Abstract
Schizophrenia (SCZ) is a severe and debilitating mental disorder, and the specific genetic factors that underlie the risk for SCZ remain elusive. The autism susceptibility candidate 2 (AUTS2) gene has been reported to be associated with autism, suicide, alcohol consumption, and heroin dependence. We hypothesized that AUTS2 might be associated with SCZ. In the present study, three polymorphisms (rs6943555, rs7459368, and rs9886351) in the AUTS2 gene were genotyped in 410 patients with SCZ and 435 controls using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and forced PCR-RFLP methods. We detected an association between SCZ and the rs6943555 genotype distribution (odds ratio (OR) = 1.363, 95% confidence interval (CI): 0.848–2.191, p = 0.001). The association remained significant after adjusting for gender, and a significant effect (p = 0.001) was observed among the females. In the present study, rs6943555 was determined to be associated with female SCZ. Our results confirm previous reports which have suggested that rs6943555 might elucidate the pathogenesis of schizophrenia and play an important role in its etiology.
Keywords: AUTS2 gene, SNP, schizophrenia, association study
1. Introduction
Schizophrenia (SCZ) is a severe and debilitating mental disorder with a prevalence of approximately 1% in the general worldwide population [1]. SCZ is globally considered to be the most serious form of mental illness. Not only does SCZ incur psychological and physiological suffering, but it also adversely affects public security. Despite the continuing efforts to identify the etiologies of SCZ, the definite causes of this disorder remain unclear [2,3]; However, epidemiological studies have demonstrated that SCZ is a familiar disorder with a complex mode of inheritance and a heritability that is as high as 80% [4].
Recently, many genome-wide association studies (GWAS) have detected new evidence related to the genetics of SCZ by focusing on single-nucleotide polymorphisms (SNPs) and copy number variation (CNV) [5,6,7,8], and the results have been consistently replicated in association studies [9,10,11,12]. Although GWA studies have provided a promising approach for the study of the genetics of complex diseases, they have not explained the majority of the underlying genetic risks for SCZ. Thus, the identification of individual candidate genes with small effects on disease risks remains important [13].
The autism susceptibility candidate 2 (AUTS2) gene maps to 7q11, spans 1.2 Mb, includes 19 exons, and encodes a nuclear protein that is expressed primarily in the developing brain [14]. Although the function of this protein remains unclear, many studies had revealed that AUTS2 is highly expressed in the neural tube and the embryonic, fetal and adult brain; these findings indicate that AUTS2 might play important roles during neuronal development, and variations of this gene might affect transcriptional regulation and be related to neurodevelopmental diseases [14,15,16].
Many studies have shown that AUTS2 is associated with autism [17,18,19], alcohol consumption [20], heroin dependence [21] and suicide [22]. The following findings are particularly noteworthy: (1) the same chromosomal rearrangements and several single genes, such as the contact in associated protein-like 2 (CNTNAP2) gene, have emerged as genetic risks for autism and SCZ [23]; (2) one study found that SCZ in males and females is associated with 10- and 20-fold increases, respectively, in the risk of suicide and that these risks increase to 30- and 140-fold among men and women, respectively, under the age of 35 years [24]; and (3) one study reviewed the evidence that developmental neuropathologies in hippocampal and prefrontal cortical pathways contribute both to the symptoms of SCZ and to the vulnerability to addictive behavior via dysfunctional interactions with the nucleus accumbens. Thus, drug addiction and SCZ might have overlapping neurobiological substrates within the hippocampus and subcortical structures [25,26,27]. Taken together, these findings indicate that it is vital to determine whether AUTS2 is also associated with SCZ. To test this hypothesis, we conducted a genetic association study in a sample from the Han population.
2. Results
The three genetic variations of the AUTS2 gene were successfully identified in 410 SCZ cases and 435 controls using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) or forced PCR-RFLP in the present study. No significant deviations from Hardy–Weinberg equilibrium (HWE) were found in the cases or controls. The allelic and genotypic frequencies of the three SNPs were shown in Table 1. The estimated risks associated with these polymorphisms in the SCZ patients and the healthy controls were tested according to three models (i.e., dominant, recessive and addictive models) that were also illustrated in Table 1. We performed association analyses for every identified SNP. When all of the samples were considered, the genotype association analysis of SNP1 (rs6943555) under the dominant model revealed a significant result (odds ratio (OR) = 1.363, 95% confidence interval (CI): 0.848–2.191, p = 0.001).
Table 1.
Allelic and genotypic frequencies of the single-nucleotide polymorphism (SNP) association analyze.
| Makers | Allele Frequency (%) | p-Value a | OR b (95% CI) | Genotype Frequency (%) | p-Value a | HWE p-Value | Models | OR b (95% CI) | p-Value a | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNPs | ID | ||||||||||||
| SNP1 | rs6943555 | A | T | 0.097 | AA | AT | TT | 0.001 | 0.496 | ||||
| SCZ | 33.0 | 77.0 | 1.191 | 7.8 | 50.5 | 41.7 | Dominant | 1.363 (0.848–2.191) | 0.001 | ||||
| CTR | 29.3 | 70.7 | (0.969–1.463) | 10.4 | 37.9 | 51.7 | Recessive | 0.734 (0.456–1.180) | 0.201 | ||||
| Addictive | 1.197 (0.971–1.475) | 0.093 | |||||||||||
| SNP2 | rs7459368 | G | A | 0.720 | GG | AG | AA | 0.809 | 0.954 | ||||
| SCZ | 17.4 | 82.6 | 1.047 | 2.7 | 29.5 | 67.8 | Dominant | 1.117 (0.495–2.523) | 0.809 | ||||
| CTR | 16.8 | 83.2 | (0.813–1.349) | 3.0 | 27.6 | 69.4 | Recessive | 0.895 (0.396–2.021) | 0.789 | ||||
| Addictive | 1.048 (0.813–1.351) | 0.719 | |||||||||||
| SNP3 | rs9886351 | G | A | 0.681 | GG | AG | AA | 0.803 | 0.114 | ||||
| SCZ | 27.4 | 72.6 | 0.956 | 8.3 | 38.3 | 53.4 | Dominant | 1.028 (0.632–1.672) | 0.803 | ||||
| CTR | 26.6 | 73.4 | (0.771–1.185) | 8.5 | 36.1 | 55.4 | Recessive | 1.067 (0.662–1.719) | 0.789 | ||||
| Addictive | 1.030 (0.825–1.286) | 0.794 | |||||||||||
SCZ: schizophrenia; CTR: control; OR: odds ratio; CI: confidence interval; SNP1: rs6943555; SNP2: rS7459368; SNP3: rS9886351; HWE: Hardy–Weinberg equilibrium. Significant p-values and HWE p-values were shown in bold font. a: p-values were based on normal chi-square statistics; b: OR refers to the risk allele odds ratio for both the cases and controls.
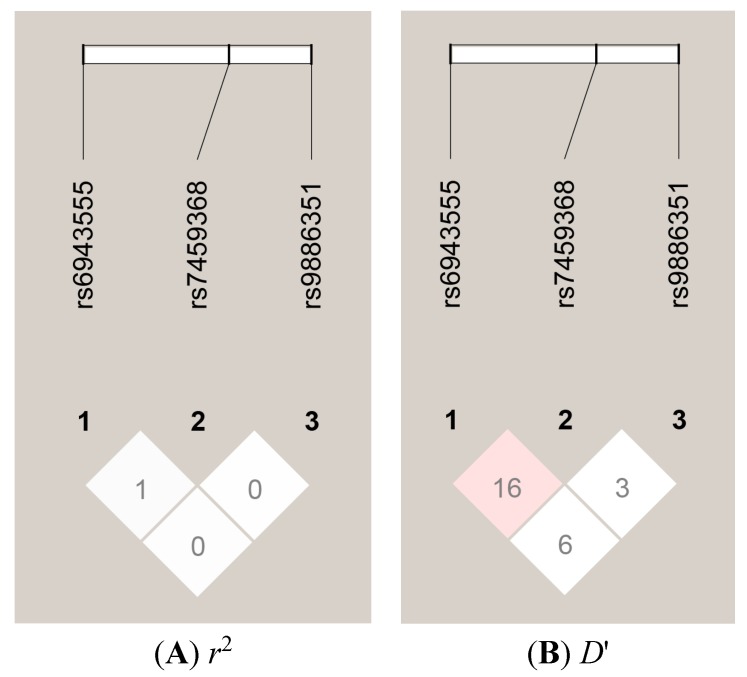
Linkage disequilibrium (LD) estimations for the three SNPs of the AUTS2 gene were performed. We observed that three SNPs exhibited very low D' and r2 values, which suggested that calculations of haplotype frequencies were not required (Figure 1). To examine whether gender influenced this association, we analyzed our data separately for the males and females based of the above results. After dividing the sample according to sex, we found that, among the females, rs6943555 AA homozygotes were over-represented among the SCZ subjects compared to the control subjects (p = 0.001) in females (Table 2).
Figure 1.
Linkage disequilibrium (LD) plots of the three single-nucleotide polymorphisms (SNPs) of the autism susceptibility candidate 2 (AUTS2) gene. The values in the squares are the pair-wise calculations of r2 (A) or D' (B). The white squares with the “0” indicate r2 = 0 (i.e., No LD between a pair of SNPs). The white squares with the “16” indicate D' = 0.16 (i.e., very low LD between a pair of SNPs).
Table 2.
Sex-specific allele and genotype association analyze.
| Marks and Sex | Allele Frequency (%) | p-Value a | OR b (95% CI) | Genotype Frequency (%) | p-Value a | ||||
|---|---|---|---|---|---|---|---|---|---|
| SNP1 | A | T | AA | AT | TT | ||||
| F | SCZ | 35.0 | 65.0 | 0.284 | 1.251 (0.881–1.538) | 8.3 | 53.4 | 38.3 | 0.001 |
| CTR | 30.0 | 70.0 | 12.3 | 35.4 | 52.3 | ||||
| M | SCZ | 31.1 | 68.9 | 0.399 | 1.140 (0.84–1.547) | 7.8 | 41.1 | 51.0 | 0.223 |
| CTR | 28.4 | 71.6 | 7.4 | 47.5 | 45.1 | ||||
| SNP2 | G | A | GG | AG | AA | ||||
| F | SCZ | 17.7 | 82.3 | 0.453 | 0.874 (0.615–1.242) | 3.4 | 28.6 | 68.0 | 0.641 |
| CTR | 15.8 | 84.2 | 2.1 | 27.6 | 70.4 | ||||
| M | SCZ | 17.2 | 82.8 | 0.764 | 1.058 (0.733–1.525) | 2.0 | 30.4 | 67.6 | 0.395 |
| CTR | 18 | 82.0 | 4.2 | 27.6 | 68.2 | ||||
| SNP3 | G | A | GG | AG | AA | ||||
| F | SCZ | 30.1 | 69.9 | 0.119 | 0.791 (0.589–1.062) | 9.2 | 40.8 | 50.0 | 0.283 |
| CTR | 25.1 | 74.9 | 7.5 | 35.3 | 57.2 | ||||
| M | SCZ | 25.2 | 74.8 | 0.318 | 1.174 (0.857–1.608) | 7.3 | 35.8 | 56.9 | 0.596 |
| CTR | 28.4 | 71.6 | 9.9 | 37.0 | 53.1 | ||||
SCZ: schizophrenia; CTR: control; SNP1: rs6943555; SNP2: rS7459368; SNP3: rS9886351; F: female; M: male. Significant p-values are shown in bold font. a: p-values were based the normal chi-square statistics. b: OR refers to the risk allele odds ratio for both the cases and controls.
3. Discussion
The purpose of this study was to investigate the association between the rs6943555, rs7459368, and rs9886351 polymorphisms of the AUTS2 gene and the presence of SCZ. Three SNPs were genotyped by PCR-RFLP or forced PCR-RFLP methods, the latter of which is a very useful, accurate and easy-to-handle strategy that overcomes the weaknesses of complicated technical demands, slow speeds and unstable reproducibilities [28].
We found that rs6943555 of the AUTS2 gene was strongly associated with SCZ and that the rs6943555 AA genotype was associated with a slight increase in the risk of SCZ. Furthermore, when the data were divided according to sex, a significant association was observed between rs6943555 and SCZ in the female population. To the best of our knowledge, this is the first report of a significant association between an AUTS2 gene polymorphism and SCZ. The following lines of evidence suggested that this observed association was unlikely to be an artifact. First, both the findings regarding rs6943555 in the entire samples and the sex-specific association analyses support the association. Second, our subjects were from the same geographical region and this geographic matching served as a good proxy for genetic matching.
The AUTS2 gene is a novel gene that is initially linked to neurodevelopmental disorders in a pair of autistic monozygotic twin sisters [29]. Following the report of this finding, many studies have found structural variants that disrupt the AUTS2 region and are related to intellectual disabilities and developmental delays [30,31]. Additional structural variants of AUTS2, some of which are intronic, have also been shown to be associated with attention deficit hyperactivity disorder (ADHD) [32] epilepsy [33,34], dyslexia [30], language delay, visual impairment, microcephaly and other conditions. Interesting, one recent study identified the intronic SNP rs6943555 in the fourth intron of the AUTS2 to be the SNP that is most strongly associated with alcohol consumption [20]. Furthermore, another study verified that rs6943555 is significantly associated with heroin dependence [21]. Previous studies have revealed that drug addiction and SCZ might have overlapping neurobiological substrates within the hippocampus and subcortical structures, which suggests that SCZ patients are at an increased risk of developing substance use disorders [26,27]. Conversely, individuals at elevated risks for substance use disorders might be particularly vulnerable to SCZ [35]. The direction of the observed association between rs6943555 and SCZ observed in this study is consistent with associations with heroin dependence [21] and suicide committed under influence of ethanol [22] that have been reported previously. The OR for the rs6943555 A allele in relation to SCZ was 1.191 in our data, and other studies have reported ORs of 1.24 for heroin dependence [21] and 1.17 for suicide under the influence of ethanol [22]. Furthermore, we found that the AA homozygote of rs6943555 was significantly over-represented in the SCZ subjects (p = 0.001), which is similar to the finding that has been reported for heroin-dependent subjects (p = 0.017).
Interestingly, we compared the allelic frequencies of rs6943555 with those from HapMap [36] and found that the frequencies in the control subjects were lower than those in the HapMap HCB (HapMap Han Chinese in Beijing) population (0.29 in the current sample and 0.37 in the HapMap HCB population). This difference might be due to the difference in the sample size of the HapMap database (n = 78) and our sample size (n = 435). Although there might not be a simple interpretation of these differences, more reliable data regarding the SNPs in the Han Chinese population will be obtained in future studies.
There are several limitations to this study. Although rs6943555 was associated with SCZ in the present study, it remains unclear whether this is a primary association or one of the effect markers of the AUTS2 gene. It is also possible that multiple variants in AUTS2 had independent effects on SCZ. Because we tested only three small LD markers, we were unable to answer this question. The cross-sectional nature of this study precluded the establishment of causality. Future research is needed to replicate our findings with longitudinal analyses. Moreover, the small SCZ sample size might have limited the detection of weaker relationships in volume, but we found clear between-group differences with respect to shape abnormalities. Thus, future research is needed to examine whether additional SNPs of the AUTS2 gene are associated with SCZ.
4. Materials and Methods
In the present study, the sample included 410 patients with SCZ (204 males and 206 females, mean age: 35.2 ± 11.4 years) and 435 healthy controls (192 males and 243 females, mean age: 37.6 ± 10.8 years) who enrolled from the Mental Health Center and Medical Examination Center of First Affiliated Hospital, Xi’an Jiaotong University. Experienced psychiatrists assessed each subject using the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID). Detailed information, including the presence of mental retardation, personality disorders, and a brief description of the subject’s psychosocial and occupational functioning during the course of the illness, were recorded for SCZ diagnoses. Patients with SCZ were interviewed independently by two experienced psychiatrists to ensure that they met the Diagnostic and Statistical Manual of Mental Disorders, 4th revision (DSM-IV) criteria for SCZ. These were interview assessments of personal histories, hospital records, and family-history reports. Research subjects with diagnoses of substance-induced psychotic disorders, learning disabilities, head injuries, epilepsy, mood disorders, mental retardation or other symptomatic psychoses were excluded from this study. The controls were confirmed to lack any mental illness and were matched with patients in terms of origin, sex, age, and education level.
All participants were volunteers and longstanding residents of Shanxi province and provided written informed consent prior to inclusion. The study protocol was approved by the institutional review board of the Xi’an Jiaotong University College of medicine, with project identification code (2011-054).
4.1. Single-Nucleotide Polymorphism (SNP) Selection
The rs6943555 SNPs of the AUTS2 gene have been reported to be associated with alcohol consumption at a genome-wide level of significance (p = 4 × 10−8 to p = 4 × 10−9) [20], heroin dependence (p = 0.017) [21], and suicide (p = 0.018) [22]. In the current study, rs6943555 was selected as the candidate SNP. Furthermore, we conducted preliminary analysis using the HapMap data to select rs6943555-adjacent tagSNPs. The selected adjacent tagSNPs met the following criteria [37]. First, we examined tagSNPs in Haploview (v4.2) using the CHB population and a minor allele frequency cut-off (MAF) > 5%. Second, a MAF > 20% with pair-wise tagging and r2 > 0.8 [35] were used as cut-off for the selection of tagSNPs. Thus, rs7459368 and rs9886351 were selected as the tagSNPs in blocks that were adjacent to rs6943555. In total, three SNPs, i.e., rs6943555, rs7459368 and rs9886351, were genotyped in this study.
4.2. Genotyping
PCR primers were designed according to the NCBI database (NC_000007.14). Three polymorphisms in the AUTS2 gene were genotyped with PCR-RFLP or forced PCR-RFLP methods, and one of the primers contained one- or two-nucleotide mismatches to enable us to use restriction enzymes to discriminate sequence variations [28]. The primers, selected restriction enzymes (Fermentas, Thermo Scientific, Beijing, China), and fragment sizes are given in Table 3.
Table 3.
Primers used to identify genetic variants of the AUTS2 gene.
| SNPs | Primers (5'–3') | Sizes (bp) | T (°C) | Enzymes/Regions | T (°C) |
|---|---|---|---|---|---|
| SNP1 | F: 5'-TGGGTGTTGGAAGAGTTTTGA-3'; R: 5'-ATACAGTATACATAAACATTGGAAAAGAGGGAA-3' | 196 | 60 | Hinf1, G▼ANTC | 37 |
| SNP2 | F: 5'-AAAGTTCTGGACAGTGGTGCTC-3'; R: 5'-TTCTGACAGTGCGTAAAGGTTG-3' | 257 | 65 | Msp1, C▼CGG | 37 |
| SNP3 | F: 5'-GGTGGAAAATAAGCCAGTATGC-3'; R: 5'-TAGGAAAATGGATTAAACGTAGGAG-3' | 221 | 65 | Hinf1, G▼ANTC | 37 |
SNP: single nucleotide polymorphism; SNP1: rs6943555; SNP2: rs7459368; SNP3: rs9886351; SNP1: G is the mismatched base; SNP3: A is the mismatched base.
The 15-μL PCR reaction volumes contained the following: 50 ng genomic DNA, 10 pmol of each primer, 7.5 μL SuperMixTaq (RUNDE, Shaanxi, China), ddH2O (produced by our lab) to reach a volume of 15 μL. The PCR protocol was 95 °C for 5 min followed by 35 cycles of 95 °C for 30 s, annealing of each primer pair at 60 °C/65 °C/65 °C for 30 s, 72 °C for 30 s and a final extension at 72 °C for 10 min. After the PCR reaction, aliquots of 10 μL PCR product from each pair of primers were digested with 10 U Hinf1 (SNP1)/Msp1 (SNP2)/Hinf1 (SNP3) for 8 h at the same temperature of 37 °C (restriction enzymes including Hinf1 and Map1 were from Fermentas, Thermo Scientific, Beijing, China). The digested products were detected by electrophoresis and were stained with ethidium bromide after approximately 1–2 h in 3% agarose gel (GENE TECH, Shanghai, China).
4.3. Statistical Analyses
The statistical power of our sample size was calculated with the G*Power program (Franz Faul, University Kiel, Kiel, Germany) according to Cohen’s method [38]. Our sample size exhibited >80% power for the detection of significant (p < 0.05) associations among the genotypes, alleles and haplotypes at an effect size index of 0.1 (corresponding to a “weak” gene effect).
In the present study, the Hardy–Weinberg equilibrium (HWE) of the genotype counts was tested with SHEsis (Yong YONG, Shanghai Jiao Tong University, Shanghai, China) [39].The differences in the frequencies of the alleles, genotypes and haplotypes between the cases and controls were evaluated with chi-square analyses. The genotypes, haplotypes and SCZ associations were estimated by computing the odds ratios (ORs) and their 95% confidence intervals (CIs) using logistic regression. Pair-wise linkage disequilibrium (LD) statistics (D' and r2) and haplotype frequencies were computed with Haploview 4.2 (Harvard University, Boston, MA, USA) [40] to construct haplotype blocks and to evaluate deviations from HWE. The haplotype blocks were defined according to the criteria of Gabriel [41]. Furthermore, stratified analyses were conducted to detect whether differences in genes and gender influenced any such associations. All statistical analyses were performed using SPSS 18.0 software (SPSS Inc., Chicago, IL, USA).
5. Conclusions
Our findings suggested that rs6943555 of the AUTS2 gene is associated with SCZ in Han Chinese patients and indicate that the AUTS2 gene might play an important role in the etiology of SCZ. Given the complex patterns offindings from association studies that have focused on SCZ and its underlying genetic heterogeneity, further studies and replications with larger samples, particularly samples of different ethnic groups, are required.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 31301949), the National Science Foundation for Post-Doctoral Scientists of China (Grant No. 2013M532056), and the National Science Foundation for Post-Doctoral Scientists of Shanxi.
Author Contributions
The author is indebted to Bao Zhang for conception, design, drafting the article and revising it critically for important intellectual content; to Yue-Hong Xu for acquisition of data; to Shu-Guang Wei and Hong-Bo Zhang for English-polishing of this document; to Dong-Ke Fu and Zu-Fei Feng for helpful discussions; to Fang-Lin Guan and Yong-Sheng Zhu analysis and interpretation of data; to Sheng-Bin Li acquisition of funding, and general supervision of the research group.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Fatemi S.H., Folsom T.D. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr. Bull. 2009;35:528–548. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldman A.L., Pezawas L., Mattay V.S., Fischl B., Verchinski B.A., Zoltick B., Weinberger D.R., Meyer-Lindenberg A. Heritability of brain morphology related to schizophrenia: A large-scale automated magnetic resonance imaging segmentation study. Biol. Psychiatry. 2008;63:475–483. doi: 10.1016/j.biopsych.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Zhang C., Fang Y., Xie B., Cheng W., Du Y., Wang D., Yu S. DNA methyltransferase 3B gene increases risk of early onset schizophrenia. Neurosci. Lett. 2009;462:308–311. doi: 10.1016/j.neulet.2009.06.085. [DOI] [PubMed] [Google Scholar]
- 4.Wirgenes K.V., Sonderby I.E., Haukvik U.K., Mattingsdal M., Tesli M., Athanasiu L., Sundet K., Rossberg J.I., Dale A.M., Brown A.A., et al. Tcf4 sequence variants and mrna levels are associated with neurodevelopmental characteristics in psychotic disorders. Transl. Psychiatry. 2012;2:e112. doi: 10.1038/tp.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Need A.C., Ge D., Weale M.E., Maia J., Feng S., Heinzen E.L., Shianna K.V., Yoon W., Kasperaviciute D., Gennarelli M., et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Donovan M.C., Craddock N., Norton N., Williams H., Peirce T., Moskvina V., Nikolov I., Hamshere M., Carroll L., Georgieva L., et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 7.Walsh T., McClellan J.M., McCarthy S.E., Addington A.M., Pierce S.B., Cooper G.M., Nord A.S., Kusenda M., Malhotra D., Bhandari A., et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 8.Williams H.J., Owen M.J., O’Donovan M.C. New findings from genetic association studies of schizophrenia. J. Hum. Genet. 2009;54:9–14. doi: 10.1038/jhg.2008.7. [DOI] [PubMed] [Google Scholar]
- 9.Lencz T., Morgan T.V., Athanasiou M., Dain B., Reed C.R., Kane J.M., Kucherlapati R., Malhotra A.K. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol. Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan P.F., Lin D., Tzeng J.Y., van den Oord E., Perkins D., Stroup T.S., Wagner M., Lee S., Wright F.A., Zou F., et al. Genomewide association for schizophrenia in the catie study: Results of stage 1. Mol. Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue W.H., Wang H.F., Sun L.D., Tang F.L., Liu Z.H., Zhang H.X., Li W.Q., Zhang Y.L., Zhang Y., Ma C.C., et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in hanchinese at 11p11.2. Nat. Genet. 2011;43:1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- 12.Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.Y., Duan J., Ophoff R.A., Andreassen O.A., et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guan F., Wei S., Feng J., Zhang C., Xing B., Zhang H., Gao C., Yang H., Li S. Association study of a new schizophrenia susceptibility locus of 10q24.32–33 in a hanchinese population. Schizophr. Res. 2012;138:63–68. doi: 10.1016/j.schres.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Bedogni F., Hodge R.D., Nelson B.R., Frederick E.A., Shiba N., Daza R.A., Hevner R.F. Autism susceptibility candidate 2 (AUTS2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr. Patterns. 2010;10:9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 16.Amarillo I.E., Li W.L., Li X., Vilain E., Kantarci S. De novo single exon deletion of AUTS2 in a patient with speech and language disorder: A review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am. J. Med. Genet. 2014;164:958–965. doi: 10.1002/ajmg.a.36393. [DOI] [PubMed] [Google Scholar]
- 17.Bakkaloglu B., O’Roak B.J., Louvi A., Gupta A.R., Abelson J.F., Morgan T.M., Chawarska K., Klin A., Ercan-Sencicek A.G., Stillman A.A., et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-David E., Granot-Hershkovitz E., Monderer-Rothkoff G., Lerer E., Levi S., Yaari M., Ebstein R.P., Yirmiya N., Shifman S. Identification of a functional rare variant in autism using genome-wide screen for monoallelic expression. Hum. Mol. Genet. 2011;20:3632–3641. doi: 10.1093/hmg/ddr283. [DOI] [PubMed] [Google Scholar]
- 19.Huang X.L., Zou Y.S., Maher T.A., Newton S., Milunsky J.M. A de novo balanced translocation breakpoint truncating the autism susceptibility candidate 2 (AUTS2) gene in a patient with autism. Am. J. Med. Genet. 2010;152:2112–2114. doi: 10.1002/ajmg.a.33497. [DOI] [PubMed] [Google Scholar]
- 20.Schumann G., Coin L.J., Lourdusamy A., Charoen P., Berger K.H., Stacey D., Desrivieres S., Aliev F.A., Khan A.A., Amin N., et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. USA. 2011;108:7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y.H., Liao D.L., Lai C.H., Chen C.H. Genetic analysis of AUTS2 as a susceptibility gene of heroin dependence. Drug Alcohol Depend. 2013;128:238–242. doi: 10.1016/j.drugalcdep.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 22.Chojnicka I., Gajos K., Strawa K., Broda G., Fudalej S., Fudalej M., Stawinski P., Pawlak A., Krajewski P., Wojnar M., et al. Possible association between suicide committed under influence of ethanol and a variant in the AUTS2 gene. PLoS One. 2013 doi: 10.1371/journal.pone.0057199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burbach J.P., van der Zwaag B. Contact in the genetics of autism and schizophrenia. Trends Neurosci. 2009;32:69–72. doi: 10.1016/j.tins.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Qin P. The impact of psychiatric illness on suicide: Differences by diagnosis of disorders and by sex and age of subjects. J. Psychiatr. Res. 2011;45:1445–1452. doi: 10.1016/j.jpsychires.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Chambers R.A., Krystal J.H., Self D.W. A neurobiological basis for substance abuse comorbidity in schizophrenia. Biol. Psychiatry. 2001;50:71–83. doi: 10.1016/s0006-3223(01)01134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green A.I., Salomon M.S., Brenner M.J., Rawlins K. Treatment of schizophrenia and comorbid substance use disorder. Curr. Drug Targets CNS Neurol. Disord. 2002;1:129–139. doi: 10.2174/1568007024606230. [DOI] [PubMed] [Google Scholar]
- 27.Smith M.J., Wang L., Cronenwett W., Goldman M.B., Mamah D., Barch D.M., Csernansky J.G. Alcohol use disorders contribute to hippocampal and subcortical shape differences in schizophrenia. Schizophr. Res. 2011;131:174–183. doi: 10.1016/j.schres.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y.Z., Zhan Z.Y., Li X.Y., Wu S.R., Sun Y.J., Xue J., Lan X.Y., Lei C.Z., Zhang C.L., Jia Y.T., et al. SNP and haplotype analysis reveal IGF2 variants associated with growth traits in chineseqinchuan cattle. Mol. Biol. Rep. 2014;41:591–598. doi: 10.1007/s11033-013-2896-5. [DOI] [PubMed] [Google Scholar]
- 29.Sultana R., Yu C.E., Yu J., Munson J., Chen D., Hua W., Estes A., Cortes F., de la Barra F., Yu D., et al. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 30.Girirajan S., Brkanac Z., Coe B.P., Baker C., Vives L., Vu T.H., Shafer N., Bernier R., Ferrero G.B., Silengo M., et al. Relative burden of large cnvs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagamani S.C., Erez A., Ben-Zeev B., Frydman M., Winter S., Zeller R., el-Khechen D., Escobar L., Stankiewicz P., Patel A., et al. Detection of copy-number variation in AUTS2 gene by targeted exonic array cgh in patients with developmental delay and autistic spectrum disorders. Eur. J. Hum. Genet. 2013;21:343–346. doi: 10.1038/ejhg.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elia J., Gai X., Xie H.M., Perin J.C., Geiger E., Glessner J.T., D’Arcy M., deBerardinis R., Frackelton E., Kim C., et al. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry. 2010;15:637–646. doi: 10.1038/mp.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mefford H.C., Muhle H., Ostertag P., von Spiczak S., Buysse K., Baker C., Franke A., Malafosse A., Genton P., Thomas P., et al. Genome-wide copy number variation in epilepsy: Novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Talkowski M.E., Rosenfeld J.A., Blumenthal I., Pillalamarri V., Chiang C., Heilbut A., Ernst C., Hanscom C., Rossin E., Lindgren A.M., et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Bakker P.I., Yelensky R., Pe’er I., Gabriel S.B., Daly M.J., Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 36.NCBI. [(accessed on 21 October 2014)]. Available online: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=6943555.
- 37.Ke X.Y., Cardon L.R. Efficient selective screening of haplotype tag SNPS. Bioinformatics. 2003;19:287–288. doi: 10.1093/bioinformatics/19.2.287. [DOI] [PubMed] [Google Scholar]
- 38.Faul F., Erdfelder E., Lang A.G., Buchner A. G*power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 39.Shi Y.Y., He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 40.Guan F.L., Zhang C., Wei S.G., Zhang H.B., Gong X.M., Feng J.L., Gao C.G., Su R., Yang H.M., Li S.B. Association of PDE4B polymorphisms and schizophrenia in Northwestern Han Chinese. Hum. Genet. 2012;131:1047–1056. doi: 10.1007/s00439-011-1120-8. [DOI] [PubMed] [Google Scholar]
- 41.Gabriel R., von Glasow R., Sander R., Andreae M., Crutzen P.J. Bromide content of sea-salt aerosol particles collected over the Indian Ocean during INDOEX 1999. J. Geophys. Res. 2002;107:8032. [Google Scholar]