Summary
Overexpression of transcription factors has been used to directly reprogram somatic cells into a range of other differentiated cell types, including multipotent neural stem cells (NSCs), that can be used to generate neurons and glia. However, the ability to maintain the NSC state independent of the inducing factors and the identity of the somatic donor cells remain two important unresolved issues in transdifferentiation. Here we used transduction of doxycycline-inducible transcription factors to generate stable tripotent NSCs. The induced NSCs (iNSCs) maintained their characteristics in the absence of exogenous factor expression and were transcriptionally, epigenetically, and functionally similar to primary brain-derived NSCs. Importantly, we also generated tripotent iNSCs from multiple adult cell types, including mature liver and B cells. Our results show that self-maintaining proliferative neural cells can be induced from nonectodermal cells by expressing specific combinations of transcription factors.
Graphical Abstract
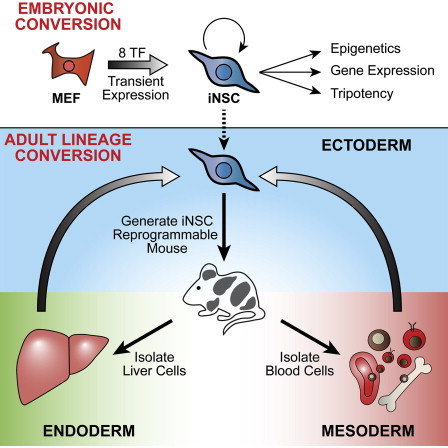
Highlights
-
•
Transgene-independent tripotent neural stem cells are induced from somatic cells
-
•
H3K27ac enhancer profiling shows iNSCs are epigenetically reprogrammed genome-wide
-
•
Adult liver and B cells are lineage converted to iNSCs using a “secondary” system
-
•
iNSCs derived from B lymphocytes have immuloglobulin loci rearrangements
In this article, Jaenisch and colleagues generate induced neural stem cells that are transcriptionally, epigenetically, and functionally similar to primary NSCs by transiently overexpressing transcription factors in fibroblasts. The authors then demonstrate lineage conversion by inducing tripotent iNSCs from multiple adult cell types, including liver cells and B cells with genetic rearrangements.
Introduction
Factor-mediated reprogramming, the process by which overexpression of a defined set of transcription factors converts one cell type into another, has important implications for regenerative medicine and demonstrates the power that transcription factors have as cell fate determinants (Jaenisch and Young, 2008). This has been shown for pluripotent stem cells, where three transcription factors (Oct4, Sox2, and Klf4) are sufficient to induce any cell type to become induced pluripotent stem cells (iPSCs) that are transcriptionally, epigenetically, and functionally indistinguishable from embryonic stem cells (ESCs) (Takahashi and Yamanaka, 2006; Wernig et al., 2008a).
Ectopic expression of key transcription factors in somatic donor cells has been used to generate many different cell types, including cells resembling blood cells (Heyworth et al., 2002; Xie et al., 2004), brown fat cells (Kajimura et al., 2009), hepatocytes (Huang et al., 2011), Sertoli cells (Buganim et al., 2012), and various cell types of the neural lineage (Vierbuchen et al., 2010; Kim et al., 2011; Son et al., 2011; Yang et al., 2013). An important advance has been the generation of neural stem cells (NSCs) from embryonic fibroblasts, as these self-renewing somatic stem cells can be expanded for use in clinical application (Lujan et al., 2012; Han et al., 2012; Ring et al., 2012). However, a number of issues remained unresolved and are the focus of this paper. In these studies, the induced NSCs (iNSCs) were still dependent on the constitutive factor expression. Importantly, because the published studies used a heterogeneous population of mouse embryonic fibroblasts (MEFs) as starting cells, it has been difficult to ascertain that indeed nonneural somatic cells gave rise to the iNSCs rather than preexisting neural cells (such as neural crest cells) present in the donor population. Finally, an extensive epigenetic analysis has not been performed on iNSCs, or any other directly converted cell type, to determine whether the chromatin has been reset to allow for cell type-specific gene expression to persist in the absence of the exogenous factors.
Here we show that transient overexpression of 8 transcription factors—Brn2, Hes1, Hes3, Klf4, Myc, Notch1, (NICD), PLAGL1, and Rfx4—in fibroblasts generates iNSCs that can differentiate into neurons, astrocytes, and oligodendrocytes and have similar genome-wide gene expression patterns and enhancer usage as primary-derived control NSC lines. We then used iPSC technology to generate a genetic system that converts multiple adult cell types to tripotent iNSCs, including liver cells and B cells that carry rearrangements in the immunoglobulin loci. Our results indicate that specific combinations of transcription factors can reset the genome of diverse cell types to that of NSCs.
Results
Induction of iNSCs from MEFs
We generated doxycycline (dox)-inducible lentiviruses (Brambrink et al., 2008) that carried factors thought to be important for NSCs (Bylund et al., 2003; Elkabetz et al., 2008; Imayoshi et al., 2010) and transduced them into fibroblasts carrying a GFP reporter in the Sox2 locus (Sox2-GFP) (Ellis et al., 2004). Initial experiments in which transduced MEFs were cultured in the presence of dox and then monitored for Sox2-GFP expression after dox withdrawal were unsuccessful. However, a modified strategy that included dox withdrawal from cells transiently grown in insulin, transferrin, selenium, and fibronectin (ITSFn) medium, which selects for NESTIN-expressing NSCs during in vitro differentiation of ESCs (Okabe et al., 1996), generated multiple lines of Sox2-GFP+ cells with a morphology resembling that of control NSCs (Figures S1A and S1B available online). Quantitative PCR (qPCR) analysis confirmed that fibroblast genes were silenced and NSC genes were activated (Figure S1C). Importantly, upon growth factor withdrawal, these cell lines differentiated and stained for neuronal marker TUJ1 and glial marker GFAP (Figure S1D).
To determine the factors responsible for generating self-maintaining iNSC lines, we used PCR to identify the proviral transgenes carried in the genome of iNSC lines. We utilized primers that either spanned introns or, for single-exon genes, amplified between the TetO lentiviral promoter and the gene of interest. We found that between 13 and 21 different proviruses had integrated into the various iNSC lines (Figure S1E).
To identify essential factors, we focused on iNSC5, the line with the fewest number of proviruses. MEFs were transduced with the 13 factors carried in iNSC5—Sox2, Hes1, Hes3, Brn2, Klf4, Rfx4, Zic1, DN-REST, NICD, Lhx2, PLAGL1, Myc, and Bmi1—without (13F) or with (14F) Foxg1, which was reported to be important for creating dox-dependent iNSCs (Lujan et al., 2012). To determine the minimal duration of factor expression needed to induce transdifferentiation, the cells were cultured in dox for different lengths of time (Figure 1A). iNSCs appeared only after 30 days on dox in both 13F and 14F cultures (Figure 1A) and displayed morphologies and Sox2-GFP expression patterns similar to brain-derived control NSCs (Figures 1B and 1C). The cells proliferated in response to epidermal growth factor (EGF)/fibroblast growth factor (FGF) signaling and were able to be expanded for more than 20 passages with similar growth properties as control NSCs (Figure 1D). qPCR revealed that iNSCs expressed NSC genes Brn2, Sox2 and Pax6, but had silenced MEF genes such as Col5a2 and Thy1 (Figures 1E).
Figure 1.
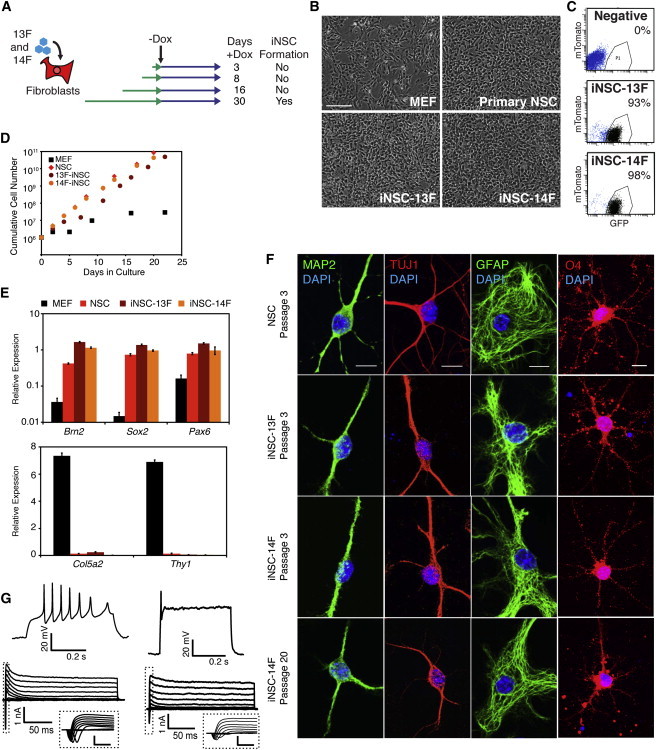
Generation of Transgene-Independent iNSCs
(A) Time course of dox induction. MEFs carrying the Sox2-GFP and Rosa26-M2rtTA alleles were transduced with the 13 factors (13F) present in iNSC5 (see Figure S1E) or the 13 factors plus Foxg1 (14F) and grown in the presence of dox for the indicated lengths of time.
(B) Morphology of MEFs (P1), a primary-derived control NSC line, iNSC-13F, and iNSC-14F (each at P3). The scale bar represents 100 μm.
(C) GFP fluorescence of iNSC-13F and iNSC-14F cultures (P3) compared with cells negative for the Sox2-GFP reporter (negative). mTomato was used to control for autofluorescence.
(D) Growth curves of MEF, control NSC, iNSC-13F, and iNSC-14F cultures. One million cells were plated on day 0 and cultured continuously for the indicated times.
(E) qPCR analysis of NSC (top) and MEF (bottom) transcript expression in MEFs, control NSCs, iNSC-13F, and iNSC-14F (all P2). Expression values were normalized to Gapdh expression for each cell type. Error bars represent SD (n = 3 technical replicates).
(F) Immunostaining for differentiation markers MAP2, TUJ1 (neurons), GFAP (astrocytes), and O4 (oligodendrocytes) after 10–14 days of differentiation. The scale bar represents 10 μm.
(G) Whole-cell patch clamp recordings from two different iNSC-14F-derived neurons after 4 weeks of differentiation. Evoked action potential(s) in response to an injected depolarizing current pulse (top) and leak subtracted current responses in voltage clamp during graded voltage steps (bottom) are shown. Insets represent initial current response (dotted line) showing voltage-gated, transient, inward (putative sodium) currents. The inset scale bar represents 1 nA and 5 ms.
See also Figure S1.
To functionally characterize the iNSCs, we assessed their capacity to differentiate into neurons, astrocytes, and oligodendrocytes by growing them in previously defined conditions (Thier et al., 2012; Lujan et al., 2012). When EGF/FGF were withdrawn from the growth medium and replaced by BDNF, NT3, and ascorbic acid, the cells differentiated into neurons that stained for TUJ1 and MAP2 (Figure 1F). Whole-cell patch clamp recordings revealed that iNSC-derived neurons differentiated for 4 weeks exhibited action potential firing and voltage-gated ion currents, suggesting excitable membrane properties typical of developing neurons (Figure 1G). The iNSCs also differentiated into GFAP+ astrocytes when the medium was supplemented with 5% serum, and growth in oligodendrocyte-inducing conditions (Glaser et al., 2007) promoted differentiation to O4+ oligodendrocytes (Figure 1F). Control NSCs were differentiated in parallel and showed similar results for each differentiation condition (Figure 1F). Finally, to determine whether iNSCs remained tripotent after numerous passages, passage (P) 20 iNSCs were subjected to the same differentiation conditions, and they retained the ability to differentiate into neurons, astrocytes, and oligoendrocytes (Figure 1F). Thus, the iNSCs possessed the self-renewal and tripotency characteristics associated with NSCs.
iNSCs Are Transcriptionally and Epigenetically Similar to Primary NSCs
Global gene expression and chromatin analysis were performed to characterize the iNSCs on a molecular level. Figure 2A shows that iNSCs had similar global gene expression patterns as ESC-derived neural precursor cells (NPCs) (Mikkelsen et al., 2008) and control NSCs derived from the same genetic background as the iNSCs. This was confirmed by hierarchical clustering and Pearson analysis, which showed that iNSC global gene expression patterns clustered with control NSCs and NPCs and were distinct from fibroblasts and differentiated neural cell types (Figure 2B).
Figure 2.
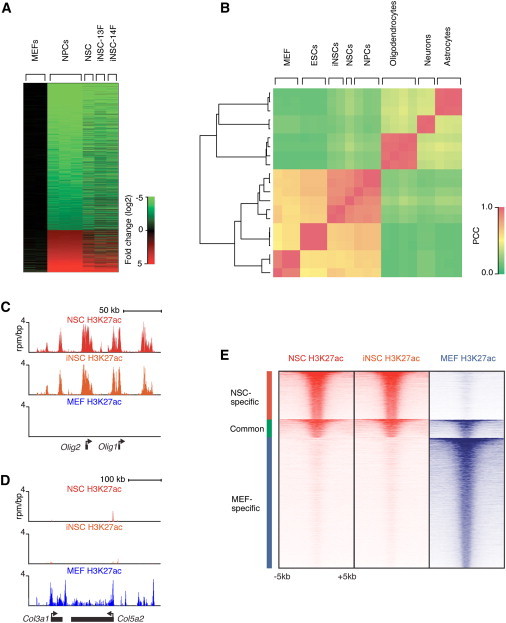
Transcriptional and Epigenetic Reprogramming of iNSCs
(A) Global gene expression analysis comparing the untransduced MEF starting population (P3), primary-derived control NSCs, iNSC-13F, and iNSC-14F (all P8). The heatmap indicates the fold change (log2) of gene expression relative to MEFs. Differentially expressed genes were determined by comparing published ESC-derived NPC and MEF data sets and arranged in rows (Mikkelsen et al., 2008).
(B) Hierarchical clustering and Pearson correlation analysis of gene expression in iNSCs (iNSC-13F and iNSC-14F), control NSCs, ESCs, ESC-derived NPCs, differentiated neural cell lineages, and MEFs (Mikkelsen et al., 2008; Cahoy et al., 2008). The color indicates the Pearson correlation coefficient (PCC).
(C and D) H3K27ac ChIP-seq profiles of the Olig1/Olig2 locus (C) or the Col3a1/Col5a2 locus (D) in control NSCs, iNSC-13F (iNSC) (both P8), and MEFs (P3).
(E) Heat map representation of enhancer-associated histone modification H3K27ac at the union of 16,706 enhancer regions from NSCs and MEFs. The enhancer regions were grouped into NSC-specific, common, and MEF-specific enhancers. Read density surrounds the center (±5 kb) of enhancer regions, rank ordered from highest to lowest average H3K27ac occupancy within each group.
See also Figure S2.
Enhancers are epigenetically marked by histone H3 lysine K27 acetylation (H3K27ac) and display unique cell type-specific profiles (Creyghton et al., 2010). To examine the epigenetic state of iNSCs, H3K27ac chromatin immunoprecipitation (ChIP)-seq was performed on MEFs, control NSCs, and iNSCs. Analysis revealed that iNSCs had a profile similar to control NSCs at many key neural loci such as Olig1/Olig2 (Figure 2C), but dissimilar from MEFs at loci expressed in fibroblasts like Col3a1/Col5a2 (Figure 2D). Finally, genome-wide analysis showed that iNSCs had a global active enhancer pattern similar to control NSCs and different from the starting population of MEFs (Figure 2E). This was confirmed for a number of loci (Figures S2A and S2B). Thus, iNSCs had transcriptionally and epigenetically reprogrammed their nucleus to a state that was very similar to control NSCs and distinct from the MEF starting population.
A Genetically Homogenous System for Efficient iNSC Induction
To assess the reproducibility of iNSC formation, we developed a “secondary” system (Wernig et al., 2008b) in which dox-inducible vectors were carried in all cells of chimeras and allowed transdifferentiation in the absence of virus transduction (Figure 3A). For this, iNSC-14F cells were reprogrammed to pluripotency using retroviral vectors (Figure S3A). The resulting clonal iPSCs (14F-iPS) carried eight transcription factors—Brn2, Hes1, Hes3, Klf4, Myc, NICD, PLAGL1, and Rfx4 (Figure S3B)—and were pluripotent, generating teratomas and contributing to chimera formation (Figures S3C and S3D). Secondary MEFs were isolated from E14.5 chimeras after removal of neural tissue and selection for puromycin resistance. When cultured in the presence of dox, these cells activated the transgenes and readily transdifferentiated into secondary iNSCs, whereas cells that had never been exposed to dox did not (Figures S3E and S3F). Secondary iNSCs displayed similar morphology, growth properties, and gene expression pattern as the original iNSC line and control NSCs (Figures S3F and S3G) and were also capable of differentiating into neurons and glia (Figure S3H).
Figure 3.
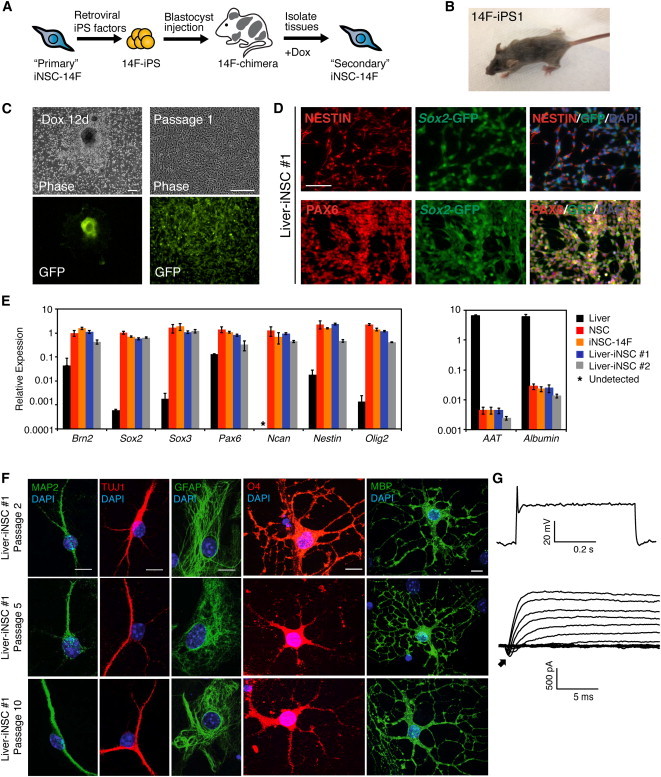
Conversion of Adult Liver Cells to iNSCs
(A) Schematic of strategy to generate an inducible system for generating iNSCs from adult tissues without additional factor transduction.
(B) Adult chimera generated from 14F-iPS1 cell line. The agouti colored hairs arose from the injected iPSCs.
(C) Adult liver cells were explanted from 14F-iPS1 chimera and subjected to iNSC-inducing conditions. Shown are the morphology and Sox2-GFP expression of an outgrowth 12 days after dox withdrawal (left) and also after the first passage (right) of Liver-iNSC #1. The scale bar represents 200 μm.
(D) Immunostaining for NESTIN, PAX6, and endogenous Sox2-GFP expression in Liver-iNSC #1 (P3). The scale bar represents 100 μm.
(E) qPCR analysis of NSC (left) and liver (right) marker gene expression in whole primary liver, control NSCs, iNSC-14F, and secondary iNSCs derived from adult liver cells (Liver-iNSC #1 and Liver-iNSC #2) (iNSCs at P2). Relative expression is normalized to Gapdh for each cell type and the error bars represent SD (n = 3 technical replicates).
(F) Immunostaining for Liver-iNSC #1-derived neurons (MAP2, TUJ1), astrocytes (GFAP), and oligodendrocytes (O4 and MBP) after 10–14 days of differentiation. Differentiation was assayed at passages 2, 5, and 10. The scale bar represents 10 μm.
(G) Whole-cell recording from a Liver-iNSC #1-derived neuron patched in a culture differentiated for 4 weeks and that showed evoked action potentials in response to injected current (top) and low-amplitude voltage-gated currents (bottom). Arrow points to low amplitude, voltage-gated, transient inward currents.
See also Figure S3.
Conversion of Adult Liver Cells to iNSCs
Although the secondary system is a robust tool for converting MEFs to iNSCs, control-treated MEF cultures contain up to 1% Sox2-GFP+ cells (Lujan et al., 2012; data not shown), which may indicate the presence of neural crest-derived cells in this heterogeneous cell population. Thus, it is possible that the iNSCs were generated from the selection of preexisting neural crest-derived cells rather than by transdifferentiation. To exclude this possibility, we next sought to convert adult liver cells to iNSCs using the secondary system. The liver of an adult 14F-iPS chimera was dissociated and cultured in the presence of dox for iNSC formation (Figures 3B and S3E). The resulting iNSC lines displayed similar morphologies and NESTIN, PAX6, and Sox2-GFP expression patterns as control NSCs and other iNSC lines (Figures 3C and 3D). qPCR analysis showed that the liver-derived iNSC lines expressed the NSC markers Brn2, Sox2, Sox3, Pax6, Ncan, Nestin, and Olig2 to a similar level as control NSCs, but did not express AAT or Albumin, which are expressed in liver cells (Figure 3E).
Differentiation analysis confirmed that liver-derived iNSCs were tripotent. The cells differentiated into TUJ1+ and MAP2+ neurons that were capable of generating action potentials (Figures 3F and 3G). In the presence of serum, they uniformly differentiated into GFAP+ astrocytes (Figure 3G). In oligodendrocyte-inducing conditions, cells assumed oligodendrocyte-like morphologies and stained for O4 and myelin basic protein (MBP) (Figure 3F). Importantly, the cells remained tripotent at P5 and P10 (Figure 3G). Thus, iNSCs could be generated from adult, nonneural crest-derived cells and had similar molecular features as lines generated from embryonic fibroblasts.
Conversion of B Lymphocytes to iNSCs
Because the liver contains numerous distinct cell types, the molecular identity of the iNSC donor cell remains unknown. To determine whether iNSCs could be induced from a well-defined cell type like B lymphocytes, spleen and bone marrow (BM) cells were isolated from a 14F-iPS chimera and cultured for iNSC formation. Multiple Sox2-GFP expressing iNSC lines were generated upon dox induction, whereas none were derived from control-treated cultures (Figure 4A). These iNSC lines had typical NSC morphologies and stained for NESTIN and PAX6 (Figure 4B; data not shown). qPCR analysis confirmed that BM- and spleen-derived iNSCs endogenously expressed the neural transcripts Brn2, Sox2, Sox3, Nestin, Ncan, and Olig2, but not blood cell-associated genes Pu.1 (Spi1) or Pax5 (Figure 4C). To test the multipotency of these cells, BM- and spleen-derived iNSCs were subjected to differentiation analysis. Similar to previously described iNSCs, these lines could produce TUJ1+ and MAP2+ neurons, GFAP+ astrocytes, and O1+ and O4+ oligodendrocytes (Figures 4D and S4A; data not shown). Finally, whole-cell patch clamp recordings showed that BM-derived iNSCs could differentiate into neurons with excitable membrane properties, such as the expression of voltage-gated ion channels and the ability to generate an action potential (Figure 4E) (note that spleen-derived iNSC neurons were not assessed for membrane properties).
Figure 4.
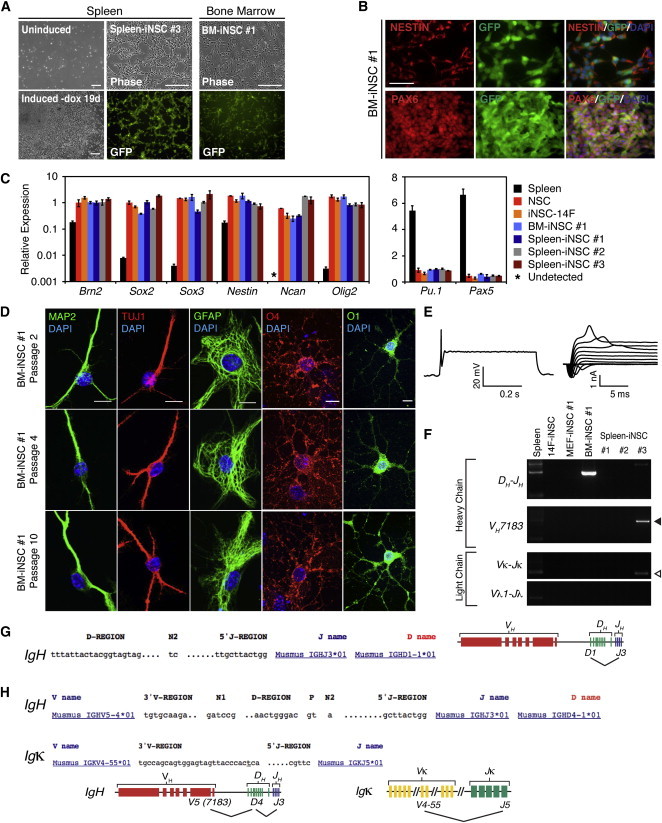
Conversion of Blood Cells to iNSCs
(A) Spleen and BM cells were isolated from 14F-iPS1 adult chimeras. (Left) Comparison of mock-treated spleen cultures (uninduced) and cultures subjected to NSC formation (19 days after dox withdrawal) (Spleen-iNSC #1 is shown). Morphology and GFP fluorescence of Spleen-iNSC #3 and BM-iNSC #1 are shown in the middle and right, respectively. The scale bar represents 200 μm.
(B) Immunostaining for NESTIN, PAX6, and endogenous Sox2-GFP expression in BM-iNSC #1 (P3). The scale bar represents 50 μm.
(C) qPCR analysis of NSC (left) and blood (right) marker gene expression in primary spleen, control NSCs, iNSC-14F, and secondary iNSCs derived from BM (BM-iNSC #1) and spleen (Spleen-iNSC #1-3) (iNSCs at P2). Relative expression is normalized to Gapdh for each cell type, and the error bars represent SD (n = 3 technical replicates).
(D) Immunostaining of BM-iNSC #1 cultures at indicated passage differentiated to neurons (MAP2 AND TUJ1), astrocytes (GFAP), or oligodendrocytes (O4 and O1). The scale bar represents 10 μm.
(E) Whole-cell recordings from a BM-iNSC #1-derived neuron patched in a culture differentiated for 4 weeks and that showed evoked action potentials in response to injected current (left) and voltage-gated currents (right).
(F) PCR analysis for heavy-chain (DH-JH, VHJ558-DJH, and VH7183-DJH) and light-chain (Vκ-Jκ and Vλ1-Jλ) recombination events in the genomic DNA of the indicated iNSC lines.
(G) Sequencing analysis of BM-iNSC #1 DH-JH rearrangement. The D and J segments involved in the IgH rearrangement are shown (left), as well as a genomic representation of the locus (right).
(H) Sequencing analysis of productive Spleen-iNSC #3 VH7183-DJH rearrangement (black arrowhead in part F) (top) and Vκ-Jκ rearrangement (white arrowhead in part F) (middle). A genomic representation of the IgH and Igκ rearrangements is shown.
See also Figure S4.
The identity of donor B lymphocytes can be retrospectively ascertained by the DNA rearrangements that occur during maturation (Jung et al., 2006). To determine whether any of the BM- or spleen-derived iNSC lines were induced from a B cell, we isolated genomic DNA and performed PCR analysis for rearrangements of the heavy and light chain immunoglobulin loci (Hanna et al., 2008) (Figure S4B). Analysis of BM-derived line BM-iNSC #1 revealed one DH-JH rearrangement that was confirmed by sequencing (Figures 4F and 4G), which is consistent with this line being derived from a Pro-B cell (Hardy and Hayakawa, 2001). Spleen-iNSC #3 had two heavy-chain rearrangements—one productive in-frame VH-DJH and a second DH-JH rearrangement—in addition to a productive Igκ light-chain locus rearrangement (Figure 4F). The PCR bands were excised, and sequencing confirmed that a donor cell for this iNSC line was a mature B lymphocyte (Figures 4H and S4C) (Hardy and Hayakawa, 2001). Thus, these results confirm that multipotent iNSCs were derived from immature and mature B cells by transdifferentiation.
Discussion
The results described here, using the efficient secondary system to induce transdifferentiation, show that multipotent self-renewing NSCs can be directly derived from a variety of adult somatic donor cells. Our data extend previous reports in three important aspects: (1) Previous studies used heterogeneous MEFs as the donor cell population, which may have contained neural-derived cells. Thus, these data did not exclude the possibility that the iNSCs were derived by outgrowth of these neural cells stimulated by the culture conditions and transduced factors. Here we unambiguously demonstrate, based on immunoglobulin loci rearrangement as a retrospective genetic marker, that iNSCs can be directly derived from B lymphocytes and from adult liver cells. (2) The maintenance of the iNSC state in previous reports was dependent on the continuous expression of the inducing transcription factors. In contrast, the iNSCs described here are independent of transgene expression. (3) An extensive chromatin and gene expression analysis showed that globally the gene expression and epigenetic states are highly similar to that of control NSCs, and thus, the endogenous NSC regulatory network has been stabilized.
Adult somatic cells have been directly reprogrammed to mature neurons (Vierbuchen et al., 2010; Marro et al., 2011; Son et al., 2011). However, since these cells are postmitotic, it is difficult to obtain large numbers of cells required for transplantation therapy. The direct conversion of readily accessible adult somatic cells to NSCs is of clinical significance since NSCs can proliferate and thus allow the generation of large quantities of neurons and glia that could be used for eventual transplantation therapy.
Experimental Procedures
iNSC Reprogramming
For the generation of iNSCs, transduced MEFs were grown for 4 days in MEF medium supplemented with 2 μg/ml dox (Sigma), then in neural induction medium (N2 medium [Okabe et al., 1996] plus 10 ng/ml EGF [R&D Systems], 10 ng/ml bFGF [Sigma], 1 μg/ml laminin [Life Technologies], and 2 μg/ml dox) for 2 to 3 weeks before the addition of ITSFn selection medium (Dulbecco’s modified Eagle’s medium/F-12 medium containing insulin [25 μg/ml], transferrin [50 μg/ml], sodium selenite [30 nM], fibronectin [5 μg/ml; Sigma], and penicillin/streptomycin [100 μg/ml; Life Technologies]) (Okabe et al., 1996), which included dox for the first 4 days. After 10 days in ITSFn (6 days after dox withdrawal), the cells were dissociated and replated onto plates coated with polyornithine (15 μg/ml; Sigma) and laminin (1 μg/ml) and subsequently cultured in neural expansion medium (N2 supplemented with 20 ng/ml EGF, 20 ng/ml bFGF, and 1 μg/ml laminin). Sox2-GFP+ cells were sorted directly into polyornithine and laminin-coated plates and split at confluence for P1. All animal procedures were performed according to NIH guidelines and were approved by the Committee on Animal Care at MIT.
Author Contributions
J.P.C. and R.J. conceived this study and wrote the paper. J.P.C., A.C.D., Z.F.P. and R.A.Y. designed, performed, and analyzed ChIP and microarray experiments. S.S. performed confocal microscopy. J.P.C., V.S.D., and M.S. designed and performed electrophysiology experiments. R.R. assisted with electrophysiology differentiations. K.G. performed microinjections. J.P.C. designed and performed all other experiments.
Acknowledgments
We thank Y. Li, M. Dawlaty, Q. Gao, and J. Hanna for advice; D. Fu and T. Lungjangwa for experimental assistance; and the Whitehead flow cytometry core facility. We are deeply indebted to C. Chen, J. Bland, and J. Huang for their help and encouragement. J.P.C. was supported by a Howard Hughes Medical Institute Gilliam Fellowship. This work was supported by NIH grants HD 045022 and R37CA084198 to R.J., HG002668 to R.Y., and a grant from the Simons Foundation (SFARI 204106 to R.J.). R.J. is a cofounder of Fate Therapeutics and an advisor to Stemgent.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Accession Numbers
The Gene Expression Ominbus (GEO) accession number for the gene expression and ChIP-seq analyses in this paper is GSE61434.
Supplemental Information
References
- Brambrink T., Foreman R., Welstead G.G., Lengner C.J., Wernig M., Suh H., Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Itskovich E., Hu Y.-C., Cheng A.W., Ganz K., Sarkar S., Fu D., Welstead G.G., Page D.C., Jaenisch R. Direct reprogramming of fibroblasts into embryonic Sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B.G., Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N.D., Tabar V., Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P., Fagan B.M., Magness S.T., Hutton S., Taranova O., Hayashi S., McMahon A., Rao M., Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Glaser T., Pollard S.M., Smith A., Brüstle O. Tripotential differentiation of adherently expandable neural stem (NS) cells. PLoS ONE. 2007;2:e298. doi: 10.1371/journal.pone.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.W., Tapia N., Hermann A., Hemmer K., Höing S., Araúzo-Bravo M.J., Zaehres H., Wu G., Frank S., Moritz S. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- Hanna J., Markoulaki S., Schorderet P., Carey B.W., Beard C., Wernig M., Creyghton M.P., Steine E.J., Cassady J.P., Foreman R. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K. B cell development pathways. Annu. Rev. Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- Heyworth C., Pearson S., May G., Enver T. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 2002;21:3770–3781. doi: 10.1093/emboj/cdf368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- Imayoshi I., Sakamoto M., Yamaguchi M., Mori K., Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J. Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R., Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- Kajimura S., Seale P., Kubota K., Lunsford E., Frangioni J.V., Gygi S.P., Spiegelman B.M. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Su S.C., Wang H., Cheng A.W., Cassady J.P., Lodato M.A., Lengner C.J., Chung C.-Y., Dawlaty M.M., Tsai L.H., Jaenisch R. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–419. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Chanda S., Ahlenius H., Südhof T.C., Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S., Pang Z.P., Yang N., Tsai M.-C., Qu K., Chang H.Y., Südhof T.C., Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B.E., Jaenisch R., Lander E.S., Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S., Forsberg-Nilsson K., Spiro A.C., Segal M., McKay R.D. Development of neuronal precursor cells and functional postmitotic neurons from embryonic stem cells in vitro. Mech. Dev. 1996;59:89–102. doi: 10.1016/0925-4773(96)00572-2. [DOI] [PubMed] [Google Scholar]
- Ring K.L., Tong L.M., Balestra M.E., Javier R., Andrews-Zwilling Y., Li G., Walker D., Zhang W.R., Kreitzer A.C., Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son E.Y., Ichida J.K., Wainger B.J., Toma J.S., Rafuse V.F., Woolf C.J., Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Thier M., Wörsdörfer P., Lakes Y.B., Gorris R., Herms S., Opitz T., Seiferling D., Quandel T., Hoffmann P., Nöthen M.M. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M., Meissner A., Cassady J.P., Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M., Lengner C.J., Hanna J., Lodato M.A., Steine E., Foreman R., Staerk J., Markoulaki S., Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yang N., Zuchero J.B., Ahlenius H., Marro S., Ng Y.H., Vierbuchen T., Hawkins J.S., Geissler R., Barres B.A., Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


