Abstract
Genome shuffling technology was used as a novel whole-genome engineering approach to rapidly improve the antimicrobial lipopeptide yield of Bacillus amyloliquefaciens. Comparative proteomic analysis of the parental ES-2-4 and genome-shuffled FMB38 strains was conducted to examine the differentially expressed proteins. The proteome was separated by 2-DE (two dimensional electrophoresis) and analyzed by MS (mass spectrum). In the shuffled strain FMB38, 51 differentially expressed protein spots with higher than two-fold spot density were detected by gel image comparison. Forty-six protein spots were detectable by silver staining and further MS analysis. The results demonstrated that among the 46 protein spots expressed particularly induced in the genome-shuffled mutant, 15 were related to metabolism, five to DNA replication, recombination and repair, six to translation and post-translational modifications, one to cell secretion and signal transduction mechanisms, three to surfactin synthesis, two to energy production and conversion, and 14 to others. All these indicated that the metabolic capability of the mutant was improved by the genome shuffling. The study will enable future detailed investigation of gene expression and function linked with surfactin synthesis. The results of proteome analysis may provide information for metabolic engineering of Bacillus amyloliquefaciens for overproduction of surfactin.
Keywords: Bacillus amyloliquefaciens, genome shuffling, surfactin, proteomics, MALDI-TOF/MS
1. Introduction
Bacillus strains produce many types of bioactive lipopeptides that are synthesized non-ribosomally by a large multifunctional enzyme complex. Of these, the lipopeptide surfactin is well characterized at the genetic level. Surfactin is biosynthesized by three non-ribosomal peptide synthetases, SrfA–C, and by the thioesterase/acyltransferase enzyme SrfD, which initiates this process. Surfactin is a powerful biosurfactant that is known to decrease the surface tension of water [1]. It exerts a detergent-like action on biological membranes, and is distinguished by its emulsifying, foaming, antiviral and anti-mycoplasma activities. Surfactin has many potential applications in plant disease biocontrol [2] and biomedicine [3]. Moreover, lipopeptides are widely used in the food [4] and cosmetics industries [5], and for enhanced oil recovery [6] and bioremediation of oil-contaminated sites [7]. However, the production of antimicrobial peptides in Bacillus is generally less than 1.0 g/L, and even as low as 0.1 g/L for some peptides. Therefore, it is particularly important to improve antimicrobial peptide production in industrially important Bacillus strains. There have been many attempts to increase lipopeptide production, but almost all have focused on the optimization of fermentation [8], isolation and purification [9], or on the regulation of lipopeptide synthesis using genetic engineering methods [10,11]. Although global techniques have been successfully applied to strain improvement, engineering more complex phenotypes requires a more combinatorial approach.
The synthesis of antimicrobial substances in Bacillus species is closely related to the formation of competent cells and sporulation, with these three pathways sharing the same metabolic network [12]. Surfactin biosynthesis and regulation of competent cell formation are also closely linked. Interestingly, the comS gene, which is involved in competent cell formation, is located in the surfactin synthase gene srfA operon. The expression of comS, srfA and several quorum sensing genes is regulated by a complex network, which includes the extracellular ComX protein and the two-component adjustment system ComPA [12]. Phosphorylated ComA (ComA–P) was shown to promote srfA operon expression by binding to the promoter region. CodY and AbrB suppressed srfA operon expression by binding to the srfA promoter [13]. The signaling protein RapC could inhibit srfA expression by dephosphorylating ComA–P. Additionally, YerP could increase the tolerance for surfactin in Bacillus subtilis [14].
Genome shuffling involves generation of mutant strains that have an improved phenotype, followed by multiple rounds of protoplast fusion to allow recombination between genomes. A strain with a high yield of a desired product can rapidly be obtained by genome shuffling without knowledge of the metabolic regulatory mechanism. We previously described the generation of a high-yield recombinant Bacillus amyloliquefaciens F2-38 (FMB38) strain that exhibited 3.5- and 10.3-fold increases in surfactin production in a shake flask and fermenter, respectively, following two rounds of genome shuffling. Comparative analysis of synthetase gene expression was conducted between the parental and shuffled strains using FQ (fluorescent quantitation) RT-PCR. ΔCt (threshold cycle) relative quantitation analysis revealed that surfactin synthetase gene (srfA) expression at the transcriptional level in the F2-38 strain was 15.7-fold greater than in the parental strain ES-2-4 [15]. However, these results only indirectly identified differences at the transcriptional level in the lipid peptide synthetase gene. Because proteins carry out molecular functions and are responsible for almost all the biochemical activitiesof the cell, a real understanding of biological systems requires the direct study of proteins. The rapid development of proteomics technology based on 2-DE, identification by MS, and bioinformatics provides a good platform for large-scale proteomic studies. In this study, we explored the molecular mechanism of high-yield surfactin using comparative proteomics methods to identify the differentially expressed proteins between the parental and mutant strains.
2. Results
2.1. Identification of Differentially Expressed Proteins
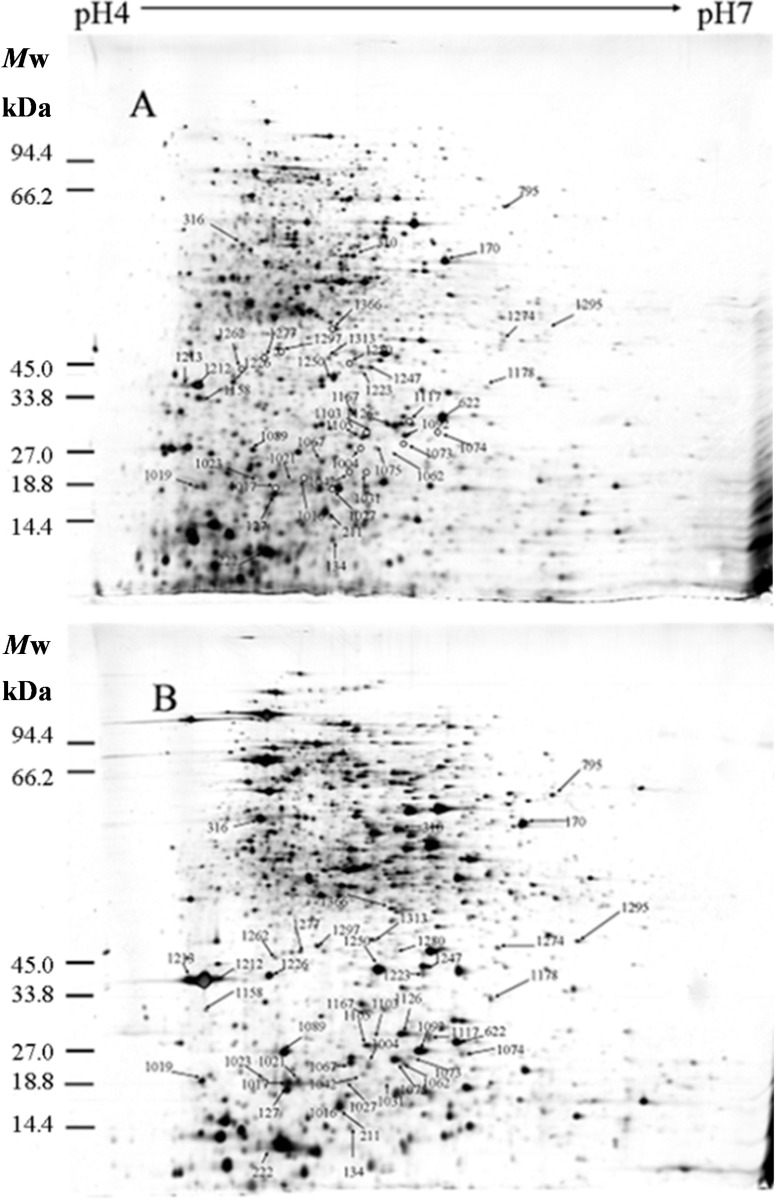
We analyzed the 2-DE profiles of soluble proteins from parental (ES-2-4) and mutant (FMB38) strains of B. amyloliquefaciens and found 51 protein spots that differed between the strains by more than two-fold (Figure 1). These 51 protein spots were identified by MS analysis and their complete peptide fingerprints were obtained. Searching of the NCBI nr database with Mascot revealed that protein spots 795 and 816, 1004 and 1056, 1062 and 1065, 1117 and 1120, and 1213 and 1220 were the same proteins, meaning that a total of 46 proteins were successfully identified. In B. amyloliquefaciens FMB38, 29 proteins had increased expression, two proteins had decreased expression, and 15 proteins appeared only in this strain (Table 1).
Figure 1.
2-DE maps of the differentially regulated cellular proteins (>2-fold change in expression) of Bacillus amyloliquefaciens FMB38. (A) ES-2-4; and (B) FMB38.
Table 1.
Identification of differentially regulated cellular proteins (>2-fold change in expression) of Bacillus amyloliquefaciens FMB38.
| Spot No. a | Protein Name b | Accession No. c | Locus d | Gene e | Theor. f Mr/pI | Exper. g Mr/pI | Protein Score h | Sequence Coverage (%) i | Fold Change j (p < 0.05) |
|---|---|---|---|---|---|---|---|---|---|
| 127 | Alkyl hydroperoxide reductase small subunit | gi|308175696 | YP_003922401 | ahpC | 20,669/4.51 | 21,013/4.65 | 104 | 44 | +15.3 |
| 134 | Conserved hypothetical protein | gi|315173048 | EFU17065 | – | 14,048/5.12 | 13,978/5.10 | 158 | 55 | −18.4 |
| 170 | DNA primase | gi|228983124 | ZP_04143383 | – | 55,979/5.93 | 56,328/5.79 | 115 | 48 | −19.8 |
| 211 | Hypothetical protein RBAM_036960 | gi|154688095 | YP_001423256 | ahpC | 20,683/4.51 | 17,985/4.62 | 126 | 50 | +34.7 |
| 222 | Hypothetical protein KSO_14324 | gi|363725374 | EHM05512 | – | 6927/4.56 | 6843/4.49 | 120 | 70 | +2.7 |
| 310 | Hypothetical protein RBAM_029780 | gi|154687379 | YP_001422540 | yurX | 48,265/5.30 | 48,965/5.33 | 109 | 32 | +29.1 |
| 316 | GroEL gene product | gi|311067075 | YP_003971998 | groEL | 57,385/4.75 | 56,789/4.78 | 190 | 43 | +6. 6 |
| 622 | Response regulator DegU | gi|157693950 | YP_003974978 | degU | 25,893/5.65 | 27,124/5.67 | 249 | 92 | −6.1 |
| 795, 816 | Vegetative catalase 1 | gi|89097371 | ZP_01170260 | – | 54,421/6.11 | 55,135/5.89 | 192 | 40 | +7.5, +4.7 |
| 1004, 1056 | Hypothetical protein RBAM_023340 | gi|154686764 | YP_001421925 | sodA | 22,365/5.21 | 23,214/5.43 | 316 | 80 | in FMB38 |
| 1016 | S-ribosylhomocysteinase | gi|154687196 | YP_001422357 | luxS | 17,913/5.27 | 18,324/5.09 | 98 | 66 | in FMB38 |
| 1017 | Thiol peroxidase | gi|154687070 | YP_001422231 | tpx | 18,262/4.99 | 17,321/4.89 | 180 | 89 | in FMB38 |
| 1019 | Hypothetical protein RBAM_026720 | gi|154687100 | YP_001422261 | yraA | 18,672/4.94 | 17,652/4.63 | 110 | 72 | +26.2 |
| 1021 | Hypothetical protein RBAM_008040 | gi|154685258 | YP_001420419 | yfkM | 18,877/4.83 | 18,896/4.85 | 163 | 85 | +17.9 |
| 1023 | Hypothetical protein RBAM_028130 | gi|154687215 | YP_001422376 | yuaE | 19,112/5.46 | 19,431/4.94 | 112 | 56 | +4.7 |
| 1027 | ATP-dependent Clp protease proteolytic subunit | gi|154687585 | YP_001422746 | clpP | 21,874/4.96 | 19,543/5.12 | 154 | 60 | in FMB38 |
| 1031 | DNA-directed DNA polymerase III α subunit | gi|325684283 | EGD26456 | dnaE | 128,931/8.83 | 19,678/5.34 | 81 | 18 | in FMB38 |
| 1042 | Methionine aminopeptidase, type I | gi|229010975 | ZP_04168170 | – | 27,381/4.89 | 190 | 49 | in FMB38 | |
| 1062, 1065 | Transaldolase | gi|154687826 | YP_001422987 | tal | 23,055/5.23 | 23,336/5.31 | 86 | 40 | +17.9, +6.5 |
| 1067 | Hypothetical protein RBAM_036480 | gi|154688047 | YP_001423208 | deoC | 23,111/4.90 | 23,352/5.08 | 94 | 40 | +24.0 |
| 1073 | Hypothetical protein KSO_05864 | gi|363723690 | EHM03828 | – | 23,280/4.62 | 242,110/5.41 | 112 | 56 | in FMB38 |
| 1074 | 2-Aminoethylphosphonate—Pyruvate transaminase | gi|229166259 | ZP_04294018 | – | 41,797/5.29 | 27,312/5.61 | 220 | 50 | in FMB38 |
| 1075 | Recombinase protein | gi|339764913 | AEK01094 | recA | 24,141/6.39 | 24,234/5.31 | 139 | 74 | +8.3 |
| 1089 | ComA | gi|154687277 | YP_001422438 | comA | 24,371/5.19 | 24,351/4.91 | 280 | 51 | +18.7 |
| 1092 | Transcriptional repressor CodY | gi|154686033 | YP_001421194 | codY | 29,038/4.90 | 28,213/5.4 | 110 | 47 | +7.7 |
| 1103 | Pyrroline-5-carboxylate reductase | gi|228921645 | ZP_04084963 | – | 29,356/5.37 | 28,531/5.31 | 77 | 45 | in FMB38 |
| 1105 | NAD synthetase NadE | gi|302671492 | YP_003831452 | nadE | 29,690/5.00 | 28,921/5.12 | 87 | 37 | +6.7 |
| 1117, 1120 | Fructose-bisphosphate aldolase | gi|154687827 | YP_001422988 | fbaA | 30,537/5.26 | 29,314/5.41 | 103 | 34 | in FMB38 |
| 1126 | Hypothetical protein RBAM_036720 | gi|154688071 | YP_001423232 | iolG | 38,449/5.14 | 37,111/5.41 | 225 | 38 | +18.6 |
| 1158 | Putative glycerol-3-phosphate acyltransferase PlsX | gi|326941622 | AEA17518 | plsX | 35,512/6.27 | 35,212/5.81 | 79 | 51 | +5.1 |
| 1167 | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | gi|157693809 | YP_001488271 | gapA | 35,822/5.03 | 35,344/5.13 | 92 | 29 | +4.0 |
| 1178 | Unnamed protein product | gi|311069921 | YP_003974844 | – | 35,900/5.10 | 35,212/5.82 | 93 | 33 | +164.7 |
| 1212 | Hypothetical protein RBAM_006650 | gi|154685120 | YP_001420281 | ydjL | 37,552/5.09 | 35,242/4.57 | 91 | 29 | +3.5, +4.9 |
| 1213, 1220 | YdjL | gi|363726006 | EHM06144 | – | 37,580/5.04 | 36,899/4.59 | 86 | 31 | +14.6 |
| 1223 | Multifunctional SOS repair factor | gi|308173657 | YP_003920362 | recA | 37,938/5.05 | 37,432/5.21 | 148 | 57 | +53.3 |
| 1226 | 30S ribosomal protein S1 | gi|328552793 | AEB23285 | rpsA | 41,960/4.82 | 41,556/4.79 | 81 | 22 | +16.8 |
| 1247 | Elongation factor Tu | gi|154684631 | YP_001419792 | tufA | 43,500/4.84 | 43,211/5.23 | 96 | 37 | +5.5 |
| 1250 | Hypothetical protein Dtox_1245 | gi|258514528 | YP_003190750 | – | 44,823/5.15 | 44,112/5.16 | 81 | 37 | +12.1 |
| 1262 | Site-specific recombinase XerD | gi|295101383 | CBK98928 | – | 46,358/9.08 | 45,212/4.81 | 77 | 43 | in FMB38 |
| 1274 | Plasmid recombination protein | gi|10956056 | NP_042279 | pre | 49,739/5.24 | 48,677/5.84 | 136 | 48 | +11.5 |
| 1277 | Hypothetical protein RBAM_007590 | gi|154685214 | YP_001420375 | yfmT | 53,319/5.26 | 52,695/4.82 | 113 | 29 | in FMB38 |
| 1280 | F0F1 ATP synthase subunit α | gi|154687798 | YP_001422959 | atpA | 54,804/5.34 | 113 | 26 | in FMB38 | |
| 1295 | Galactose-1-phosphate uridylyltransferase | gi|363725414 | EHM05552 | – | 56,804/6.27 | 56,123/6.16 | 301 | 54 | +9.2 |
| 1297 | Phosphopyruvate hydratase | gi|154687527 | YP_001422688 | eno | 46,645/4.68 | 46,131/4.85 | 300 | 61 | in FMB38 |
| 1313 | Dak2 domain fusion protein ylov | gi|312135039 | YP_004002377 | – | 60,930/4.99 | 43,123/5.10 | 77 | 23 | +7.0 |
| 1366 | M6 family metalloprotease | gi|172058939 | YP_001815399 | – | 86,115/5.33 | 48,531/5.21 | 91 | 21 | in FMB38 |
a Spot numbers assigned by the software refer to the proteins labeled in Figure 1; b Protein name in the National Center for Biotechnology Information (NCBI) database for Bacillus amyloliquefaciens; c Accession number in the NCBI database for Bacillus amyloliquefaciens; d The specific location of a gene or DNA sequence on of the Bacillus amyloliquefaciens chromosome; e Gene designation in the NCBI database for Bacillus amyloliquefaciens; f Theoretical molecular mass (Mr) and isoelectric point (pI) were obtained from the protein database in the NCBI database for Bacillus amyloliquefaciens; g Experimental molecular mass (Mr) and isoelectric point (pI) were obtained from the 2-DE gels; h MASCOT protein score from MS; i Percentage of amino acids in reference proteins covered by matching peptides from MS; and j Fold change: positive values represent over-expressed proteins, negative values represent under-expressed proteins, “in FMB38” indicates that the protein appeared only in high-yield strain FMB38.
2.2. Isoelectric Point and Molecular Weight Analysis of Theoretically and Experimentally Identified Proteins
Isoelectric points and molecular weights of the identified proteins were determined using analysis software (ImageMaster 2D). Samples were compared with the migration distances of the molecular weight marker, and the pH of the IPG (immobilized pH gradient) strips was used to determine the isoelectric point of the protein spots in the silver stained gel. These values were compared with proteins in the database to assist with identification. Overall, the gel estimated molecular weight and isoelectric point of each protein closely matched the theoretical values, but there were some proteins for which the estimated molecular weight (spots 1031, 1074, 1313, and 1366) or isoelectric point (spots 1023, 1031, 1073, 1074, 1075, and 1262) differed greatly from the theoretical value (Table 1).
2.3. Cellular Localization Analysis of Experimentally Identified Proteins
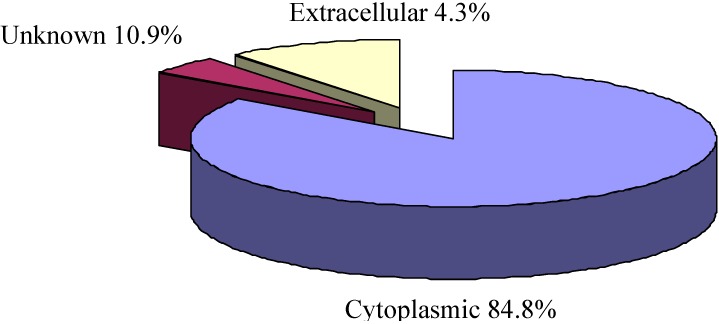
PSORTb version 3.0.1 (http: //www.psort.org/psortb/index.html) was used to predict the cellular localization of the 46 identified proteins (Table 2). Thirty-nine proteins localized to the cytoplasm, two proteins were extracellular, and five proteins had an unknown cellular location (Table 2). The separated proteins are mainly the proteins in the cytoplasm, accounting for 84.8% of the total proteins (Figure 2).
Table 2.
Cellular localization and function of differentially regulated cellular proteins (>2-fold change in expression) of Bacillus amyloliquefaciens FMB38.
| Spot No. a | Protein Name b | COG c | Cellular Localization d | Biological Process e | Molecular Functional Annotation f |
|---|---|---|---|---|---|
| Energy Production and Conversion | |||||
| 1277 | Hypothetical protein RBAM_007590 | C | Cytoplasmic | Unknown | Oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor |
| 1280 | F0F1 ATP synthase subunit α | C | Cytoplasmic | ATP hydrolysis coupled proton transport, plasma membrane ATP synthesis Coupled proton transport | Hydrogen ion transporting ATP synthase activity, rotational mechanism; hydrolase activity |
| Cell Division and Chromosome Partitioning | |||||
| 1274 | Plasmid recombination protein | D | Cytoplasmic | DNA recombination | DNA binding |
| Amino Acid Transport and Metabolism | |||||
| 1074 | 2-Aminoethylphosphonate—Pyruvate transaminase | E | Cytoplasmic | Organic phosphonate catabolic process | 2-Aminoethylphosphonate-pyruvate transaminase activity, pyridoxal phosphate binding |
| 1103 | Pyrroline-5-carboxylate reductase | E | Cytoplasmic | Proline biosynthetic process | Nucleotide binding, oxidoreductase activity, acting on the CH–OH group of donors, NAD or NADP as acceptor, pyrroline-5-carboxylate reductase activity |
| 1212 | Hypothetical protein RBAM_006650 | ER | Cytoplasmic | Unknown | Nucleotide binding, oxidoreductase activity, zinc ion binding |
| 1213, 1220 | YdjL | ER | Cytoplasmic | Unknown | Nucleotide binding, oxidoreductase activity, zinc ion binding |
| Nucleotide Transport and Metabolism | |||||
| 1067 | Hypothetical protein RBAM_036480 | F | Cytoplasmic | Deoxyribonucleotide catabolic process | Deoxyribose-phosphate aldolase activity |
| Carbohydrate Transport and Metabolism | |||||
| 1062, 1065 | Transaldolase | G | Cytoplasmic | Pentose-phosphate shunt | Sedoheptulose-7-phosphate: d-glyceraldehyde-3-phosphate glyceronetransferase activity |
| 1117, 1120 | Fructose-bisphosphate aldolase | G | Cytoplasmic | Fructose 1,6-bisphosphate metabolic process, glycolysis, sporulation resulting in formation of a cellular spore | Fructose-bisphosphate aldolase activity, zinc ion binding |
| 1167 | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating) | G | Cytoplasmic | Glycolysis | NAD binding; NADP binding, glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity |
| 1178 | Unnamed protein product | G | Cytoplasmic | Unknown | Glyceraldehyde-3-phosphate dehydrogenase/erythrose-4-phosphate dehydrogenase |
| 1297 | Phosphopyruvate hydratase | G | Cytoplasmic | Glycolysis | Magnesium ion binding, phosphopyruvate hydratase activity |
| Coenzyme Metabolism | |||||
| 1105 | NAD synthetase NadE | H | Unknown | NAD biosynthetic process, response to stress, sporulation resulting in formation of a cellular spore | ATP binding, NAD+ synthase (glutamine-hydrolyzing) activity, NAD+ synthase activity |
| Lipid Metabolism | |||||
| 1158 | Putative glycerol-3-phosphate acyltransferase PlsX | I | Unknown | Phospholipid biosynthetic process | Transferase activity, transferring acyl groups other than amino-acyl groups |
| Translation, Ribosomal Structure, and Biogenesis | |||||
| 1042 | Methionine aminopeptidase, type I | J | Cytoplasmic | Protein initiator methionine removal, proteolysis | Ferrous iron binding, metalloaminopeptidase activity |
| 1226 | 30S ribosomal protein S1 | J | Cytoplasmic | Translation | RNA binding, structural constituent of ribosome |
| 1247 | Elongation factor Tu | JE | Cytoplasmic | Response to antibiotic | GTP binding, GTPase activity, protein binding, translation elongation factor activity |
| DNA Replication | |||||
| 170 | DNA primase | L | Cytoplasmic | DNA replication, synthesis of RNA primer | ATP binding; DNA binding, DNA helicase activity, DNA primase activity, zinc ion binding |
| 1031 | DNA-directed DNA polymerase III α subunit | L | Cytoplasmic | Transcription | Nucleotidyltransferase, transferase |
| 1075 | Recombinase protein | L | Cytoplasmic | DNA recombination, DNA repair, SOS response | ATP binding, DNA-dependent ATPase activity, single-stranded DNA binding |
| 1223 | Multifunctional SOS repair factor | L | Cytoplasmic | DNA recombination, DNA repair, SOS response | ATP binding, DNA-dependent ATPase activity, damaged DNA binding, single-stranded DNA binding |
| 1262 | Site-specific recombinase XerD | L | Cytoplasmic | DNA integration, DNA recombination | DNA binding |
| Cell Motility and Secretion | |||||
| 1027 | ATP-dependent Clp protease proteolytic subunit | NO | Cytoplasmic | Protein metabolic process | ATP binding, ATP-dependent peptidase activity, protein binding |
| Posttranslational Modification | |||||
| 127 | Alkyl hydroperoxide reductase small subunit | O | Cytoplasmic | Unknown | Cytochrome-c peroxidase activity, peroxiredoxin activity |
| 211 | Hypothetical protein RBAM_036960 | O | Cytoplasmic | Unknown | Peroxidase activity; peroxiredoxin activity |
| 316 | GroEL gene product | O | Cytoplasmic | Cellular protein metabolic process | ATP binding |
| Inorganic Ion Transport and Metabolism | |||||
| 795, 816 | Vegetative catalase 1 | P | Cytoplasmic | Response to oxidative stress | Catalase activity, heme binding |
| 1004, 1056 | Hypothetical protein RBAM_023340 | P | Extracellular | Superoxide metabolic process | Metal ion binding, superoxide dismutase activity |
| Secondary Metabolite Biosynthesis, Transport, and Catabolism | |||||
| 1073 | Hypothetical protein KSO_05864 | Q | Cytoplasmic | Peptidyl-pyrromethane cofactor linkage, porphyrin-containing compound biosynthetic process | Hydroxymethylbilane synthase activity |
| General Function Prediction | |||||
| 310 | Hypothetical protein RBAM_029780 | R | Unknown | Iron-sulfur cluster assembly | Unknown |
| 1019 | Hypothetical protein RBAM_026720 | R | Cytoplasmic | Unknown | Hydrolase activity, acting on glycosyl bonds |
| 1021 | Hypothetical protein RBAM_008040 | R | Cytoplasmic | Unknown | Hydrolase activity, acting on glycosyl bonds |
| 1126 | Hypothetical protein RBAM_036720 | R | Cytoplasmic | Inositol Catabolic Process | Inositol 2-dehydrogenase activity, nucleotide binding |
| 1313 | Dak2 domain fusion protein ylov | R | Cytoplasmic | Glycerol metabolic process | Glycerone kinase activity |
| Signal Transduction Mechanisms | |||||
| 622 | Response regulator DegU | TK | Cytoplasmic | Transcription, DNA-dependent | Sequence-specific DNA binding, sequence-specific DNA binding transcription factor activity, two-component response regulator activity |
| 1016 | S-ribosylhomocysteinase | T | Cytoplasmic | Quorum Sensing | iron ion binding, lyase activity |
| 1089 | ComA | TK | Cytoplasmic | Transcription, DNA-dependent | Sequence-specific DNA binding, sequence-specific DNA binding transcription factor activity, two-component response regulator activity |
| Others | |||||
| 134 | Conserved hypothetical protein | – | Unknown | Growth of symbiont in host, protein omooligomerization | ATP binding, ATPase activity, protein binding |
| 222 | Hypothetical protein KSO_14324 | – | Unknown | Unknown | Electron carrier activity, heme binding |
| 1017 | Thiol peroxidase | – | Cytoplasmic | Cellular response to oxidative stress | Thioredoxin peroxidase activity |
| 1023 | Hypothetical protein RBAM_028130 | – | Cytoplasmic | Unknown | Unknown |
| 1092 | Transcriptional repressor CodY | – | Cytoplasmic | Transcription, DNA-dependent | DNA binding, GTP binding, sequence-specific DNA binding, transcription factor activity |
| 1250 | Hypothetical protein Dtox_1245 | – | Cytoplasmic | Unknown | Unknown |
| 1295 | Galactose-1-phosphate uridylyltransferase | – | Cytoplasmic | Galactose Metabolic Process | UDP-glucose: hexose-1-phosphate uridylyltransferase activity |
| 1366 | M6 family metalloprotease | – | Extracellular | Proteolysis | Metallopeptidase activity |
a Spot numbers assigned by the software refer to the proteins labeled in Figure 1; b Protein name in the National Center for Biotechnology Information (NCBI) database for Bacillus amyloliquefaciens; c Cellular localization of proteins; d Clusters of orthologous groups; e Biological process was assigned according to the protein knowledge base (http://www.uniprot.org) [16] for Bacillus amyloliquefaciens; f Molecular functional annotation was assigned according to the protein knowledge base (http://www.uniprot.org) [16] for Bacillus amyloliquefaciens; and g not in clusters of orthologous groups (COG).
Figure 2.
Cellular localization of the differentially expressed proteins identified in Bacillus amyloliquefaciens FMB38 predicted by the PSORTb database.
2.4. Classification and Functional Analysis of Differential Proteins
Experimentally identified proteins were functionally characterized using clusters of orthologous groups (COG) analysis (Table 2). The isolated proteins were mainly divided into the following categories: Energy production and conversion (C); Cell division and chromosome distribution (D); Amino acid transport and metabolism (E); Nucleic acid transport and metabolism (F); Carbon transport and metabolism (G); Coenzyme metabolism (H); Lipid metabolism (I); Translation, ribosomal structure, and biosynthesis (J); DNA replication, recombination, and repair (L); Cell motility and secretion (N); Post-translational modification, protein folding, chaperone proteins (O); Inorganic ion transport and metabolism (P); Secondary metabolites biosynthesis, transport, and catabolism (Q); General function prediction (R); Signal transduction mechanisms (T); and not included in the COG classification (−). To determine the mechanism of increased antimicrobial peptide yield from B. amyloliquefaciens FMB38, biological process and molecular function data were obtained from the UniProKB (http://www.uniprot.org) (accessed on 1 October 2014) database [16].
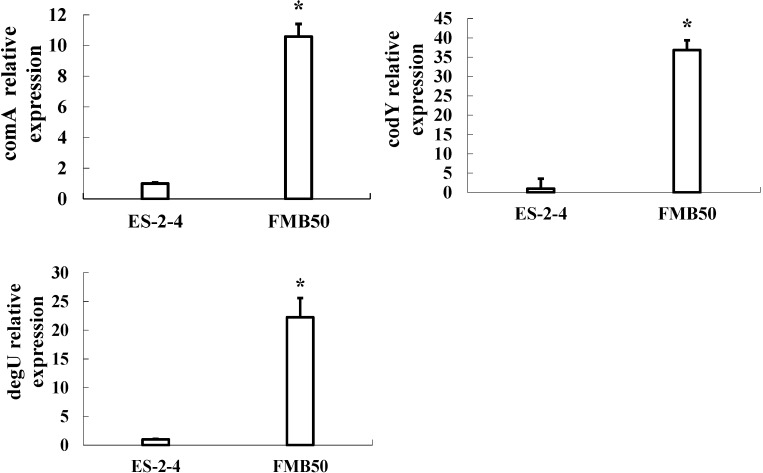
2.5. Gene Expression Verification by qRT-PCR
Expression of the three genes encoding differentially expressed proteins related to surfactin synthesis (comA, codY, and degU) was analyzed by qRT-PCR analysis of mRNA from FMB38. The mRNA expression profiles of these genes are shown in Figure 3. The mRNA levels of comA, codY and degU were upregulated in FMB38. The upregulated expression of comA and codY mRNA in FMB38 agreed with their protein levels, although degU showed upregulation at the transcriptional level, but was downregulated at the protein level. This disparity was probably due to a methodological error, different regulation of the rates of mRNA and protein turnover, or coverage of several protein isoforms or several members of a gene family [16].
Figure 3.
qRT-PCR analysis of mRNA expression of comA, degU and codY genes. Asterisks indicate a statistically significant difference (p < 0.05) between the parental strain ES-2-4 and recombination strain FMB38.
3. Discussion
3.1. Proteins Related to Surfactin Synthesis
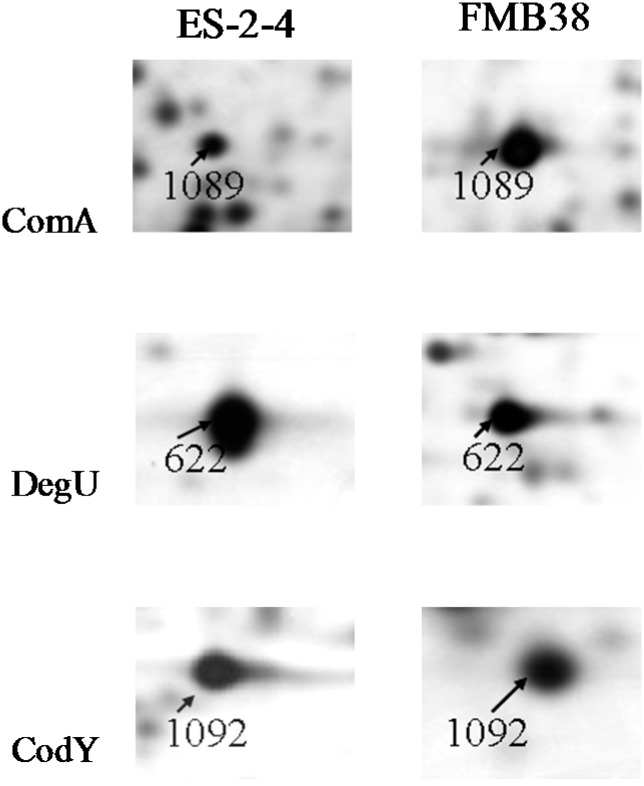
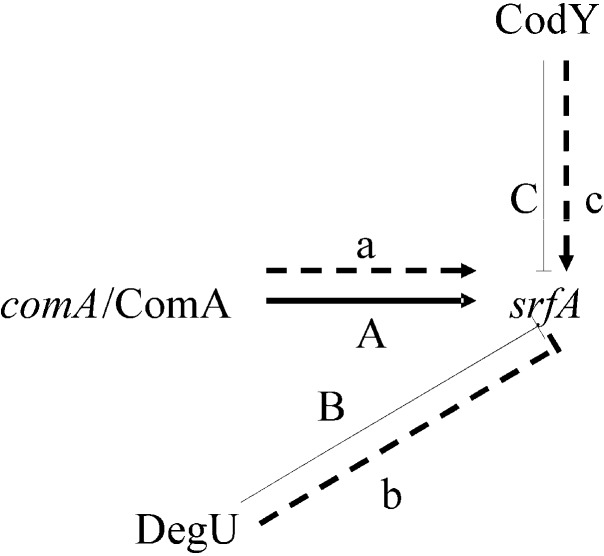
Of the 46 identified differentially expressed proteins, ComA, DegU, and CodY were directly related to surfactin synthesis. ComA and CodY were upregulated in FMB38, while DegU was downregulated. The three spots corresponding to these proteins on the 2-DE gels are shown in Figure 4. Marahiel et al. [17] reported that ComA acts as a transcriptional regulatory protein, and can directly bind to the srfA-D promoter, thus promoting the synthesis and secretion of surfactin (Figure 5A). DegU/DegS is an important regulatory protein, and plays a role in many physiological activities of the cell, including the secretion of extracellular proteases, cell migration, and competence. As a protein kinase regulatory protein, both phosphorylated and unphosphorylated DegU are also involved in the physiological activity of the cell. Phosphorylated DegU stimulates the synthesis of extracellular protease and inhibits the expression of sigD. However, in its unphosphorylated state, DegU can stimulate comK expression, thereby triggering cell competence. In B. subtilis, DegU/DegS and ComA/ComP act as a molecular switch that controls the occurrence of cellular changes in different physiological states. The physiological state in the later phases of cell growth is closely linked to the synthesis and secretion of antimicrobial substances. Hahn et al. [18] reported that DegU inhibited srfA operon expression (Figure 5B); However, the specific negative regulatory mechanism remains unclear. Hamoen et al. [13], Stein [12], and Serror et al. [19] showed that CodY acted as a transcriptional inhibitor of srfA expression (Figure 5C). Duitman et al. [20] constructed a codY-knockout vector to transform B. subtilis ATCC 6633. The resulting strain, BV12I38, showed no effect of CodY on surfactin synthesis. In the present study, upregulated ComA and downregulated DegU were consistent with a surfactin production increase, which was stimulated by ComA (Figure 5A), but inhibited by DegU (Figure 5B). The upregulated CodY in FMB38 contradicted the surfactin production increase (Figure 5C), contrary to previous reports. This was possibly because the entire network of metabolic regulation relies on signal factors, which both promote each other and are mutually antagonistic. Therefore, the CodY regulatory mechanism in surfactin synthesis in B. amyloliquefaciens requires further study.
Figure 4.
Enlargement of ComA, DegU, and CodY protein spots on 2-DE gels.
Figure 5.
Simplified scheme showing some of the regulators of surfactin synthesis and the roles of ComA, DegU, and CodY in positive (→) and negative (┤) regulation. The solid lines and capital letters show the regulation has been confirmed in the literature, the dash lines and small letters show the regulation speculated in this study.
3.2. Metabolism-Related Proteins
We identified 12 differentially expressed proteins related to the metabolism of carbohydrates, lipids, amino acids, nucleic acids, and coenzymes. A further two proteins were involved in inorganic ion transport and metabolism, and one protein was associated with co-metabolism, biosynthesis, transport, and catabolism.
The synthesis levels of key enzymes involved in glycolysis and the pentose phosphate pathway of glucose metabolism were higher in the genome-shuffled strain. The putative implications of these increases are described below. Fructose-bisphosphate aldolase is an important enzyme in the glycolysis pathway of all organisms. It catalyzes the cleavage of 1,6-diphosphate-d-fructose to 3-phospho-d-glyceraldehyde and α-dihydroxyacetone phosphate, and can also catalyze the reverse reaction in gluconeogenesis. Glyceraldehyde-3-phosphate dehydrogenase catalyzes glyceraldehyde-3-phosphate dehydrogenation and phosphorylation to generate 1, 3-diphosphoglycerate with a high-energy phosphate bond. The released hydrogen and electron are then transferred to NAD+ by the dehydrogenase coenzyme, to form NADH. The phosphate radical comes from inorganic phosphate. Phosphopyruvate hydratase catalyzes the reaction with 2-phosphoglycerate to generate phosphoenolpyruvate. Glucose can be degraded in the glycolytic pathway to generate ATP and provide raw materials for the synthesis reaction. Transaldolase in the non-oxidative stage of the pentose phosphate pathway can catalyze the reaction between glyceraldehyde-3-phosphate and 7-sedoheptulose monophosphate to generate 4-phosphate erythrose and fructose 6-phosphate. NADP produced in the pentose phosphate pathway could provide reducing power for the biosynthesis reaction, while pentose phosphate generated in this pathway could then participate in nucleic acid metabolism.
Pyrroline-5-carboxylate reductase is an important enzyme involved in the reduction of pyrroline-5-carboxylate to proline. Because of this function, it was widely believed that pyrroline-5-carboxylate reductase played an important regulatory function in a series of pathological and physiological processes such as cell apoptosis. 2-Aminoethylphosphonate-pyruvate transaminase belongs to the Valley-Grass transaminase family, and catalyzes the synthesis of aspartate, which is one of the amino acids that constitute surfactin. We speculate that the increased surfactin production by strain FMB38 is a result of increases in this enzyme. The acetoin reductase 2,3-butanediol dehydrogenase (YdjL) encoded by YdjL plays an important role in amino acid transport and metabolism [21]. Biosynthesis of many amino acids is closely linked to certain metabolic pathways such as glycolysis and the pentose phosphate pathway. Therefore, substances closely related to amino acid biosynthesis could be regarded as starting materials. Under conditions where the expression of enzymes related to amino acid transport and metabolism are increased, surfactin synthesis will also be enhanced in the recombinant strain. Upregulation of NAD synthetase (NadE) improves NAD generation, and thus would greatly improve the metabolic activities in of enzymes using NAD as a coenzyme in the recombination strain. Thus, we speculate that the increase in antibacterial lipopeptide yield accompanies the increased synthesis of key enzymes of the glycolysis and pentose phosphate pathways. Additionally, the abundance of aspartic acid transaminase and other key enzymes in amino acid metabolism likely enhances surfactin production. Furthermore, expression levels of key enzymes in lipid metabolism, coenzyme metabolism, and inorganic ion metabolism processes were also raised.
3.3. Proteins Related to Energy Generation and Transformation
Two of the identified proteins were related to energy production and conversion: the hypothetical protein RBAM_007590, and the ATP synthase α subunit (F0F1 ATP synthase subunit α). Both of these proteins were upregulated in the recombination strain. The ATP synthase α subunit is involved in proton transport processes coupled with ATP hydrolysis and with ATP synthesis. The enhancement of synthesis of enzymes associated with energy production and conversion may cause the increase of the antibacterial lipopeptide yield indirectly.
3.4. Proteins Related to DNA Replication, Recombination and Repair
The five DNA replication-related proteins, DNA primase, DNA-directed DNA polymerase III α subunit, recombinase protein, multifunctional SOS repair factor and Site-specific recombinase XerD, are upregulated.
DNA primase is a DNA-dependent RNA polymerase, whose function is to synthesis an RNA primer firstly in the process of DNA replication and carry on to extend DNA fragments in this primer [22], different from the RNA polymerase in the DNA transcription. DNA polymerase III is the necessary enzyme for DNA replication in the cells, having polymerase, exonuclease and endonuclease activities. Replication of the DNA molecule is the synthesis process of progeny DNA with the parental DNA molecule as a template. This process is completed with the chromosome replication in cell mitosis and the first meiosis interphase. The transmission of genetic information is accomplished by the replication of the DNA molecule. Through DNA molecules replication, the genetic information is passed from parent to offspring, thereby ensuring the continuity of the genetic information. The abundances increase of DNA replication-related enzymes could ensure the transmission of genetic information more powerful in the recombination strain.
SOS response is the emergency effects in case of cell DNA injury or copy system inhibited. The SOS response is caused by the interaction of RecA protein and LexA repressor. RecA protein not only play an important role in homologous recombination, but it is also the SOS response factor initially launched. It can be showed that, in the high-yielding strain, DNA serious injury caused by UV, NTG, and ion beam mutagenesis will lead to a series of complex inductive effects, called emergency response. Emergency response induces the excision and reorganization repair enzymes, elevate the levels of these enzymes within the cell and further strengthen excision repair and recombination repair capacity. In addition, SOS reaction can induce the development of DNA polymerase’s lack of proofreading function, speed up repair, and avoid death, while at the same time increase the mutation rate.
3.5. Proteins Related to Translation and Post-Translational Modifications
There are three proteins related to translation and post-translational modification respectively. The former includes Tu elongation factor Tu, 30S ribosomal protein S1, Methionine aminopeptidase, type I, while the latter includes alkyl hydroperoxide reductase small subunit, groEL gene product and hypothetical protein RBAM_036960. These six proteins are upregulated in the recombination strain.
Translation elongation factor EF-Tu belongs to the protein elongation factor family and is related to protein synthesis. There are three elongation factor EF-Tu, EF-Ts and EF-G in prokaryotes. The complexes are formed by EF-Tu and the second tt-tRNA integrated with A site of ribosomes, then GTP are hydrolyzed to release, regenerate and form complexes by EF-TS for the next cycle. The protein extending factors Tu/Ts/G are necessary for prokaryotic protein synthesis.
Ribosomes are the place of protein biosynthesis. Ribosome size is represented by the sedimentation coefficient S: The larger S value, the larger the particle and the greater the molecular weight. There are approximately 20,000 ribosomes in a vigorous growth bacterial, wherein the protein accounts for 10% of the total cellular proteins and rRNA account for 80% of the total cellular RNA. In 1968, the self-assembly of E. coli small subunit in vitro was researched, and it was found that 30S small subunit with natural activity be formed by adding in 16s rRNA and 21 proteins. In prokaryote 70S ribosome, the 30S subunit (small subunit) contains 22 kinds of the ribosomal protein, and the 50S subunit (large subunit) contains 34 kinds of the ribosomal protein, accounting for 35% of the ribosome. Methionine amino peptidase are involved in protein N-terminus processing. Protein biosynthesis occupies an important place in cell metabolism. Protein translation process is very complex, involving almost all types of intracellular RNA and dozens of protein factors. Ribosomes are the factories for protein synthesis, methionine amino peptidase and EF-Tu (translation elongation factor) and play an important role in protein processing, and so the upregulation of three proteins in the recombination strain will more effectively ensure the process of protein translation.
GroEL belongs to a highly conserved protein whose function is to assist the biological macromolecules correct folding, assembly, degradation and transport as well as improving cell stressor tolerance. When bacteria encounter environmental stress (such as heat shock), they will be high expressed within the cytoplasm to enhance bacterial tolerance, and therefore these proteins are also known as heat shock proteins [22]. GroEL belongs to the chaperonin 60 (Hsp60) protein family which includes cpn60, E. coli GroES/GroEL and Helicobacter pylori GroES/GroEL, etc. These proteins are highly conserved, but also produce cross-reactivity. Alkyl peroxide reductase is a key H2O2-degrading enzyme [23]. It is widely known that most of the protein peptide chain showed the expected biological activity only after certain processing to cause the maturation of the protein. Upregulation of proteins related to post-translational modification can accelerate the maturation process of the proteins and make them bioactive.
3.6. Proteins Related to Cell Secretion and Signal Transduction Mechanisms
There are three proteins involved in cell secretion and signal transduction mechanisms. The prokaryotes are mainly regulated at the transcriptional level. The activator protein bind the sequences close to the promoter, the affinity enhancement of RNA polymerase with the promoter, and RNA polymerase activity augmentation. The repressor protein can hinder gene transcription by binding manipulate sequence. As previously mentioned DegU (downregulated) and ComA (upregulated) as a transcription factor, the expression change and surfactin production increase in the high-yield strain is closely related. S-ribosylhomocysteinase upregulated in the recombination strain has lyase activity, which is both combined with the iron ions and related to quorum sensing [24]. In addition, the upregulation of ClpP [25] (ATP-dependent Clp protease proteolytic subunit) can improve the cell motility and secretion in FMB 38.
3.7. Hypothetical and Unknown Proteins
Fourteen spots subjected to mass spectrometry are identified as hypothetical or unknown proteins. The proteomics research of B. amyloliquefaciens is rarely reported and its protein database imperfect. Therefore, further research is needed to obtain the related protein function message in surfactin synthesis process. For the protein function indirectly noted in B. amyloliquefaciens, we can refer to its function in B. subtillis or other microorganisms to speculate.
All these indicated that the metabolic capability of the mutant was improved by the genome shuffling. The study will enable future detailed investigation of gene expression and function linked with surfactin synthesis. The results of proteome analysis may provide information for metabolic engineering of B. amyloliquefaciens for overproduction of surfactin.
4. Experimental Section
4.1. Strains and Culture Conditions
B. amyloliquefaciens ES-2 is an endophytic bacterium isolated from the Chinese medicinal plant Scutellaria baicalensis Georgi [26]. B. amyloliquefaciens ES-2-4 was obtained by 20 keV N+ ion beam implantation [27]. B. amyloliquefaciensFMB38 was the genome-shuffled mutant strain of B. amyloliquefaciens ES-2-4 [15]. These strains are available from the Key Laboratory of Food Processing and Quality Control of the Food Science and Technology College at Nanjing Agricultural University, Nanjing, China. B. amyloliquefaciens ES-2-4 was cultured in standard potato dextrose agar (PDA) media at 37 °C. All microbial strains were maintained in BPY supplemented with 20% (v/v) glycerol and stored at −70 °C. Seed medium (BPY) (beef extract 5.0 g/L, peptone 10.0 g/L, yeast extract paste 5.0 g/L, glucose 10.0 g/L, NaCl 5.0 g/L) and fermentation medium (glucose 42.0 g/L, l-sodium glutamate 4.0 g/L, MgSO4 0.5 g/L, KCl 0.5 g/L, KH2PO4 1.0 g/L, FeSO4 0.15 mg/L, MnSO4 5.0 mg/L, CuSO4 0.16 mg/L) were adjusted to pH 7.0.
4.2. Protein Sample Preparation
Cells were statically cultured at 32 °C for 36 h. After harvesting by centrifugation at 6000× g for 5 min at 4 °C, pelleted cells were washed three times with 20 mM Tris-HCl (pH 6.8). Cells were subsequently resuspended in lysis solution containing 7 M urea, 2 M thiourea, 4% (w/v) CHAPS (3-[(3-Cholamidopropyl) dimethylammonio] propanesulfonate), 40 mM DTT (Dithiothreitol), and 2% pH 3–10 ampholytes. Samples were disrupted by sonication in an ultrasonic cell pulverizer (Xin-zhi Biotechnology Co., Ningbo, China), equipped with a cup horn, for 45 min on ice. Following ultra-sonication, Nuclease Mix (GE Healthcare, Little Chalfont, UK) was added to a final concentration of 1% (v/v). The mixture was incubated for 1 h at room temperature and then centrifuged for 30 min at 13,000× g at 4 °C. Protein concentrations of the resulting supernatants were determined using a 2-D Quant kit (GE Healthcare), with bovine serum albumin as a standard. The proteins were stored at −70 °C until required for 2-DE.
4.3. 2-DE and Staining
For in-gel rehydration of each sample, 300 µg of protein was dissolved in 130 µL of rehydration buffer containing 7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 18 mM DTT, 2% Bio-Lyte (Bio Rad, Hercules, CA, USA), and 0.002% (w/v) bromophenol blue. IEF was performed on an Ettan IPGphor 3 IEF system (GE Healthcare) with 24 cm linear immobilized pH gradient (IPG) strips (pH 4–7, GE Healthcare). The loaded IPG strips were focused at 20 °C and 50 V for 10 h, 250 V for 3 h, 500 V for 3 h, 1000 V for 1 h, and 8000 V for 1 h, followed by 8000 V until a total of 80 kVh was reached. Following separation in the first dimension, the strips were equilibrated in a solution containing 6 M urea, 75 mM Tris-HCl (pH 8.8), 30% (w/v) glycerol, 2% (w/v) SDS, and 2% (w/v) DTT for 15 min at room temperature. The IPG strips were then equilibrated with the rehydration buffer described above, in which the DTT was replaced with 2.5% (w/v) iodoacetamide, for 15 min at room temperature. The strips were then transferred to 12.5% (w/v) SDS-polyacrylamide gels. Second dimension electrophoresis was carried out in an Ettan DALTII system (GE Healthcare) with a constant power of 5 W per gel for the first 30 min, followed by 12 W per gel for 6–7 h until the bromophenol blue dye front reached the bottom of the gels. Gels were fixed in a 40% (v/v) methanol and 10% (v/v) acetic acid solution overnight, and then stained with 0.25% (w/v) silver nitrate. At least three biological replicates were performed for each treatment.
4.4. Image Acquisition and Data Analysis
The silver-stained 2-DE gels were scanned at 400 dots per inch using an ImageScanner (GE Healthcare), and resulting images were analyzed using ImageMaster 2D Elite software (version 2.00; GE Healthcare) for spot detection, quantification, and comparative and statistical analyses. Images were cropped and optimized, and then gel-to-gel matching of the standard protein maps was performed. The spot detection parameters were optimized by checking different protein spots in certain regions of the gel, then automatically detected as described above, followed by visual inspection for removal or addition of undetected spots. Spot detection was refined by manual spot curation when needed. Spots that were present on at least two gels of one treatment or control were identified as expressed protein spots. The abundance of each protein spot was estimated by normalizing the spot volumes as a percentage of the total volume of all the spots in the gel to correct for any variability due to loading, gel staining, and destaining. Triplicate gels were used for each sample. The volume of each spot on the three replicate gels was normalized to the total spot volume from the reference gel, quantified, and then subjected to one-way ANOVA. Only those spots showing reproducible and significant changes were considered to be differentially expressed proteins.
4.5. Protein In-Gel Digestion
Detected spots and four control spots in unstained gel areas were excised (approximately 1 mm3 cubes) from silver nitrate-stained gels. For in-gel protein digestion, the gel-bound proteins were washed at room temperature with a 1:1 solution of 50 mM ACN (acetonitrile): NH4HCO3, once for 10 min and once for 30 min. Gels were dehydrated in 20 μL ACN for 20 min, and then dried in a vacuum centrifuge (Eppendorf, Hamburg, Germany) for 30 min at 30 °C. Proteins were reduced by incubation in 50 μL of 10 mM DTT/25 mM NH4HCO3 at 56 °C for 1 h, and were then alkylated in 50 μL of 55 mM iodoacetamide/25 mM NH4HCO3 for 45 min at room temperature in darkness. The liquid was discarded and gel pieces were washed twice in 25 mM NH4HCO3, dehydrated in ACN, and dried in a vacuum centrifuge for 30 min at 30 °C. Gel pieces were then rehydrated in 4 μL of 25 mM NH4HCO3 containing 40 ng trypsin, and incubated at 4 °C for 1 h. Excess liquid was discarded and gel plugs were incubated at 37 °C overnight, with tubes inverted to keep gel pieces wet for sufficient enzymatic cleavage. Then, 8 μL of 5% (v/v) TFA (trifluoroacetic acid) was added and samples were incubated at 37 °C for 1 h. Supernatants were collected and the proteins were extracted twice by incubating the gel pieces in 8 μL of 2.5% TFA/50% ACN at 37 °C for 1 h. Supernatants were mixed and completely dried in a vacuum centrifuge. Resulting peptides were stored at 4 °C until further use.
4.6. Protein Identification by MALDI-TOF and Database Searches
Dried peptides were dissolved in 2 μL of 0.5% TFA. The matrix material was dissolved until saturated in TA solution (ACN:0.1% TFA:acetone = 3:6:1). The matrix and the analyte solution were mixed at a ratio of 1:1, then 1 μL of the mixture was deposited onto the stainless steel sample target, and the solvent was allowed to evaporate at ambient temperature. MALDI-TOF analyses of trypsin digests were performed on a Biflex IV MALDI-TOF-MS (Bruker, Billerica, MA, USA) equipped with a N2 laser (337 nm, 3 ns pulse length) in positive ion mode at an accelerating voltage of 19 kV. Peptide data were collected in reflectron mode. Each spectrum was the accumulation of approximately 200 laser shots. External calibration was performed using peptide calibration standards.
Data were compared with the National Center for Biotechnology Information (NCBI) nr database using the MASCOT search program (Matrix Science, Boston, MA, USA). The following parameters were allowed: taxonomy restrictions to other firmicutes, one missed cleavage, 120 ppm mass tolerance in MS, carbamidomethyl (C) as a fixed modification, and oxidation (M) as a variable modification. The confidence in the peptide mass fingerprinting matches (p < 0.05) was based on the MOWSE score and confirmed by accurate overlap of the matched peptides with the major peaks of the mass spectrum. Only significant hits, as defined by the MASCOT probability analysis (p < 0.05), were accepted.
4.7. RT-PCR Analysis
Total RNA was extracted from B. amyloliquefaciens cultures using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and then treated with RNase-free DNase (Biomiga, Santiago, PA, USA). First-strand cDNA was synthesized from total RNA using an RT-PCR kit (Fermentas, Vilnius, Lithuania). Real-time PCR was performed using a real-time PCR SYBR Green Master Mix kit (Toyobo Biologics, Osaka, Japan) on a Rotor-Gene 3000 RT-PCR System (Corbett Research, Sydney, Australia). Each 25 µL PCR reaction contained 10 µL of real-time PCR mix, 1 µL of template DNA (100 ng), and 2 µL of 10 µM primers (Table 3). The PCR conditions were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Each reaction was performed in triplicate. Following threshold-dependent cycling, melting was performed from 45–95 °C at a 0.1 °C/s melt rate with a smooth curve setting averaging one point. Primer specificity was verified by melt curve analysis. Band intensities were normalized to the 16S rDNA transcript band for 2−ΔΔCt relative quantification. The B. amyloliquefaciens nucleotide sequences for these genes were obtained from the NCBI GenBank database. Primer pairs were designed from these sequences with Primer Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA; Table 3).
Table 3.
Primers used for qRT-PCR.
| Gene | Forward Primer (5'→3') | Reverse Primer (5'→3') |
|---|---|---|
| 16S rDNA | CCTACGGGAGGCAGCAG | ATTACCGCGGCTGCTGG |
| comA | TCAAAGTGAGCAGGATCGGTTAA | CTTCTGTACGGGAGCCGACAT |
| codY | GGCAGGCAAACCCGTAAACT | ACTGGCGGTCTTCCAGCATT |
| degU | CACCCGAAAGTAACCCACAAT | AGCACTTCACATTCCCGTCTC |
5. Conclusions
In conclusion, the functions of the identified differential proteins appeared to be interconnected. The results revealed that the improvement in lipopeptide yield in the genome-shuffled strain was not the result of a single metabolic pathway or a small number of proteins, but a combination of gene transcription and translation processes, sugar, protein, and energy metabolism, and many other related factors. Of the 46 identified proteins, ComA, DegU, and CodY were directly related to surfactin synthesis, and we determined that ComA and CodY stimulate surfactin synthesis, while DegU acts as an inhibitor. The integration of proteomic information could offer more rational strategies for the genetic modification of B. amyloliquefaciens cells with an enhanced surfactin synthesis capacity.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30871753), the National Research Program of China (No. 2011BAD23B05), and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author Contributions
Zhaoxin Lu and Junfeng Zhao designed and carried out the experiments, and wrote the manuscript; Lin Cao, Chong Zhang, Lei Zhong and Jing Lu performed the experiment and analyzed the data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Carrillo C., Teruel J.A., Aranda F.J., Ortiz A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim. Biophys. Acta. 2003;1611:91–97. doi: 10.1016/S0005-2736(03)00029-4. [DOI] [PubMed] [Google Scholar]
- 2.Ongena M., Jacques P. Bacillus lipopeptides: Versatile weapons for plant disease bio-control. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Kowall M., Vater J., Kluge B., Stein T., Franke P. Separation and characterization of surfactin isoforms produced by Bacillus subtilis OKB 105. J. Colloid Interface Sci. 1998;204:1–8. doi: 10.1006/jcis.1998.5558. [DOI] [PubMed] [Google Scholar]
- 4.Bie X.M., Lu Z.X., Lu F.X., Zeng X.X. Screening the main factors affecting extraction of the antimicrobial substance from Bacillus sp. fmbJ using Plackett–Burman method. World J. Microbiol. Biotechnol. 2005;21:925–928. doi: 10.1007/s11274-004-6722-z. [DOI] [Google Scholar]
- 5.Kanlayavattanakul M., Lourith N. Lipopeptides in cosmetics. Int. J. Cosmet. Sci. 2010;32:1–8. doi: 10.1111/j.1468-2494.2009.00543.x. [DOI] [PubMed] [Google Scholar]
- 6.Schaller K.D., Fox S.L., Bruhn D.F., Noah K.S., Bala G.A. Characterization of surfactin from Bacillus subtilis for application as an agent for enhanced oil recovery. Appl. Biochem. Biotechnol. 2004;115:827–836. doi: 10.1385/ABAB:115:1-3:0827. [DOI] [PubMed] [Google Scholar]
- 7.Mulligan C.N., Yong R.N., Gibbs B.F. Heavy metal removal from sediments by biosurfactants. J. Hazard. Mater. 2001;85:111–125. doi: 10.1016/S0304-3894(01)00224-2. [DOI] [PubMed] [Google Scholar]
- 8.Jacques P., Hbid C., Destain J., Razafindralambo H., Paquot M. Optimization of biosurfactant lipopeptide production from Bacillus subtilis S499 by Plackett-Burman design. Appl. Biochem. Biotechnol. 1999;77:223–233. doi: 10.1385/ABAB:77:1-3:223. [DOI] [Google Scholar]
- 9.Dimitrov K., Gancel F., Montastruc L., Nikov I. Liquid membrane extraction of bioactive amphiphilic substances:Recovery of surfactin. Biochem. Eng. J. 2008;42:248–253. doi: 10.1016/j.bej.2008.07.005. [DOI] [Google Scholar]
- 10.Cao G.Q., Zhang X.H., Zhong L., Lu Z.X. A modified electro-transformation method for Bacillus subtilis and its application in the production of antimicrobial lipopeptides. Biotechnol. Lett. 2010;33:1047–1051. doi: 10.1007/s10529-011-0531-x. [DOI] [PubMed] [Google Scholar]
- 11.Sun H.G., Bie X.M., Lu F.X., Lu Y.P., Wu Y. Enhancement of surfactin production of Bacillus subtilis fmbR by replacement of the native promoter with the Pspac promoter. Can. J. Microbiol. 2009;55:1003–1009. doi: 10.1139/W09-044. [DOI] [PubMed] [Google Scholar]
- 12.Stein T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005;56:845–857. doi: 10.1111/j.1365-2958.2005.04587.x. [DOI] [PubMed] [Google Scholar]
- 13.Hamoen L.W., Venema G., Kuipers O.P. Controlling competence in Bacillus subtilis: Shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 14.Tsuge K., Ohata Y., Shoda M. Gene yerP, involved in surfactin self-resistance in Bacillus subtilis. Anti-Microb. Agents Chemother. 2001;45:3566–3573. doi: 10.1128/AAC.45.12.3566-3573.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J.F., Li Y.H., Zhang C., Yao Z.Y., Zhang L. Genome shuffling of Bacillus amyloliquefaciens for improving antimicrobial lipopeptide production and an analysis of relative gene expression using FQ RT-PCR. J. Ind. Microbiol. Biotechnol. 2012;39:889–896. doi: 10.1007/s10295-012-1098-9. [DOI] [PubMed] [Google Scholar]
- 16.Zhang J.W., Ma H.Q., Chen S., Ji M., Perl A. Stress response proteins’ differential expression in embryogenic and non-embryogenic callus of Vitis. vinifera L. cv. Cabernet Sauvignon-A proteomic approach. Plant. Sci. 2009;177:103–113. doi: 10.1016/j.plantsci.2009.04.003. [DOI] [Google Scholar]
- 17.Marahiel M.A., Nakano M.M., Zuber P. Regulation of peptide antibiotic production in Bacillus. Mol. Microbiol. 1997;7:631–636. doi: 10.1111/j.1365-2958.1993.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 18.Hahn J., Dubnau D. Growth stage signal transduction and the requirements for srfA induction in development of competence. J. Bacteriol. 1991;173:7275–7282. doi: 10.1128/jb.173.22.7275-7282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serror P., Sonenshein A.L. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 1996;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duitman E.H., Wyczawski D., Boven L.G., Venema G., Kuipers O.P. Novel methods for genetic transformation of natural Bacillus subtilis isolates used to study the regulation of the mycosubtilin and surfactin synthetases. Appl. Environ. Microbiol. 2007;73:3490–3496. doi: 10.1128/AEM.02751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson W.L. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanedioldehydrogenase. Appl. Environ. Microbiol. 2008;74:6832–6838. doi: 10.1128/AEM.00881-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbons H.S., Broomall S.M., McNew L.A., Daligault H., Chapman C. Genomic signatures of strain selection and enhancement in Bacillus atrophaeus var. globigii, a historical biowarfare simulant. PLoS One. 2011;6:e17836. doi: 10.1371/journal.pone.0017836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhyay A., Redding A.M., Joachimiak M.P., Arkin A.P., Borglin S.E. Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 2007;189:5996–6010. doi: 10.1128/JB.00368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borriss R., Chen X., Rueckert C., Blom J., Becker A. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM7T and FZB42T: A proposal for Bacillus amyloliquefaciens sub sp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on their discriminating complete genome sequences. Int. J. Syst. Evol. Microbiol. 2011;61:1786–1801. doi: 10.1099/ijs.0.023267-0. [DOI] [PubMed] [Google Scholar]
- 25.Chen X.H., Koumoutsi A., Scholz R., Eisenreich A., Schneider K. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- 26.Sun L.J., Lu Z.X., Bie X.M., Lu F.X., Yang S.Y. Isolation and characterization of a co-producer of fengycins and surfactins, endophytic Bacillus amyloliquefaciens ES-2, from Scutellaria. Baicalensis Georgi. World J. Microbiol. Biotechnol. 2006;22:1259–1266. doi: 10.1007/s11274-006-9170-0. [DOI] [Google Scholar]
- 27.Fang C.J., Lu Z.X., Sun L.J., Lu F.X., Bie X.M. Study on mutation breeding and fermentation of antimicrobial lipopeptides yielding bacterium with 20 keV N+ ion beam implantation. J. Radiat. Res. Radiat. Process. 2006;24:333–336. [Google Scholar]