Summary
The Rhesus Site is a resource for information of the ‘Rhesus’ blood group. It is intended for specialists and non-specialists. The website details research in the field relevant for transfusion medicine, immunohematology, and molecular research. Link areas guide to important publications and to methodological resources for Rhesus. Many data originally presented at The Rhesus Site have been formally published later. The ‘RhesusBase’ section represents the largest database for RHD alleles; the ‘RhesusSurveillance’ section details the results of the largest prospective observational study on anti-D immunization events in D-positive patients. Visitors to the website are encouraged to explore the intricacies of the most complex blood group gene locus.
Keywords: Rh-Hr blood group system, Antigen D, Rh blood group, Databases as topic, Immunization
Introduction
In the 1980s, with the accumulation of much new information on the RH blood group system, its complexity became apparent. RH is commonly designated in Germany and elsewhere as ‘Rhesus’ blood group. With the advent of monoclonal antibodies, the characterization of partial D variants made huge progress. The few research laboratories with rare, coveted and well sheltered polyclonal antisera were joined by many laboratories freely sharing their monoclonal antibody resources and associated data. Partial D phenotypes were found to lack specific epitopes defined by monoclonal antibodies [1], distinguishing them from weak D believed to express normal D antigen in reduced amounts. A plethora of ‘new’ partial Ds that did not fit the D category classification were described and used for the characterization of monoclonal anti-D, such as DFR, DBT, DHMi, and DHMii [2]. With every workshop on monoclonal antibodies to red blood cells [3, 4, 5, 6], the number of D epitopes increased, and the D antigen seemed to become an extremely complex entity. Still, the RH antigens resisted many attempts to characterize their molecular basis: discussions continued whether all RH antigens derived from a single gene or whether there were three genes coding for C, D, or E. It was not until 1990 that the sequence of the Rh protein was elucidated [7, 8]. This event opened up a real explosion of knowledge on RH in the 1990s, with milestones succeeding in an ever accelerating path: sequence of RHCE [7, 8]; demonstration that C and E were encoded by the same gene [9]; demonstration that D was encoded by a separate gene [10]; characterization of RHD [11, 12]; and hybrid alleles as basis for many D variants [13].
In 1994, the immunohematology research group in Ulm began focusing on RH. Initial contributions were the frequency of the D category VI (today known as DVI) in the German population [14]; characterization of the partial D DNU [15]; and of a third molecular basis for DVI [16], which proved that the low antigen density of many DVI could not be explained by the loss of D epitopes. Based on these contributions, the German Guidelines for Blood Group Determination and Blood Transfusion were changed in 1996 [17]: the use of monoclonal anti-D with distinct characteristics was implemented for determination of the D antigen; the concept of ‘Du’ carriers considered as D-positive blood donors and D-negative transfusion recipients was replaced by distinct recommendations for weak D (D-positive transfusion strategy) and partial D carriers (D-negative transfusion strategy, especially for DVI). Many international guidelines have followed this example set by the German Guidelines in 1996.
These events prompted us in 1998 to establish ‘The Rhesus Site’. The name is based on the commonplace designation of the RH blood group in Germany and many other countries. At this time, hosting of webpages at universities, let alone at blood centers, was in its infancy. Hence, the webpages constituting The Rhesus Site were established as an adjunct to the homepage of the laboratory and research head at the University of Ulm webserver, where it has remained to this day.
Features of The Rhesus Site
The Rhesus Site has been designed to propagate knowledge of RH and to enable visitors to perceive major features of the RH blood group system. Its special focus is on the work done by the Ulm group with regular updates until 2009. It features four sections, which are separated by horizontal bars on the first page of The Rhesus Site: i) research projects of the Ulm group (fig. 1), ii) links to online publications of the Ulm group, iii) links to other Rhesus-related resources, and iv) links to publications of the Ulm group in German.
Fig. 1.
The graphic appearance of The Rhesus Site, when visitors access its URL (www.uni-ulm.de/wflegel/RH/). Beneath the heading and a horizontal bar, the first section has links to 4 major areas of scientific research in the Rh blood group system.
Presentation of the Research Projects
When accessing The Rhesus Site online, the first page presents the four major areas of our research (fig. 1). The pair of tiles at the top leads to two important areas in their own right: the RhesusBase and the Rhesus Immunization Registry (‘RhesusSurveillance’). The next pair of tiles links to didactic material for the Rhesus gene organization on chromosome 1 [18] and a schematic 3D model of the Rh protein.
The next pair of tiles within the first section (not shown) features the numerous serologically D-negative alleles of the RHD gene and guidelines for the weak D nomenclature. The final pair of tile presents two collaborative efforts of the Ulm group: the EU funded BloodGen consortium from 2003 to 2008 and the flow cytometry workshop section of the 4th International Workshop on Monoclonal Antibodies to Red Blood Cells and Related Antigens in 2001. Since the BloodGen consortium dissolved after 2009, the link to its extensive documentation is lost.
Three of these research projects proved to be of great practical relevance and are described separately in detail: i) the RhesusBase, ii) the RhesusSurveillance, and iii) guidelines for weak D nomenclature.
The 3 Link Areas
The 3 link areas had been designed to allow visitors an easy, direct access to publications on the Rhesus blood group. The intended audience included non-experts. Hence much effort was placed establishing direct links to full-text versions of references, thereby easing the access for visitors not experienced with literature databases. In the late 1990s this idea of open access was not generally shared.
The first link area of The Rhesus Site presents links to our work, loosely grouped into online publications, online reviews, and online theses. Rather than being merely a publication list, the studies are sorted in a didactic way and link to online full-text versions of the respective publications, thus permitting the most rapid access even for non-experts. Some of these full-text versions are hosted on The Rhesus Site webserver. Examples are the publication on blood group frequencies [14], a review on RH phenotype prediction by DNA typing [19], and a review of the molecular biology of partial D and weak D [20]. The links to ‘thesis work’ may be helpful even for experts, since these works are notoriously difficult to retrieve online.
The second link area is dedicated to an array of different resources, hosted both inside and outside the local webserver. External links include the OMIM database at the National Institutes of Health and the Blood Group Antigen Mutation Database as well as animations and animated slide shows related to blood groups.
There are two internal links: One leads to a part of The Rhesus Site dedicated to methods in immunohematology (fig. 2). Protocols for antibody elution, RBC cryopreservation and DNA extraction as well as flow cytometry are documented; the ‘KryoBeads’ or ‘droplet freezing’ method is even demonstrated in video clips, which were later revised, published [21] and can be found on YouTube since 2011. The other internal link leads to a comparison of the German D typing recommendations of 1996 and 2000.
Fig. 2.
Methods section of The Rhesus Site. The links lead to detailed descriptions of major methods in the RH field. Features include video clips depicting RBC freezing and a protocol to determine the D antigen densities, which was used during an international workshop.
The third and final link area is dedicated to publications in German and largely focused on use by medical technologists. The links lead to full text versions of the publications, and several of these publications are directly hosted on the local webserver. Examples are a publication in MTA Dialog on the molecular biology of partial D and weak D [22]. Another link, explaining the determination of the D antigen in a transfusion recipient, leads to a hypertext adaptation of an article in the MTA Zeitschrift [23].
Guidelines for ‘Weak D Nomenclature’
A nomenclature based on ‘weak D types’ was introduced in 1999, when we described 16 new RHD alleles [24]. This single publication almost doubled the number of RHD alleles known at that time. Today, there are 86 weak D types, some with subtypes, such that the total number approaches 100 and represent the RHD allele group with the largest number of alleles as listed in the RhesusBase. These weak D types have become part of clinical practice worldwide. In 1999, a webpage with guideline for weak D nomenclature was added to The Rhesus Site and is a frequently used feature for requesting a weak D number for a newly identified RHD allele. There were 4 main requirements to assign a weak D type (table 1).
Table 1.
Principles for weak D nomenclature
| The full length coding sequence (CDS) by nucleotide sequencing |
| A non-exofacial membrane location of the involved amino acid substitution(s) |
| Weak expression of the D antigen |
| Documentation of the results in the public record |
i) A new RHD allele as defined by the nucleotide sequencing of the full length RHD coding sequence, either as cDNA or from genomic DNA covering all 10 RHD exons.
ii) At least one amino acid substitution located in the membrane or below the membrane of the red blood cell. Usually, no amino acid substitution will be present at or above the membrane as this is likely to cause a partial D phenotype.
iii) Proof of a weak D phenotype by a peer approved method. The preferred method is D antigen density determination of less than about 5,000 antigens per red blood cell using a flow cytometry method. As an alternative, weak reactivity by standard serology with monoclonal antibodies of IgM or IgG type in tube or gel is also acceptable.
iv) Deposition of the RHD allele in a nucleotide sequence databases. The rules for these database entries have changed over the years, such that more documentation can be presented online. Examples for recently deposited database entries can be found for weak D types 3, 10, 25 and 86 (GenBank accession numbers KF680198, KC915029, JX495049 and KJ722727; National Center for Biotechnology Information, Bethesda, MD, USA).
The RhesusBase
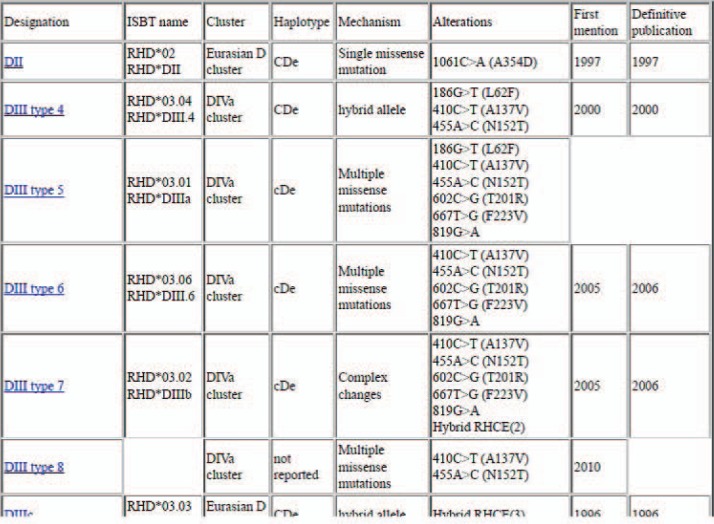
The RhesusBase has become the most widely used feature of The Rhesus Site. It provides an up-to-date complete listing of the known RHD alleles as well as their molecular structure, phenotypic information and geographical or ethnical occurrence.
The RhesusBase started out in 1998 as a set of html-files covering RHD gene variants (alleles). For each RHD allele the following information was gathered, evaluated and presented in a structured way: nucleotide and amino acid changes relative to the ‘standard RHD’ sequence; initial description (with references); serologic data; distribution and frequency; haplotype and RHD allele cluster; and additional comments, often unavailable in the published literature. These html-files were written using a simple text editor, such as Microsoft Word. In separate frames, link lists are presented to allow rapid screening of the allele files, sorted by various criteria.
These link lists included subsets of the alleles based on their phenotype (weak D, D category, other partial D, incompletely characterized weak or partial D, D-negative and DEL, normal D-positive), molecular type (hybrid gene, single nucleotide exchange, multiple nucleotide exchanges, deletions or insertions, splice site mutations, premature stop codons) and sorted listings of the alleles according to name or mutation and a listing of RHCE alleles with RHD fragments. All information was presented in this manually maintained assortment of html-files, the addition of a single allele included writing a new html-file with the allele data and editing all relevant link lists. Thus, many data had to be entered multiple times, and there was a possibility of broken links (if the name of the html-file had been changed) or alleles missing from a link list (if the update of this link list had been omitted).
In 2011, a major rework designated RhesusBase V2 was instituted, and the manual data entry was replaced by entering the data into a single database with all information. A major advantage of this re-design was that any information needs to be entered only once: the html-files of an allele and the link lists are now generated automatically. As a result, maintaining the link lists became easy allowing for a larger number of criteria and data sets. Therefore, alleles are now listed according to haplotype, allele cluster, name, ISBT name, mutation, year of publication, year of first description, input date, last update, and geographic information (fig. 3). As part of this re-design, the quality assurance of data entry was much improved, including an automated check comparing whether the entered mutation fitted the wild type nucleotide and whether the reported amino acid change corresponded to the reported nucleotide change. These checks prevented incorrect data entries and even helped to detect errors in some publications. The ‘serology’ feature is increasingly used to add serologic information, although the coverage is much less complete than the allele listing. Likewise, with RhesusBase V2 the technological basis for entering RHCE data has been established, but these data are not shown yet.
Fig. 3.
View of the RhesusBase. In the left frame (‘Gene shown’), a grouping criterion can be selected. In this example, ‘D category’ was chosen. In the right frame showing a table (‘D category’), all related RHD alleles are listed, which link to files detailing the information known for any given allele. The original RhesusBase (version 1) has been replaced by the current version RhesusBase V2, which is hosted by the University of Ulm webserver.
One of the most important achievements of the RhesusBase is a rapid orientation on the knowledge of a new allele. If an investigator detects a mutation, she or he can just go to the ‘allele by mutation’ listing and scroll until the mutation is found. If the mutation is not listed, no allele with this mutation has been entered in the RhesusBase and the mutation is either new or has been described only recently. If the mutation is denoted, the alleles carrying these mutations are listed and may be accessed by a hyperlink.
For each allele, data available include molecular structure, key changes form the standard allele, ISBT proposed designation, nucleotide changes relative to ‘standard RHD‘, amino acid changes relative to standard RhD protein, typical haplotype in which the allele occurs, allele cluster, references, ISBT group, phenotypic grouping, references for serology, references for observations, and possibly comments of the curator. In contrast to other allele databases, the RhesusBase V2 is intended to also cover serologic and geographic information.
The RhesusBase representation on the web is not a database but an accumulation of html-files; therefore, there is no possibility for automated querying. By design, this is not seen as a problem, as the RhesusBase was instituted to help scientists orient among RHD alleles; for this task, the current interactive format seems sufficient. The main limitation of the RhesusBase – as well as other manually maintained data sources – is the timeliness of data. The RhesusBase is intended to be more than a mirror of the data present in automated databases or by automated literature database searches, hence entering the alleles includes appraisal of the data for any allele, which is a time-consuming but valuable process.
Rhesus Immunization Registry
Rhesus Surveillance is another important information resource on The Rhesus Site. The link leads to the Rhesus Immunization registry (RIR). RIR was constituted on February 10, 1998 as a tool to gauge the possible impact of the change of the D typing strategy introduced by the German Guidelines in 1996. The objective was to allow any immunohematologist in Germany to send D-positive samples with anti-D to a molecularly skilled laboratory, where the molecular structure of the allele would be identified and the anti-D specificity confirmed or ruled out.
RIR was originally intended to cover all German-speaking countries. In practice all samples from any country were accepted and, until the RIR was closed in 2009, no sample fulfilling the inclusion criteria was ever rejected (table 2).
Table 2.
Rhesus Immunization Registry (RIR) 1998–2009
| Study inclusion criteria |
| The proband is Rhesus D-positive or it was not possible to exclude an antigen D or RHD-specific nucleotide sequences |
| The proband carries an anti-D antibody or it was not possible to exclude an anti-D |
| The proband's residence is in a German-speaking country (in practice, sample from anywhere worldwide had been accepted, but were registered separately) |
| The proband's identification shall be in accordance with chapter 2.3. of the ‘Guidelines’ (initials, birthday) in order to exclude duplicate entries |
| The residence (city, county or state) and the exact and complete transfusion and pregnancy history have to be submitted |
| Study exclusion criteria |
| Rh-negative probands |
| The probands has a proven negative transfusion and pregnancy history |
The RIR was closed in March 2009. Until then, 124 samples had been collected and analyzed regarding the RHD molecular basis, type of antibody (alloantibody vs. autoantibody) and titer of antibody (fig. 4). One immediate success of the RIR was the identification of DNB [25], a partial D allele that was typed as D-positive by most typing strategies and was rather frequent in the Austria and Switzerland. Several additional publications in journals, such as Blood and Transfusion, followed and the publication of study results continues.
Fig. 4.
Rhesus Immunization Registry (RIR) – results page. The contributing serologist, type and titer of anti-D, possible immunization risk, and RHD allele involved are listed. For example, the 2 original samples of DNB were entries 2 and 3 [25].
With more than 100 presumed anti-D immunization events in D-positive individuals, the RIR is a source of clinically relevant information: When a partial D is described, it is often difficult to discern its practical role: Does the partial D occur in a relevant frequency? Is there a relevant likelihood that its carrier will be transfused with D-positive blood, or do current typing strategies ensure a D-negative transfusion strategy? And finally, if the carrier is transfused with D-positive blood, is there a relevant risk for anti-D production? The answers vary among partial Ds. For example, DVII is rather frequent [26] and often typed and transfused as D-positive without much clinical consideration or consequence. DVI is much rarer [14] and should not be mistaken as D-positive if German standard typing strategies are applied. Still, there are more cases of DVI with anti-D listed in RIR than of DVII with anti-D, indicating that the likelihood of immunization is much higher for a DVI recipient than for a DVII recipient. For an evidence-based decision process to improve D typing strategies, RIR provides valuable data and represents the largest dataset to date.
Conclusion
Social media activities in the transfusion medicine community fostered research collaborations, one of which was completed in 2001 [27, 28]. For a decade, The Rhesus Site was an instrument for research collaborations and result dissemination and was regularly updated between 1998 and 2009.
The Rhesus Site remains an important resource for anyone interested in the Rhesus blood group. It hosts or links to resources relevant for the Rhesus blood group field. The type of information ranges from publications on Rhesus for non-experts in the field to very specialized resources that are highly relevant to expert discussions. There are ongoing research projects, originally presented at The Rhesus Site, still to be published.
While some features of The Rhesus Site were successful, but now completed or published, others are still thriving: for example, the RhesusBase was initially just one among several features of The Rhesus Site. Over the years, the RhesusBase has developed into a key international resource for research, development and diagnostics, wherever up to date knowledge of RHD gene variants (alleles) is needed.
Web Resources
The Rhesus Site (www.uni-ulm.de/wflegel/RH/)
Weak D nomenclature (www.uni-ulm.de/wflegel/RH/WeakD/)
GenBank (www.ncbi.nlm.nih.gov/nuccore/KJ722727.1)
RhesusBase (www.uni-ulm.de/fwagner/RH/RB2/)
Rhesus Immunization Registry (www.uni-ulm.de/wflegel/RH/RIR/#title).
Authorship Contribution
WAF designed the concept of The Rhesus Site and its content; he maintained the webpages until 2009. FFW is maintaining the RhesusBase. Both authors wrote the manuscript.
Statement of Disclaimer
The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Disclosure Statement
FFW receives royalties for RHD genotyping. WAF receives royalties and holds intellectual property rights for RHD genotyping applications.
Acknowlegdement
We express our very great appreciation to all scientists who provided samples and data for the Rhesus Immunization Registry or who provided data and suggestions for the RhesusBase. We are particularly grateful to the editors and publishers of TRANSFUSION MEDICINE AND HEMOTHERAPY, Clinical Laboratory, Transfusion Medicine, MTA Dialog and Deutsches Ärzteblatt for giving us early on the permission to host full-text copies of our publications on The Rhesus Site webserver. Without the provision of free webspace and external FTP access by the University of Ulm, the whole project of The Rhesus Site would not have been possible; we thank the administrators of the Kommunikations- und Informationszentrum (KIZ), whose help was critical for the success, particularly in the beginning of the project in the late 1990s. We thank the former and the current head of the Department of Transfusion Medicine of the University of Ulm, Bernhard Kubanek and Hubert Schrezenmeier, for continuously supporting The Rhesus Site project and its IT solution at the University of Ulm, even after both authors had left the Institut Ulm of the DRK Blutspendedienst Baden-Württemberg – Hessen in 2003 or 2009, while retaining their academic affiliations with the University of Ulm.
References
- 1.Lomas C, Tippett P, Thompson KM, Melamed MD, Hughes-Jones NC. Demonstration of seven epitopes on the Rh antigen D using human monoclonal anti-D antibodies and red cells from D categories. Vox Sang. 1989;57:261–264. doi: 10.1111/j.1423-0410.1989.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 2.Jones J, Filbey D. Selection of monoclonal antibodies for the identification of D variants: ability to detect weak D and to split epD2, epD5 and epD6/7. Vox Sang. 1996;70:173–179. doi: 10.1111/j.1423-0410.1996.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 3.First International Workshop on Monoclonal Antibodies Against Red Blood Cell and Related Antigens. Paris, September 21–24, 1987. Vol. II: Other blood group systems. Rev Fr Transfus Immunohematol. 1988;31:115–486. [PubMed] [Google Scholar]
- 4.Proceedings of the Second International Workshop and Symposium on Monoclonal Antibodies Against Human Red Blood Cells and Related Antigens. J Immunogenet. 1990;17:213–357. 1–4 April, 1990, Lund, Sweden. [PubMed] [Google Scholar]
- 5.Proceedings of the 3rd International Workshop and Symposium on Monoclonal Antibodies against Red Blood Cells and Related Antigens. Transfus Clin Biol. 1996;3:329–541. Nantes, 25–27 September 1996. [PubMed] [Google Scholar]
- 6.Proceedings of the Fourth International Workshop on Monoclonal Antibodies Against Human Red Blood Cell and Related Antigens. Under the Aegis of the Institut National de la Transfusion Saguine and of the Etablissement Francais du Sang. Transfus Clin Biol. 2002;9:1–108. July 19–20. Paris, France. [PubMed] [Google Scholar]
- 7.Chérif-Zahar B, Bloy C, Le Van Kim C, Blanchard D, Bailly P, Hermand P, Salmon C, Cartron JP, Colin Y. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci U S A. 1990;87:6243–6247. doi: 10.1073/pnas.87.16.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avent ND, Ridgwell K, Tanner MJ, Anstee DJ. cDNA cloning of a 30 kDa erythrocyte membrane protein associated with Rh (Rhesus)-blood-group-antigen expression. Biochem J. 1990;271:821–825. doi: 10.1042/bj2710821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mouro I, Colin Y, Chérif-Zahar B, Cartron JP, Le Van Kim C. Molecular genetic basis of the human Rhesus blood group system. Nat Genet. 1993;5:62–65. doi: 10.1038/ng0993-62. [DOI] [PubMed] [Google Scholar]
- 10.Colin Y, Chérif-Zahar B, Le Van Kim C, Raynal V, Van Huffel V, Cartron JP. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- 11.Le van Kim C, Mouro I, Chérif-Zahar B, Raynal V, Cherrier C, Cartron JP, Colin Y. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci U S A. 1992;89:10925–10929. doi: 10.1073/pnas.89.22.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arce MA, Thompson ES, Wagner S, Coyne KE, Ferdman BA, Lublin DM. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood. 1993;82:651–655. [PubMed] [Google Scholar]
- 13.Mouro I, Le Van Kim C, Rouillac C, van Rhenen DJ, Le Pennec PY, Bailly P, Cartron JP, Colin Y. Rearrangements of the blood group RhD gene associated with the DVI category phenotype. Blood. 1994;83:1129–1135. [PubMed] [Google Scholar]
- 14.Wagner FF, Kasulke D, Kerowgan M, Flegel WA. Frequencies of the blood groups ABO, Rhesus, D category VI, Kell, and of clinically relevant high-frequency antigens in south-western Germany. Infusionsther Transfusionsmed. 1995;22:285–290. doi: 10.1159/000223144. [DOI] [PubMed] [Google Scholar]
- 15.Avent ND, Jones JW, Liu W, Scott ML, Voak D, Flegel WA, Wagner FF, Green C. Molecular basis of the D variant phenotypes DNU and DII allows localization of critical amino acids required for expression of Rh D epitopes epD3, 4 and 9 to the sixth external domain of the Rh D protein. Br J Haematol. 1997;97:366–371. doi: 10.1046/j.1365-2141.1997.632710.x. [DOI] [PubMed] [Google Scholar]
- 16.Wagner FF, Gassner C, Muller TH, Schonitzer D, Schunter F, Flegel WA. Three molecular structures cause rhesus D category VI phenotypes with distinct immunohematologic features. Blood. 1998;91:2157–2168. [PubMed] [Google Scholar]
- 17.Wissenschaftlicher Beirat der Bundesärztekammer. Richtlinien zur Blutgruppenbestimmung und Bluttransfusion (Hämotherapie). Kapitel 2.5.5: Bestimmung des Rh-Merkmals D. Köln, Deutscher Ärzteverlag. 1996.
- 18.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–3668. [PubMed] [Google Scholar]
- 19.Flegel WA, Wagner FF, Müller TH, Gassner C. Rh phenotype prediction by DNA typing and its application to practice. Transfus Med. 1998;8:281–302. doi: 10.1046/j.1365-3148.1998.00173.x. [DOI] [PubMed] [Google Scholar]
- 20.Flegel WA, Wagner FF. Molecular biology of partial D and weak D: implications for blood bank practice. Clin Lab. 2002;48:53–59. [PubMed] [Google Scholar]
- 21.Schmid P, Huvard MJ, Lee-Stroka AH, Lee JY, Byrne KM, Flegel WA. Red blood cell preservation by droplet freezing with polyvinylpyrrolidone or sucrose-dextrose and by bulk freezing with glycerol. Transfusion. 2011;51:2703–2708. doi: 10.1111/j.1537-2995.2011.03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flegel WA, Wagner FF. Molekularbiologie von partial D und weak D. MTA Dialog. 2002;3:662–665. [Google Scholar]
- 23.Flegel WA, Northoff H, Wagner FF. Rhesus-D-Bestimmung beim Transfusionsempfänger. MTA Zeitschrift. 1998;13:8. [Google Scholar]
- 24.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood. 1999;93:385–393. [PubMed] [Google Scholar]
- 25.Wagner FF, Eicher NI, Jørgensen JR, Lonicer CB, Flegel WA. DNB: a partial D with anti-D frequent in Central Europe. Blood. 2002;100:2253–2256. doi: 10.1182/blood-2002-03-0742. [DOI] [PubMed] [Google Scholar]
- 26.Wagner FF, Hillesheim B, Flegel WA. D-category VII depends on amino acid substitution Leu(110) Pro (in German) Beitr Infusionsther Transfusionsmed. 1997;34:220–223. [PubMed] [Google Scholar]
- 27.Chambers LA. Special interest groups at the AABB website. Transfusion. 2000;40:396–397. doi: 10.1046/j.1537-2995.2000.40040396.x. [DOI] [PubMed] [Google Scholar]
- 28.Flegel WA, Khull SR, Wagner FF. Primary anti-D immunization by weak D type 2 RBCs. Transfusion. 40:428–434. doi: 10.1046/j.1537-2995.2000.40040428.x. [DOI] [PubMed] [Google Scholar]