Abstract
Platinum-based chemotherapy toxicity is always one of the serious problems from which lung cancer patients suffer. The genetic polymorphism of WISP1 was revealed to be associated with susceptibility and platinum-based chemotherapy response in our previous studies. In this study, we aimed to investigate the relationship of WISP1 genetic polymorphisms with platinum-based chemotherapy toxicity in lung cancer patients. A total of 412 lung cancer patients were enrolled in this study, and 28 polymorphisms of the WISP1 gene were genotyped by SequenomMassARRAY. We found that WISP1 polymorphisms (rs2929965, rs2929969, rs2929970, rs2929973 and rs754958) were related to the overall chemotherapy toxicity of lung cancer in subgroup analyses. Rs16904853, rs2929970, rs2977549 and rs2977551 (p = 0.021, 0.028, 0.024, 0.048, respectively) polymorphisms were significantly associated with hematologic toxicity. Rs2929946, rs2929970, rs2977519, rs2977536, rs3739262 and rs754958 (p = 0.031, 0.046, 0.029, 0.016, 0.042, 0.035, respectively) polymorphisms were significantly associated with the gastrointestinal toxicity of lung cancer. Genotypes of WISP1 may be novel and useful biomarkers for predicting platinum-based chemotherapy toxicity in lung cancer patients.
Keywords: WISP1, lung cancer, genetic polymorphism, chemotherapy toxicity
1. Introduction
In recent years, lung cancer has become the highest morbidity and mortality cancer among cancers in the world [1]. It consists of non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). Most lung cancer patients were diagnosed at advance stages. They were not suitable for surgery, and chemotherapy was the best choice for them. Platinum-based chemotherapy is widely used for different cancers, especially cisplatinum, for the treatment of lung cancer [2]. However, chemotherapy resistance and toxicity are still serious problems from which lung cancer patients suffer. It was reported that chemotherapy efficacy and toxicity were related to many genes in multiple pathways, such as DNA repair, apoptosis, transportation, as well as the Wnt pathway [3,4,5,6,7]. Wnt-1 inducible signaling protein 1 (WISP1) is a target gene of the canonical Wnt signaling pathway [8]. Its expression was revealed to be associated with pulmonary fibrosis, ventilator-induced lung injury and lung cancer [9,10].
WISP1 belongs to the CCN protein family, which consists of cysteine-rich 61 (CYR61/CCN1), connective tissue growth factor (CTGF/CCN2), nephroblastoma overexpressed (NOV/CCN3), WISP1, WISP2 and WISP3 [11,12,13]. It maps to chromosome 8q24.1–8q24.3 and has five exons and four introns [14]. It is a secreted matricellular protein and has four modules, namely insulin growth factor binding protein (IGFBP), Van Willebrand factor C (VWC), thrombospondin type I repeat domain (TSP) and C-terminal domain (CT) [11]. It is involved in diverse biological effects, such as cell proliferation, differentiation and survival, and is related to multiple pathologic processes, especially in lung diseases [9,15,16,17].
Nowadays, single-nucleotide polymorphisms (SNPs) play vital roles in the occurrence and development of diseases. Numerous recent studies have revealed that polymorphic genetic mutations contributed to the toxicity and chemotherapy resistance of lung cancer. Moreover, the polymorphisms of WISP1 were discovered to be associated with diverse lung diseases [10,18]. In our previous study, WISP1 polymorphisms were revealed to be related to susceptibility and the platinum-based chemotherapy response of lung cancer in Chinese patients [19].
In order to further investigate the role of WISP1 polymorphisms in the chemotherapy toxicity of lung cancer, we selected 28 SNPs of the WISP1 gene and genotyped in lung cancer patients to explore the association between WISP1 polymorphisms and platinum-based chemotherapy toxicity.
2. Results
2.1. Clinical Characteristics and Toxicity Outcomes of Subjects and Genotyping
A total of 412 lung cancer patients were enrolled in our study. Their clinical characteristics are summarized in Table 1. There were 288 NSCLC patients and 124 SCLC patients. All 412 patients received at least two cycles of platinum-based chemotherapy, and severe toxicity occurred in 163 (39.6%) of them. Seventy one (22.1%) patients suffered hematologic toxicity; 72 (22.3%) patients suffered gastrointestinal toxicity; and 20 (4.8%) patients suffered both hematologic and gastrointestinal toxicity. All 28 SNPs of the WISP1 gene were genotyped by Sequenom’s MassARRAY system. As shown in Table 1, the call rate of the SNPs ranged from 95.63% to 99.76%, and the MAF (minor allele frequency) of each SNP was more than 5%. The genotype distribution of 28 SNPs in the severe toxicity group and non-severe toxicity group were in accordance with the Hardy-Weinberg equilibrium.
Table 1.
The polymorphisms of WISP1 examined in this study.
| Polymorphisms | Alleles | Call Rate (%) | MAF † |
|---|---|---|---|
| rs10956696 | C/T | 98.79 | 0.10 |
| rs10956697 | A/C | 96.60 | 0.33 |
| rs11778573 | G/T | 98.30 | 0.41 |
| rs16893344 | C/T | 98.30 | 0.13 |
| rs16904853 | C/T | 95.63 | 0.47 |
| rs2013146 | C/T | 98.54 | 0.35 |
| rs2929946 | A/G | 97.82 | 0.08 |
| rs2929965 | C/T | 97.57 | 0.42 |
| rs2929969 | A/G | 97.57 | 0.32 |
| rs2929970 | A/G | 99.76 | 0.37 |
| rs2929973 | G/T | 97.33 | 0.34 |
| rs2929986 | C/T | 97.57 | 0.37 |
| rs2977519 | A/T | 95.87 | 0.31 |
| rs2977529 | A/T | 99.27 | 0.19 |
| rs2977530 | A/G | 98.54 | 0.45 |
| rs2977536 | C/G | 98.30 | 0.33 |
| rs2977537 | A/G | 98.30 | 0.47 |
| rs2977549 | C/T | 98.79 | 0.38 |
| rs2977551 | T/C | 98.79 | 0.38 |
| rs3739262 | C/T | 97.33 | 0.15 |
| rs4330674 | T/C | 99.76 | 0.15 |
| rs62514003 | T/C | 98.79 | 0.14 |
| rs62514004 | G/A | 96.36 | 0.12 |
| rs72731505 | C/T | 98.06 | 0.38 |
| rs72731507 | A/G | 99.76 | 0.09 |
| rs754958 | C/T | 98.79 | 0.32 |
| rs7828685 | G/A | 98.78 | 0.29 |
| rs7843546 | C/T | 99.75 | 0.49 |
† Minor allele frequency.
2.2. Association between WISP1 Polymorphisms and the Severe Toxicity of Platinum-Based Chemotherapy in Lung Cancer Patients
The 28 polymorphisms of WISP1 were not statistically significantly related to increased risk of overall severe toxicity in all three models (Table S1). However, there were five SNPs related to the toxicity in subgroup analyses. As shown in Figure 1, WISP1 rs2929965 polymorphism was related to the overall toxicity of NSCLC patients in the additive model; WISP1 rs2929969 and rs2929973 were related to the overall toxicity of SCLC in additive and dominant models and related to the toxicity of NSCLC in the dominant model. Additionally, they were also related to overall toxicity in lung cancer patients ≤55 years old in the recessive model. WISP1 rs2929970 was related to overall toxicity in patients >55 years old and in NSCLC and SCLC patients in the dominant model, as well as in patients ≤55 years old in the recessive model. WISP1 rs754958 was related to the overall toxicity of SCLC patients in additive and dominant models and related to the overall toxicity of NSCLC in the dominant model. In conclusion, lung cancer patients ≤55 years old carrying the A allele of (rs2929969, rs2929970) or the G allele of rs2929973; patients >55 years old carrying the G allele of rs2929970; NSCLC patients carrying the T allele of (rs2929965, rs2929973, rs754958) or the G allele of (rs2929969, rs2729970); SCLC patients carrying the A allele of (rs2929965, rs2929970), or the G allele of rs2929973, or the C allele of rs754958 suffered more risk of Grade 3 or 4 toxicity overall for platinum-based chemotherapy.
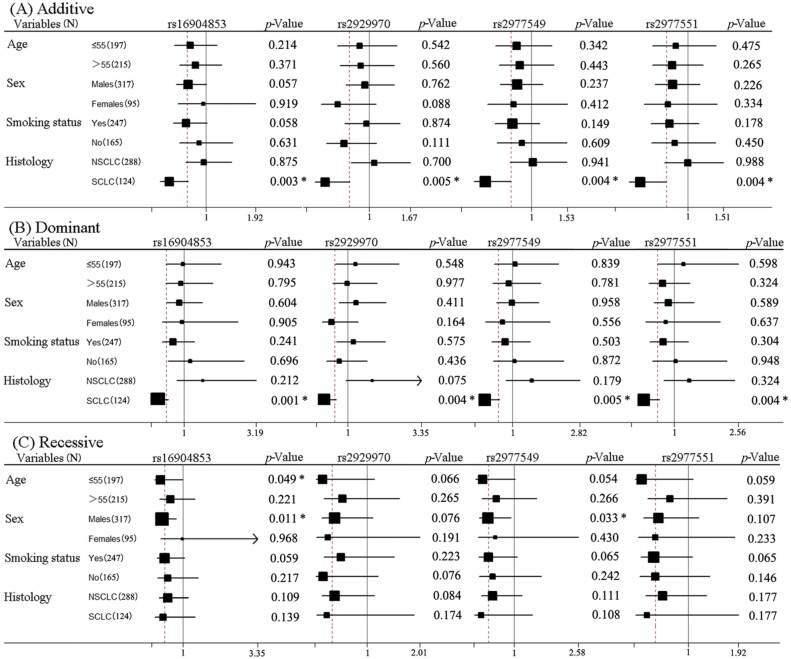
Figure 1.
Stratification analyses of the associations of WISP1 rs2929965, rs2929969, rs2929970, rs2929973 and rs754958 polymorphisms with the overall chemotherapy toxicity of lung cancer. Each box and horizontal line represents the values of OR and 95% CI. NSCLC, non-small cell lung carcinoma; SCLC, small cell lung carcinoma. * p < 0.05.
2.3. Association between WISP1 Polymorphisms and the Hematologic Toxicity of Platinum-Based Chemotherapy in Lung Cancer Patients
As shown in Table 2, WISP1 polymorphisms (rs16904853, rs2929970, rs2977549, rs2977551) were significantly associated with the hematologic toxicity of platinum-based chemotherapy of lung cancer patients in the recessive model. The results of subgroup analyses of the four SNPs are demonstrated in Figure 2. WISP1 rs16904853 and rs2977549 were associated with the hematologic toxicity of SCLC in additive and dominant models and associated with the toxicity of patients ≤55 years old in the recessive model. WISP1 rs16904853 was also associated with the hematologic toxicity of the lung cancer in males in the recessive model. WISP1 rs2929970 and rs2977551 were associated with the hematologic toxicity of SCLC in additive and dominant models. All of the results showed that individuals carrying the C allele of (rs16904853, rs2977549), or the A allele of rs2929970, or the T allele of rs2977551 had an increased risk of hematologic toxicity for platinum-based chemotherapy.
Table 2.
Association of rs16904853, rs2929970, rs2977549 and rs2977551 with hematologic toxicity in lung cancer patients.
| SNPs | Non-3 or 4 Grade Toxicity (11/12/22 †) | 3 or 4 Grade Toxicity (11/12/22) | MAF | Additive | Dominant | Recessive | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non-3 or 4 Grade Toxicity | 3 or 4 Grade Toxicity | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |||
| rs16904853 | 83/152/75 | 26/51/11 | 0.49 | 0.41 | 0.74 (0.52–1.05) | 0.087 | 0.87 (0.52–1.47) | 0.607 | 0.45 (0.23–0.89) | 0.021 * |
| rs2929970 | 123/145/47 | 34/49/5 | 0.38 | 0.34 | 0.82 (0.57–1.17) | 0.276 | 1.02 (0.63–1.65) | 0.944 | 0.34 (0.13–0.89) | 0.028 * |
| rs2977549 | 58/136/124 | 7/45/36 | 0.40 | 0.34 | 0.78 (0.55–1.10) | 0.153 | 0.92 (0.57–1.49) | 0.745 | 0.39 (0.17–0.88) | 0.024 * |
| rs2977551 | 122/142/52 | 38/44/7 | 0.39 | 0.33 | 0.76 (0.54–1.08) | 0.128 | 0.84 (0.52–1.36) | 0.486 | 0.43 (0.19–0.99) | 0.048 * |
† Wild-type/heterozygote/homozygote; * p < 0.05.
Figure 2.
Stratification analyses of the associations of WISP1 rs16904853, rs2929970, rs2977549 and rs2977551 polymorphisms with the hematologic toxicity of lung cancer. Each box and horizontal line represents the values of OR and 95% CI. NSCLC, non-small cell lung carcinoma; SCLC, small cell lung carcinoma. * p < 0.05.
2.4. Association between WISP1 Polymorphisms and the Gastrointestinal Toxicity of Platinum-Based Chemotherapy in Lung Cancer Patients
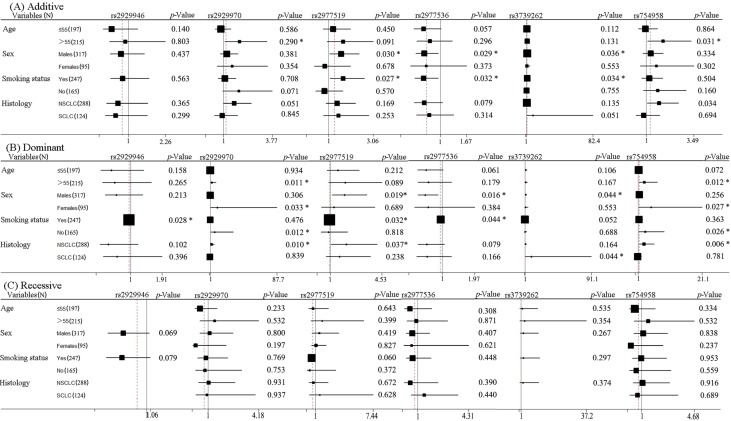
As shown in Table 3, WISP1 polymorphisms (rs2929946, rs2929970, rs2977519, rs2977536, rs754958) were significantly associated with gastrointestinal toxicity in lung cancer patients in the dominant model. WISP1 rs2977536 and rs3739262 were significantly associated with gastrointestinal toxicity in the additive model. The results of subgroup analyses of the six SNPs are demonstrated in Figure 3. WISP1 rs2929946 was associated with the gastrointestinal toxicity of smoking patients in the dominant model. WISP1 rs2929970 was associated with the gastrointestinal toxicity of patients >55 years old in additive and dominant models, and it was related to the toxicity of female patients, nonsmoking patients and NSCLC patients in the dominant model. WISP1 rs2977519 and rs2977536 were associated with the gastrointestinal toxicity of male patients and smoking patients in additive and dominant models, and rs29277519 was also associated with the toxicity of NSCLC in the dominant model. WISP1 rs3739262 was associated with the gastrointestinal toxicity of male patients in additive and dominant models, and it was also related to the toxicity of smoking patients in the additive model and of SCLC in the dominant model. WISP1 rs754958 was associated with the gastrointestinal toxicity of patients >55 years old and NSCLC in additive and dominant models, and it was also associated with the toxicity of female patients and nonsmoking patients in the dominant model. The subgroup analyses indicating that individuals with the A allele of (rs2929946, rs2977549), or the G allele of rs2929970, or the C allele of rs2977536, or the T allele of (rs2977549, rs3739262, rs754958) presented a greater risk of gastrointestinal toxicity for platinum-based chemotherapy.
Table 3.
Association of rs2929946, rs2929970, rs2977519, rs2977536, rs3739262 and rs754958 with gastrointestinal toxicity in lung cancer patients.
| SNPs | Non-3 or 4 Grade Toxicity (11/12/22 †) | 3 or 4 Grade Toxicity (11/12/22) | MAF | Additive | Dominant | Recessive | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Non-3 or 4 Grade Toxicity | 3 or 4 Grade Toxicity | OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | |||
| rs2929946 | 42/59/254 | 2/7/82 | 0.48 | 0.06 | 0.55(0.28–1.10) | 0.093 | 0.42(0.19–0.92) | 0.031 * | 5.41(0.43–67.28) | 0.190 |
| rs2929970 | 130/140/42 | 27/54/10 | 0.20 | 0.41 | 1.27(0.88–1.84) | 0.201 | 1.72(1.01–2.93) | 0.046 * | 0.87(0.40–1.86) | 0.715 |
| rs2977519 | 158/130/31 | 36/48/8 | 0.15 | 0.35 | 1.39(0.95–2.02) | 0.089 | 1.75(1.06–2.91) | 0.029 * | 1.01(0.43–2.40) | 0.980 |
| rs2977536 | 125/152/32 | 48/30/8 | 0.16 | 0.27 | 0.64(0.43–0.96) | 0.030 * | 0.53(0.32–0.89) | 0.016 * | 0.70(0.29–1.66) | 0.417 |
| rs3739262 | 232/77/6 | 59/28/3 | 0.03 | 0.19 | 1.64(1.02–2.63) | 0.042 * | 1.70(0.99–2.91) | 0.055 | 2.34(0.52–10.50) | 0.266 |
| rs754958 | 34/120/153 | 8/49/33 | 0.36 | 0.36 | 1.32(0.91–1.91) | 0.150 | 1.74(1.04–2.91) | 0.035 * | 0.86(0.37–2.02) | 0.732 |
† Wild-type/heterozygote/homozygote; * p < 0.05.
Figure 3.
Stratification analyses of the associations of WISP1 rs2929946, rs2929970, rs2977519, rs2977536, rs3739262 and rs754958 polymorphisms with the gastrointestinal toxicity of lung cancer. Each box and horizontal line represents the values of OR and 95% CI. NSCLC, non-small cell lung carcinoma; SCLC, small cell lung carcinoma. * p < 0.05.
3. Discussion
In the current study, we examined the relationship of WISP1 polymorphisms and platinum-based chemotherapy toxicity in Chinese lung cancer patients. Our results showed that WISP1 SNPs (rs2929965, rs2929969, rs2929970, rs2929973, rs754958) contributed to the overall toxicity in different subgroups; WISP1 SNPs (rs16904853, rs2929970, rs2977549, rs2977551) were related to hematologic toxicity, and WISP1 SNPs (rs2929946, rs2929970, rs2977519, rs2977536, rs3739262, rs754958) were related to the gastrointestinal toxicity of platinum-based chemotherapy in lung cancer patients.
The Wnt signaling pathway regulates the transcription of many genes that are involved in cell proliferation, differentiation and the prevalence of cancers [20,21,22]. The canonical Wnt-β-catenin pathway is one of the three major Wnt signaling pathways, and it is involved in diverse cancers, such as hepatocellular carcinoma and esophageal squamous carcinoma [23,24]. WISP1 is a downstream gene of the canonical Wnt-β-catenin pathway, and its mutations were reported to be associated with multiple diseases, including asthma, hypertension and spinal osteoarthritis [18,25,26]. Our previous studies revealed that WISP1 genetic polymorphisms were related to susceptibility and the platinum-based chemotherapy response of lung cancer, and we hypothesized that WISP1 polymorphisms may also be associated with the chemotherapy toxicity of lung cancer [19]. In this study, we found 12 WISP1 polymorphisms contributing to platinum-based chemotherapy toxicity. Five SNPs (rs2929969, rs2929970, rs2929973, rs2977549, rs2977551) were located in the 3’UTR of WISP1. rs2929970 and rs2929973 have been reported to be related to many diseases [18,25,26]. Polymorphisms in the 3’UTR of genes would be able to regulate gene expression. Four SNPs (rs2799519, rs3739262, rs2977536, rs2929946) were in the intron 1 of the WISP1 gene. Mutations at exon-intron junctions, the branch point sequence and the polypyrimidine tract would disrupt the highly conserved and receptor sites. Especially, the first intron is significantly related to gene expression. Three SNPs (rs16904853, rs2929965, rs754958) were located in other introns; these polymorphisms may play important roles in the splicing process or the structure of proteins [27,28,29,30,31]. However, the detailed mechanisms about the functions of WISP1 polymorphisms need further investigations.
Lung cancer patients ≤55 years old carrying the A allele of (rs2929969, rs2929970) or the G allele of rs2929973, patients >55 years old carrying the G allele of WISP1 rs2929970, NSCLC patients carrying the T allele of (rs2929965, rs2929973, rs754958) or the G allele of (rs2929969, rs2929970), SCLC patients carrying the A allele of (rs2929969, rs2929970), or G allele of rs2929973, or the C allele of rs754958 presented more risk of overall severe toxicity of platinum-based chemotherapy. Lung cancer patients ≤55 years old carrying the C allele of rs16904853, male patients carrying the C allele of (rs16904853, rs2977549), SCLC patients carrying the C allele of (rs16904853, rs2977549), or the A allele of rs2929970, or the T allele of rs2977551 had more risk of hematologic toxicity of platinum-based thermotherapy. Lung cancer patients >55 years old carrying the G allele of rs2929970 or the T allele of rs754958, male patients carrying the T allele of (rs2977519, rs3739262) or the C allele of rs2977536, female patients carrying the G allele of rs2929970 or the T allele of rs754958, smoking patients carrying the A allele of rs2929946, or the T allele of (rs2977519, rs3739262), or the C allele of rs2977536, nonsmoking patients carrying the G allele of rs2929970 or the T allele of rs754958, NSCLC patients carrying the G allele of WISP1 rs2929970 or the T allele of (rs2977519, rs754958) exhibited more risk of gastrointestinal toxicity of platinum-based chemotherapy.
All of these SNPs were discovered for the first time to be associated with the chemotherapy toxicity of lung cancer. In our previous studies, the genetic polymorphisms of WISP1, eukaryotic translation initiation factor 3 (eIF3a) and copper transport protein 1 (CTR1) were shown to be associated with platinum-based chemotherapy response or toxicity in lung cancer patients [32,33,34,35]. Most previous studies about the relationships of WISP1 polymorphisms and diseases were focused on the polymorphisms of WISP1 rs2929973 and rs2929970. These two polymorphisms were located in the 3'UTR of the WISP1 gene. Sunita et al. indicated that WISP1 rs2929973 was associated with asthma and individuals carrying the G allele of rs2929973 conferring lower forced expiratory volume in the first second [18]. Yamada Y et al. reported that WISP1 rs2929970 was associated with hypertension in men, and the men carrying the G allele of rs2929970 had higher blood pressure [25]. Moreover, Urano et al. demonstrated that WISP1 rs2929970 was associated with spinal osteoarthritis in postmenopausal Japanese women, and the women carrying AA genotypes had significantly higher endplate sclerosis [26]. In our study, WISP1 rs2929973 was related to the chemotherapy toxicity of SCLC; WISP1 rs2929970 was associated with the overall toxicity, hematologic toxicity and gastrointestinal toxicity of lung cancer. Perhaps, the two SNPs play very important roles in the expression or function of the WISP1 gene, and they are associated with multiple diseases.
However, there were several limitations to our study. Considering multiple-testing correction, which calculated the p-value of the SNPs by FDR-BH (Benjamini and Hochberg (1995) step-up False Discovery Rates control) correction, and that no SNPs remained significant, we consider that perhaps the sample size for the study was not large enough. The functional relevance of the identified polymorphisms that are associated with chemotherapy toxicity was not determined in our study. Finally, the validation of our results requires replication studies with other independent subjects.
In conclusion, our results showed that WISP1 polymorphisms of (rs16893344, rs2929970, rs2977549, rs2977551, rs2929946, rs2977519, rs2977536, rs3739262, rs754958, rs2929965, rs2929969, rs2929973) were significantly associated with the chemotherapy toxicity of lung cancer patients. Thus, we thought that the genotypes of WISP1 may be used to predict the platinum-based chemotherapy toxicity in lung cancer patients.
4. Materials and Methods
4.1. Study Population and Treatments
We enrolled 412 lung cancer patients in this study. All individuals were provided written informed consent in compliance with the code of ethics of the World Medical Association (Declaration of Helsinki) before this study was initiated. Eligible subjects were from The Affiliated Cancer Hospital or Xiangya Hospital of Central South University (Changsha, Hunan, China) between November, 2011, and May, 2013. The patients that were eligible for the study had to meet the following criteria: (1) histologic or cytologic confirmed of lung cancer; (2) no prior chemotherapy; (3) Eastern Cooperative Oncology Group 0–2; (4) all patients were treated with platinum-based chemotherapy for at least two periods; and (5) organ function before chemotherapeutic treatment: liver function test (aspartate transaminase ≤1.5 × normal upper limit, alanine transaminase ≤1.5 × normal upper limit); blood test (leukocyte count ≥1.5 × 109/L, neutrophil count ≥1.5 × 109/L, platelet count ≥100 × 109/L); kidney test (serum creatinine ≤1.5 × normal upper limit, creatinine clearance ≥60 mL/minute). Exclusion criteria included: (1) pregnancy or lactation; (2) active infection; (3) symptomatic brain or leptomeningeal metastases; and (4) previous or other concomitant malignancies. The study protocol was approved by the Ethics Committee of Xiangya School of Medicine, Central South University, with Registration Number CTXY-110008-1. We applied for this study for clinical admission in the Chinese Clinical Trial Register (Registration Number: ChiCTR-RNC-12002892). The chemotherapy regimens are listed in Table 4.
Table 4.
Clinical characteristic of lung cancer patients.
| Patients Characteristics | N (%) |
|---|---|
| Total No. of patients | 412 |
| Lung cancer | |
| NSCLC | 288 (69.9) |
| SCLC | 124 (30.1) |
| Age | |
| ≤55 | 197 (47.8) |
| >55 | 215 (52.2) |
| Sex | |
| Male | 317 (76.9) |
| Female | 95 (23.1) |
| Smoking status | |
| Non-smoker | 165 (40.0) |
| Smoker | 247 (60.0) |
| ECOG PS † = 0–2 | 412 (100) |
| Stage | |
| I–II | 20 (4.8) |
| III–IV | 264 (64.1) |
| LD †† | 56 (13.6) |
| ED ††† | 65 (15.8) |
| Histology | |
| Adenocarcinoma | 145 (35.2) |
| Squamous cell | 143 (34.7) |
| Small cell | 124 (30.1) |
| Chemotherapy regimens | |
| Platinum/gemcitabine | 228 (55.3) |
| Platinum/paclitaxel | 55 (13.3) |
| Platinum/navelbine | 8 (1.9) |
| Platinum/etoposide | 113 (27.4) |
| Platinum/irinotecan | 8 (1.9) |
| Severe toxicity | |
| Total no. | 163 (39.6) |
| Hematologic toxicity | 91 (22.1) |
| Gastrointestinal toxicity | 92 (22.3) |
† Eastern Cooperative Oncology Group, performance status; †† Limited Disease; ††† Extensive Disease.
4.2. Data Collection
Clinical data of all patients were collected, including age, sex, smoking status, tumor histology, clinical stage and ECOG PS (Eastern Cooperative Oncology Group, Performance Status). According to National Cancer Institute Common Toxicity Criteria Version 3.0, the incidence of Grade 3 or 4 toxicity during first-line chemotherapy was collected. Toxicities that were associated with treatment that we collected included hematologic toxicity (anemia, leukopenia, neutropenia and thrombocytopenia) and gastrointestinal toxicity (nausea, vomiting and diarrhea). The investigators were blinded to the polymorphism status of the patients. Severe toxicity was defined as any Grade 3 or 4 toxicity; it consisted of Grade 3 or 4 hematologic toxicity and Grade 3 or 4 gastrointestinal toxicity. Toxicity outcomes were grouped into three: (1) any Grade 3 or 4 toxicity; (2) any Grade 3 or 4 hematologic toxicity; (3) any Grade 3 or 4 gastrointestinal toxicity.
4.3. SNP Selecting, DNA Extraction and Genotyping
We selected 28 SNPs of the WISP1 gene from the HapMap database according to the following criteria: (1) the minor allele frequency (MAF) of the SNP was >5% in the Chinese population; (2) haplotype tagger SNPs were selected by Haploview version 4.2 (Cambridge, MA, USA) using the pair-wise tagging with default settings (pair-wise r2 threshold = 0.8); and (3) SNPs in the promoter region, exon region and 3’UTR. Additionally, 2 SNPs that reported to be associated with cancer risk or clinical outcome were selected.
Genomic DNA of all subjects were isolated from the peripheral blood sample using the FlexiGene DNA Kit (Qiagen, Hilden, Germany) and stored at 4 °C until use. Genotyping was conducted by Sequenom’s MassARRAY system (Sequenom, San Diego, CA, USA).
4.4. Statistical Analysis
Toxicity outcomes were dichotomized by the presence or absence of Grade 3 or 4 toxicity. χ2 test and Student’s t-test were used to determine the differences in sex, age, smoking status, histology, stage and ECOG between these two groups. Unconditional logistic regression was performed to estimate the association of toxicity outcome with WISP1 polymorphisms by calculating odds ratios (OR) and their 95% confidence intervals (CI) adjusted by the covariates. The p-value was two-sided, and p < 0.05 was considered statistically significant. All association analyses were conducted by three models. The additive model is for the additive effects of SNPs, and the direction of the regression coefficient represents the effect of each extra minor allele. Dominant and recessive models are tests for the minor allele with two of the classes pooled. That is, if A is a minor allele and a is the major allele, the dominant model means (AA, Aa) versus aa, and the recessive model means AA versus (Aa, aa). The aforementioned statistical analyses were performed using PLINK [36] and SPSS 18.0 (SPSS Inc., Chicago, Illinois, USA).
5. Conclusions
We considered that genotypes of WISP1 may be used to predict the platinum-based chemotherapy toxicity in lung cancer patients.
Acknowledgments
We first thank all of support of the funds from the National High-tech R&D Program of China (863 Program) (2012AA02A517), the National Natural Science Foundation of China (81173129, 81202595, 81373490), the Program for the Special Scientific Research Foundation of Doctor Disciplines at the University of Ministry of Education of China (20110162110034), the Hunan Provincial Natural Science Foundation of China (12JJ7006) and the Fundamental Research Funds for the Central Universities of Central South University (2014zzts323). We also thank all of the patients who participated in the study.
Supplementary Materials
Supplementary materials can be found at: http://www.mdpi.com/1422-0067/15/11/21011/s1.
Author Contributions
Juan Chen, Jiye Yin and Zhaoqian Liu conceived of and designed the experiments. Juan Chen performed the experiments. Juan Chen and Jiye Yin analyzed the data. Juan Chen, Xiangping Li, Ying Wang, Chenyue Qian, Yi Zheng, Ling Xiao, Ting Zou, Zhan Wang and Junyan Liu contributed clinical samples and data. Juan Chen wrote the manuscript. Juan Chen, Wei Zhang, Zhaoqian Liu and Honghao Zhou reviewed/edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Spira A., Ettinger D.S. Multidisciplinary management of lung cancer. N. Engl. J. Med. 2004;350:379–392. doi: 10.1056/NEJMra035536. [DOI] [PubMed] [Google Scholar]
- 3.Akiri G., Cherian M.M., Vijayakumar S., Liu G., Bafico A., Aaronson S.A. Wnt pathway aberrations including autocrine wnt activation occur at high frequency in human non-small-cell lung carcinoma. Oncogene. 2009;28:2163–2172. doi: 10.1038/onc.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul I., Chacko A.D., Stasik I., Busacca S., Crawford N., McCoy F., McTavish N., Wilson B., Barr M., O’Byrne K.J., et al. Acquired differential regulation of caspase-8 in cisplatin-resistant non-small-cell lung cancer. Cell Death Dis. 2012;3:e449. doi: 10.1038/cddis.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olaussen K.A., Planchard D., Adam J., Soria J.C. DNA repair pathways and non-small cell lung cancer: Clinical perspectives. Bull. Cancer. 2011;98:305–322. doi: 10.1684/bdc.2011.1327. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.Y., Kang H.G., Yoo S.S., Kang Y.R., Choi Y.Y., Lee W.K., Choi J.E., Jeon H.S., Shin K.M., Oh I.J., et al. Polymorphisms in DNA repair and apoptosis-related genes and clinical outcomes of patients with non-small cell lung cancer treated with first-line paclitaxel-cisplatin chemotherapy. Lung Cancer. 2013;82:330–339. doi: 10.1016/j.lungcan.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Kim S.H., Lee G.W., Lee M.J., Cho Y.J., Jeong Y.Y., Kim H.C., Lee J.D., Hwang Y.S., Kim I.S., Lee S., et al. Clinical significance of ercc2 haplotype-tagging single nucleotide polymorphisms in patients with unresectable non-small cell lung cancer treated with first-line platinum-based chemotherapy. Lung Cancer. 2012;77:578–584. doi: 10.1016/j.lungcan.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 8.Xu L., Corcoran R.B., Welsh J.W., Pennica D., Levine A.J. Wisp-1 is a wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 2000;14:585–595. [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P.P., Li W.J., Wang Y., Zhao S., Li D.Y., Feng L.Y., Shi X.L., Koeffler H.P., Tong X.J., Xie D. Expression of cyr61, ctgf, and wisp-1 correlates with clinical features of lung cancer. PLoS One. 2007;2:e534. doi: 10.1371/journal.pone.0000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H.H., Li Q., Liu P., Liu Y., Li J., Wasserloos K., Chao W., You M., Oury T.D., Chhinder S., et al. Wnt1-inducible signaling pathway protein 1 contributes to ventilator-induced lung injury. Am. J. Respir. Cell Mol. Biol. 2012;47:528–535. doi: 10.1165/rcmb.2012-0127OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berschneider B., Konigshoff M. Wnt1 inducible signaling pathway protein 1 (wisp1): A novel mediator linking development and disease. Int. J. Biochem. Cell Biol. 2011;43:306–309. doi: 10.1016/j.biocel.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Watari H., Xiong Y., Hassan M.K., Sakuragi N. Cyr61, a member of ccn (connective tissue growth factor/cysteine-rich 61/nephroblastoma overexpressed) family, predicts survival of patients with endometrial cancer of endometrioid subtype. Gynecol. Oncol. 2009;112:229–234. doi: 10.1016/j.ygyno.2008.09.039. [DOI] [PubMed] [Google Scholar]
- 13.Pennica D., Swanson T.A., Welsh J.W., Roy M.A., Lawrence D.A., Lee J., Brush J., Taneyhill L.A., Deuel B., Lew M., et al. Wisp genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc. Natl Acad. Sci. USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies S.R., Watkins G., Mansel R.E., Jiang W.G. Differential expression and prognostic implications of the ccn family members wisp-1, wisp-2, and wisp-3 in human breast cancer. Ann. Surg. Oncol. 2007;14:1909–1918. doi: 10.1245/s10434-007-9376-x. [DOI] [PubMed] [Google Scholar]
- 15.Venkatesan B., Prabhu S.D., Venkatachalam K., Mummidi S., Valente A.J., Clark R.A., Delafontaine P., Chandrasekar B. Wnt1-inducible signaling pathway protein-1 activates diverse cell survival pathways and blocks doxorubicin-induced cardiomyocyte death. Cell. Signal. 2010;22:809–820. doi: 10.1016/j.cellsig.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inkson C.A., Ono M., Bi Y., Kuznetsov S.A., Fisher L.W., Young M.F. The potential functional interaction of biglycan and wisp-1 in controlling differentiation and proliferation of osteogenic cells. Cells Tissues Organs. 2009;189:153–157. doi: 10.1159/000151377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkatachalam K., Venkatesan B., Valente A.J., Melby P.C., Nandish S., Reusch J.E., Clark R.A., Chandrasekar B. Wisp1, a pro-mitogenic, pro-survival factor, mediates tumor necrosis factor-alpha (tnf-alpha)-stimulated cardiac fibroblast proliferation but inhibits tnf-alpha-induced cardiomyocyte death. J. Biol. Chem. 2009;284:14414–14427. doi: 10.1074/jbc.M809757200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma S., Tantisira K., Carey V., Murphy A.J., Lasky-Su J., Celedon J.C., Lazarus R., Klanderman B., Rogers A., Soto-Quiros M., et al. A role for wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am. J. Respir. Crit. Care Med. 2010;181:328–336. doi: 10.1164/rccm.200907-1009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J., Yin J.Y., Li X.P. Clin. Lung Cancer. 2014. submitted for publication.
- 20.Lien W.H., Fuchs E. Wnt some lose some: Transcriptional governance of stem cells by wnt/beta-catenin signaling. Genes Dev. 2014;28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novo M.C., Osugui L., Dos Reis V.O., Longo-Maugeri I.M., Mariano M., Popi A.F. Blockage of wnt/beta-catenin signaling by quercetin reduces survival and proliferation of b-1 cells in vitro. Immunobiology. 2014 doi: 10.1016/j.imbio.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Salaroli R., Ronchi A., Buttarelli F.R., Cortesi F., Marchese V., Bella E.D., Renna C., Baldi C., Giangaspero F., Cenacchi G. Wnt activation affects proliferation, invasiveness and radiosensitivity in medulloblastoma. J. Neurooncol. 2014 doi: 10.1007/s11060-014-1621-0. [DOI] [PubMed] [Google Scholar]
- 23.Yuan R., Wang K., Hu J., Yan C., Li M., Yu X., Liu X., Lei J., Guo W., Wu L., et al. Ubiquitin-like protein fat10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying beta-catenin degradation. Cancer Res. 2014 doi: 10.1158/0008-5472.CAN-14-0284. [DOI] [PubMed] [Google Scholar]
- 24.Lee K.B., Ye S., Park M.H., Park B.H., Lee J.S., Kim S.M. P63-mediated activation of the beta-catenin/c-myc signaling pathway stimulates esophageal squamous carcinoma cell invasion and metastasis. Cancer Lett. 2014;353:124–132. doi: 10.1016/j.canlet.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Yamada Y., Ando F., Shimokata H. Association of polymorphisms of sorbs1, gck and wisp1 with hypertension in community-dwelling japanese individuals. Hypertens. Res. 2009;32:325–331. doi: 10.1038/hr.2009.23. [DOI] [PubMed] [Google Scholar]
- 26.Urano T., Narusawa K., Shiraki M., Usui T., Sasaki N., Hosoi T., Ouchi Y., Nakamura T., Inoue S. Association of a single nucleotide polymorphism in the wisp1 gene with spinal osteoarthritis in postmenopausal japanese women. J. Bone Miner. Metab. 2007;25:253–258. doi: 10.1007/s00774-007-0757-9. [DOI] [PubMed] [Google Scholar]
- 27.Baralle D., Baralle M. Splicing in action: Assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buratti E., Baralle M., Baralle F.E. Defective splicing, disease and therapy: Searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matoulkova E., Michalova E., Vojtesek B., Hrstka R. The role of the 3' untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–576. doi: 10.4161/rna.20231. [DOI] [PubMed] [Google Scholar]
- 30.Zamorano J., Erdine S., Lopez A.P., Kim J.H., Al Khadra A., Westergaard M., Sutradhar S., Yunis C. Design and rationale of a real-life study to compare treatment strategies for cardiovascular risk factors: The crucial study. Postgrad. Med. 2010;122:7–15. doi: 10.3810/pgm.2010.03.2117. [DOI] [PubMed] [Google Scholar]
- 31.Klett C.P., Bonner T.I. Identification and characterization of the rat m1 muscarinic receptor promoter. J. Neurochem. 1999;72:900–909. doi: 10.1046/j.1471-4159.1999.0720900.x. [DOI] [PubMed] [Google Scholar]
- 32.Xu X., Duan L., Zhou B., Ma R., Zhou H., Liu Z. Genetic polymorphism of copper transporter protein 1 is related to platinum resistance in chinese non-small cell lung carcinoma patients. Clin. Exp. Pharmacol. Physiol. 2012;39:786–792. doi: 10.1111/j.1440-1681.2012.05741.x. [DOI] [PubMed] [Google Scholar]
- 33.Xu X., Han L., Duan L., Zhao Y., Yang H., Zhou B., Ma R., Yuan R., Zhou H., Liu Z. Association between eif3alpha polymorphism and severe toxicity caused by platinum-based chemotherapy in non-small cell lung cancer patients. Br. J. Clin. Pharmacol. 2013;75:516–523. doi: 10.1111/j.1365-2125.2012.04379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X., Han L., Yang H., Duan L., Zhou B., Zhao Y., Qu J., Ma R., Zhou H., Liu Z. The a/g allele of eif3a rs3740556 predicts platinum-based chemotherapy resistance in lung cancer patients. Lung Cancer. 2013;79:65–72. doi: 10.1016/j.lungcan.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Xu X., Ren H., Zhou B., Zhao Y., Yuan R., Ma R., Zhou H., Liu Z. Prediction of copper transport protein 1 (ctr1) genotype on severe cisplatin induced toxicity in non-small cell lung cancer (nsclc) patients. Lung Cancer. 2012;77:438–442. doi: 10.1016/j.lungcan.2012.03.023. [DOI] [PubMed] [Google Scholar]
- 36.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]