre-directed recombination.
Finally, inducible (Cre-ER) versions of these lines have been generated using the same or similar promoter elements (Imai et al., 2001; Rivers et al., 2008; Kawanami et al., 2009; Kang et al., 2010). However, even if the same genomic sequences are used, the unique integration site of each transgenic line and mouse genetic background will likely alter the Cre expression pattern from what is reported here. We would recommend that investigators employing these and other lines evaluate the expression patterns to assess the specific Cre recombination profiles. Nevertheless, it will be worthwhile to examine whether increased temporal resolution can further restrict the Cre activity and thus increase their specificity for defined target populations.
Experimental Procedures
Mouse Models
All studies were approved by Stanford University’s Administrative Panel on Laboratory Animal Care Committee. Mouse lines were obtained from JAX Mice: Fabp4-Cre, stock #005069 (He et al., 2003); Prrx1-Cre, stock #005584 (Logan et al., 2002); PdgfRα-Cre, stock #013148 (Roesch et al., 2008); and Rosa-EYFP, stock #006148 (Srinivas et al., 2001). Hemizygous male Cre mice for each line were crossed to homozygous female Rosa-EYFP indicator mice, and both male and female progeny were analyzed.
Preparation of SVF and BMSC Cultures
Animals were euthanized by asphyxiation with CO2 and cervical dislocation, and adipose depots were excised. Tissues were minced and incubated in 1 mg/ml collagenase (Worthington) in Dulbecco’s modified Eagle’s medium (DMEM)/F12 for 1 hr at 37°C with 5% CO2. After inactivation of the digest with an equal volume of DMEM/10% fetal bovine serum (FBS), samples were passed through a 100 μm cell strainer. For cell culture, samples were centrifuged at 300 g for 5 min, and the resulting SVF was resuspended in complete media (DMEM/10% FBS/1% penicillin/streptomycin) and plated for imaging or differentiation. For BMSCs, the femurs and tibia were flushed with complete media. The flushed cells were triturated briefly, resuspended in additional complete media, and then cultured for imaging and differentiation. Adipogenic induction was carried out as described (Costa et al., 2011).
Flow Cytometry and Fluorescence-Activated Cell Sorting
Antibodies used include SCA1 Pacific Blue (BioLegend, 108120), CD45 PE-Cy7 (eBioscience, 25-0421-82), CD11b APC-Cy7 (BioLegend, 101226), and CD31 APC (eBioscience, 17-0311-82). SVF was prepared as for cell culture, except that following centrifugation, the resulting cell pellet was resuspended in 1 ml ACK Lysing Buffer (Invitrogen) to remove red blood cells. After inactivation with Hank’s balanced salt solution (HBSS)/2% FBS, samples were centrifuged again and resuspended in HBSS/2% FBS for antibody labeling for flow cytometry or fluorescence-activated cell sorting (FACS). Samples were analyzed on a BD LSR Fortessa analyzer or sorted on a BD FACS Aria II. Cell gating was based on comparison with unstained and fluorescence-minus-one-stained controls. Single live cells were discriminated by forward-scatter and side-scatter analysis and 7-AAD labeling (BD Bioscience), respectively. Cells were sorted into serum-free DMEM media for gene expression analysis or into complete media for cell culture. Flow cytometry standard files were analyzed using FlowJo version 10.
Histological Analysis
Antibodies used include chicken anti-GFP (Aves, GFP-1020), rabbit anti-Perilipin (Sigma, P1998), and rat anti-CD31 (BD Bioscience, 553370). Mice were transcardially perfused with PBS followed by 4% formaldehyde in PBS. Tissues were excised and fixed overnight, cryoprotected in 30% sucrose in PBS, then frozen in OCT (Sakura), and stored at −80°C. Cryosections (10 μm) were prepared using a Leica cryostat and mounted on SuperfrostPlus sides (Fisher). After drying, sections were blocked with 5% bovine serum albumin, followed by labeling with primary antibody for 1 hr at room temperature. After five washes with PBS/0.1% Triton X-100, sections were incubated with appropriate secondary antibodies (goat antichicken Alexa 488, A11039; goat antirat Alexa 647, A21247; goat antirabbit 647, A21244; all from Invitrogen) for 30 min at room temperature. Following additional washes, coverslips were mounted with DAPI antifade reagent (Vector Labs) and sealed with clear nail polish. Images were obtained using inverted Leica epifluorescence microscope, pseudocolored using ImageJ, and merged in Photoshop CS4 (Adobe Creative Suites).
Acknowledgments
We thank Patty Lovelace of the Stanford Shared FACS Facility for assistance with the flow cytometry experiments. K.C.K. was supported by NIH grant F32 DK093191. K.C.K. and H.D. were supported by NIH grant T32 DK007217. K.C.K. and M.J.C. were supported by the Stanford School of Medicine Dean’s Postdoctoral Fellowship. K.C.K, M.J.C. and H.D. were supported by the Lucile Packard Foundation for Children’s Health, Stanford NIH-NCATS-CTSA UL1 TR001085, and the Child Health Research Institute of Stanford University. The work was funded by an NIH Director’s New Innovator Award (DP2 OD006740) to B.J.F.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Abel E.D., Peroni O., Kim J.K., Kim Y.B., Boss O., Hadro E., Minnemann T., Shulman G.I., Kahn B.B. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–733. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- Barlow C., Schroeder M., Lekstrom-Himes J., Kylefjord H., Deng C.X., Wynshaw-Boris A., Spiegelman B.M., Xanthopoulos K.G. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic Acids Res. 1997;25:2543–2545. doi: 10.1093/nar/25.12.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R., Rodeheffer M.S. Characterization of the adipocyte cellular lineage in vivo. Nat. Cell Biol. 2013;15:302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry R., Jeffery E., Rodeheffer M.S. Weighing in on adipocyte precursors. Cell Metab. 2014;19:8–20. doi: 10.1016/j.cmet.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan J., Tilmann C., Capel B. Pdgfr-alpha mediates testis cord organization and fetal Leydig cell development in the XY gonad. Genes Dev. 2003;17:800–810. doi: 10.1101/gad.1052503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Quintero-Estades J.A., Danielian P.S., Nedelcu S., Berman S.D., Lees J.A. Rb regulates fate choice and lineage commitment in vivo. Nature. 2010;466:1110–1114. doi: 10.1038/nature09264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau Y.Y., Bandiera R., Serrels A., Martínez-Estrada O.M., Qing W., Lee M., Slight J., Thornburn A., Berry R., McHaffie S. Visceral and subcutaneous fat have different origins and evidence supports a mesothelial source. Nat. Cell Biol. 2014;16:367–375. doi: 10.1038/ncb2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J., Chandrakanthan V., Xaymardan M., Asli N.S., Li J., Ahmed I., Heffernan C., Menon M.K., Scarlett C.J., Rashidianfar A. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell. 2011;9:527–540. doi: 10.1016/j.stem.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.J., So A.Y., Kaasik K., Krueger K.C., Pillsbury M.L., Fu Y.H., Ptacek L.J., Yamamoto K.R., Feldman B.J. Circadian rhythm gene period 3 is an inhibitor of the adipocyte cell fate. J. Biol. Chem. 2011;286:9063–9070. doi: 10.1074/jbc.M110.164558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., Barak Y., Hevener A., Olson P., Liao D., Le J., Nelson M., Ong E., Olefsky J.M., Evans R.M. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc. Natl. Acad. Sci. USA. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert J.M., McConnell S.K. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Imai T., Jiang M., Chambon P., Metzger D. Impaired adipogenesis and lipolysis in the mouse upon selective ablation of the retinoid X receptor alpha mediated by a tamoxifen-inducible chimeric Cre recombinase (Cre-ERT2) in adipocytes. Proc. Natl. Acad. Sci. USA. 2001;98:224–228. doi: 10.1073/pnas.011528898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe A.W., Yi L., Even Y., Vogl A.W., Rossi F.M. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-fat diet. Stem Cells. 2009;27:2563–2570. doi: 10.1002/stem.190. [DOI] [PubMed] [Google Scholar]
- Joe A.W., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.H., Fukaya M., Yang J.K., Rothstein J.D., Bergles D.E. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson L., Lindahl P., Heath J.K., Betsholtz C. Abnormal gastrointestinal development in PDGF-A and PDGFR-(alpha) deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development. 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- Kawanami A., Matsushita T., Chan Y.Y., Murakami S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochem. Biophys. Res. Commun. 2009;386:477–482. doi: 10.1016/j.bbrc.2009.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.H., Petkova A.P., Mottillo E.P., Granneman J.G. In vivo identification of bipotential adipocyte progenitors recruited by β3-adrenoceptor activation and high-fat feeding. Cell Metab. 2012;15:480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A., Smyth G., Rourk M., Cederquist C., Rosen E.D. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Bi P., Shan T., Yang X., Yin H., Wang Y.X., Liu N., Rudnicki M.A., Kuang S. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9:e1003626. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Martens K., Bottelbergs A., Baes M. Ectopic recombination in the central and peripheral nervous system by aP2/FABP4-Cre mice: implications for metabolism research. FEBS Lett. 2010;584:1054–1058. doi: 10.1016/j.febslet.2010.01.061. [DOI] [PubMed] [Google Scholar]
- Morán-Salvador E., Titos E., Rius B., González-Périz A., García-Alonso V., López-Vicario C., Miquel R., Barak Y., Arroyo V., Clària J. Cell-specific PPARγ deficiency establishes anti-inflammatory and anti-fibrogenic properties for this nuclear receptor in non-parenchymal liver cells. J. Hepatol. 2013;59:1045–1053. doi: 10.1016/j.jhep.2013.06.023. [DOI] [PubMed] [Google Scholar]
- Morikawa S., Mabuchi Y., Kubota Y., Nagai Y., Niibe K., Hiratsu E., Suzuki S., Miyauchi-Hara C., Nagoshi N., Sunabori T. Prospective identification, isolation, and systemic transplantation of multipotent mesenchymal stem cells in murine bone marrow. J. Exp. Med. 2009;206:2483–2496. doi: 10.1084/jem.20091046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullican S.E., Tomaru T., Gaddis C.A., Peed L.C., Sundaram A., Lazar M.A. A novel adipose-specific gene deletion model demonstrates potential pitfalls of existing methods. Mol. Endocrinol. 2013;27:127–134. doi: 10.1210/me.2012-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong W.K., Sugii S. Adipose-derived stem cells: fatty potentials for therapy. Int. J. Biochem. Cell Biol. 2013;45:1083–1086. doi: 10.1016/j.biocel.2013.02.013. [DOI] [PubMed] [Google Scholar]
- Rivers L.E., Young K.M., Rizzi M., Jamen F., Psachoulia K., Wade A., Kessaris N., Richardson W.D. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat. Neurosci. 2008;11:1392–1401. doi: 10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeheffer M.S., Birsoy K., Friedman J.M. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Roesch K., Jadhav A.P., Trimarchi J.M., Stadler M.B., Roska B., Sun B.B., Cepko C.L. The transcriptome of retinal Müller glial cells. J. Comp. Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Guertin D.A. Adipocyte lineages: tracing back the origins of fat. Biochim. Biophys. Acta. 2014;1842:340–351. doi: 10.1016/j.bbadis.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Gurmaches J., Hung C.M., Sparks C.A., Tang Y., Li H., Guertin D.A. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T.J., Huang T.L., Tran T.T., Zhang H., Townsend K.L., Shadrach J.L., Cerletti M., McDougall L.E., Giorgadze N., Tchkonia T. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. Proc. Natl. Acad. Sci. USA. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan T., Liu W., Kuang S. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB J. 2013;27:277–287. doi: 10.1096/fj.12-211516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Zeve D., Suh J.M., Bosnakovski D., Kyba M., Hammer R.E., Tallquist M.D., Graff J.M. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–586. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T., Thomou T., Zhu Y., Karagiannides I., Pothoulakis C., Jensen M.D., Kirkland J.L. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013;17:644–656. doi: 10.1016/j.cmet.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.T., Yamamoto Y., Gesta S., Kahn C.R. Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab. 2008;7:410–420. doi: 10.1016/j.cmet.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs S., Harrington A., Liaw L., Small D. Selective expression of an aP2/Fatty Acid Binding Protein 4-Cre transgene in non-adipogenic tissues during embryonic development. Transgenic Res. 2006;15:647–653. doi: 10.1007/s11248-006-9000-z. [DOI] [PubMed] [Google Scholar]
- Zhang X.Q., Afink G.B., Svensson K., Jacobs J.J., Günther T., Forsberg-Nilsson K., van Zoelen E.J., Westermark B., Nistér M. Specific expression in mouse mesoderm- and neural crest-derived tissues of a human PDGFRA promoter/lacZ transgene. Mech. Dev. 1998;70:167–180. doi: 10.1016/s0925-4773(97)00190-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


