Abstract
Exposure to Paraquat and RNA interference knockdown of Mn or mitochondrial superoxide dismutase (Sod2) are known to result in significant lifespan reduction, locomotor dysfunction, and mitochondrial degeneration in Drosophila melanogaster. Both perturbations increase the flux of the progenitor ROS, superoxide, but the molecular underpinnings of the resulting phenotypes are poorly understood. Improved understanding of such processes could lead to advances in the treatment of numerous age-related disorders. Superoxide toxicity can act through protein carbonylation. Analysis of carbonylated proteins is attractive since carbonyl groups are not present in the twenty canonical amino acids and are amenable to labeling and enrichment strategies. Here, carbonylated proteins were labeled with biotin hydrazide and enriched on streptavidin beads. On-bead digestion was used to release carbonylated protein peptides, with relative abundance ratios versus controls obtained using the iTRAQ MS-based proteomics approach. Western blotting and biotin quantitation assay approaches were also investigated. By both western blotting and proteomics, Paraquat exposure, but not Sod2 knockdown, resulted in increased carbonylated protein relative abundance. For Paraquat exposure versus control, the median carbonylated protein relative abundance ratio (1.53) determined using MS-based proteomics was in good agreement with that obtained using a commercial biotin quantitation kit (1.36).
Keywords: cytochrome c oxidase, Paraquat, protein carbonylation, Sod2 knockdown, superoxide
1. Introduction
Damage caused by endogenous ROS is closely associated with aging [1], cancer [2], cardiovascular disease [3], diabetes [4], and neurodegenerative disorders [5]. The mitochondrial electron transport chain (ETC) is often regarded as the chief source of the progenitor ROS, superoxide, O2•−, but the relative importance of the numerous cellular sources is far from certain [6]. Mitochondrial O2•− production sites have been studied extensively, but most work relies on ETC inhibitors that produce artificial conditions [7]. However, progress is being made toward the determination of native O2•− production rates [8]. O2•− toxicity in the membrane environment is likely to proceed through protonation of O2•− to produce HO2•, a powerful lipid peroxidation agent [9], while in aqueous environments the exceptionally reactive species HO• can be produced in the iron-catalyzed reaction of O2•− with H2O2 [10]. Synergistically, elevated O2•− levels have been shown to result in the release of protein-bound iron in yeast [11]. Toxicity is reduced by the superoxide dismutases, which catalyze the disproportionation of O2•− to H2O2 and O2 [12]. While O2•− will undergo spontaneous disproportionation at a lower rate in the absence of superoxide dismutases, the importance of rapid destruction and low steady-state concentrations of O2•− is illustrated by the inactivation of catalase by O2•− [13].
Although numerous products of ROS damage to biomolecules are being actively studied, here we focus on carbonylated proteins due to the introduction of a functional group that is both relatively stable and amenable to labeling and enrichment strategies. Protein carbonyls can result from direct oxidation by ROS [14] and addition of lipid peroxidation products [15]. Similarly to ROS in general, increases in protein carbonylation have been observed on aging [16] and during age-related conditions such as Alzheimer’s disease [17], diabetes [18], kidney disease [19], and Parkinson’s disease [20]. The impact and analysis of protein carbonylation have been extensively reviewed [21–24], with a recent example listing 179 carbonylated proteins associated with aging [25]. The present study is based on our working hypothesis that protein carbonylation is not a random process, but is localized to sites with features such as transition metals able to catalyze HO• generation or hydrophobic pockets that promote interactions with lipid peroxidation products.
In this work, using Drosophila melanogaster as a model system, we have investigated the impact on protein carbonylation levels resulting from two contrasting approaches, one exogenous and one endogenous, intended to increase O2•− concentrations. Oxidative stress was increased first by application of Paraquat (methyl viologen; 1,1’-dimethyl-4,4’-bipyridinium dichloride) and second by the RNA interference knockdown of the antioxidant enzyme, Mn or mitochondrial superoxide dismutase (SOD2), which is localized to the mitochondrial matrix. Paraquat, which is a redox cycling agent capable of producing O2•− on reduction by a suitable electron donor (typically an oxidoreductase enzyme that uses NADH or NADPH as electron donor [26]), has been used to induce oxidative stress in cultured mammalian cells [27], isolated mitochondria [28], D. melanogaster [29], Caenorhabditis elegans [30], and mice [31]. Paraquat exposure in D. melanogaster is known to result in significantly reduced lifespan and locomotor dysfunction [32] and mitochondrial degeneration [33]; furthermore, Paraquat application has been used to model Parkinson’s disease [34]. Paraquat-induced O2•− generation would be expected throughout the cell, with perhaps greatest flux in mitochondria [35,36]. In contrast, removal of SOD2 would be expected to produce much higher steady-state O2•− concentrations localized to the mitochondrial matrix. As with Paraquat exposure, silencing of Sod2 in D. melanogaster has been shown to result in significant lifespan reduction, locomotor dysfunction, and mitochondrial degeneration [37,38]. However, normal lifespan in these flies can be restored by hypoxia [39] and in an investigation of Sod2 knockout in yeast grown under aerobic conditions, while a number of carbonylated proteins were identified, no increase in aggregate carbonylation levels was observed [40].
Hydrazide chemistry is commonly used for labeling protein carbonyls [41]. There have been numerous reports in recent years of carbonylated proteins being labeled with biotin hydrazide reagents and then enriched using avidin affinity [42–48]. However, streptavidin, which has similar affinity for biotin, is often used in place of avidin due to reduced non-specific binding [49]. Streptavidin, which has been shown to be resistant to denaturation with high-concentration urea solutions [50], is especially suitable for SPE enrichment approaches since harsh washing conditions can be employed. Soreghan et al. [51] employed streptavidin affinity-based enrichment of biotin-labeled carbonylated proteins in a high-throughput proteomics investigation of carbonylation in mouse brain. Here we use immobilized streptavidin for the isolation of biotinylated carbonylated proteins. Tryptic digestion was performed on-bead as efficient elution of bound proteins proved to be challenging. The resulting peptide samples from Paraquat-exposed, Sod2 knockdown, and associated control flies were labeled with iTRAQ reagents [52] to allow relative carbonylated protein abundances to be obtained. For both the endogenous and exogenous oxidative stress systems we were able to collect abundance ratios that were in close agreement across full biological replicates, suggesting the robustness and repeatability of our analytical method.
2. Materials and methods
2.1. Materials
Biotin hydrazide, calcium chloride, iodoacetamide, sodium cyanoborohydride, SDS, and triethylammonium bicarbonate were purchased from Sigma (St. Louis, MO). Ammonium bicarbonate was supplied by J.T. Baker (Phillipsburg, NJ) and DMSO, DTT, and HPLC-grade water and methanol were obtained from Fisher Scientific (Pittsburgh, PA). Sequencing-grade modified trypsin was from Promega (Madison, WI) and iTRAQ reagent kits were purchased from AB SCIEX (Framingham, MA).
2.2. Fruit fly oxidative stress systems
Flies for all experiments were reared under standard conditions [53]. RNA interference Sod2 knockdown was achieved using da-Gal4 to drive ubiquitous expression of the UAS-Sod2-IR24 transgene as previously described by Martin et al. [38]. Two control populations of flies were produced for the Sod2-knockdown study, the first expressing the da-Gal4 driver alone and the second expressing only the UAS-Sod2-IR24 transgene. w1118 flies (Stock Number 5905; Drosophila Stock Center; Bloomington, IN) were used in Paraquat-exposure studies, which were performed essentially as described by Goddeeris et al. [54].
2.3. Aging or Paraquat exposure and initial preparation
Mixed populations of males and females were grown to adulthood, collected under brief CO2 anesthesia, placed into food vials (25 flies per vial), allowed to recover overnight, and then aged (Sod2-knockdown studies) or exposed to Paraquat until ~50% were dead. Age-matched populations of control flies were handled in parallel. Batches of ~500 living flies per group were frozen at −20°C overnight and then homogenized with 15 strokes of a Dounce homogenizer in 5 mL ice-cold buffer (100 mM sucrose, 100 mM KCl, 50 mM Tris-HCl, 1 mM KH2PO4, 0.1 mM EGTA, adjusted to pH 7.3) containing the protease inhibitors aprotinin, leupeptin, and pepstatin A at 500 ng/mL. Homogenates were centrifuged at 300 × g for 20 min at 4°C to remove large debris. Supernatants were stored at −20°C until further processing could be performed. Protein concentrations in the homogenates were determined using the Lowry protein assay (DC Protein Assay; Bio-Rad; Hercules, CA).
2.4. Biotinylation of protein carbonyls
Biotin hydrazide was added to Paraquat-exposed, Sod2-knockdown, and associated control fly homogenates (500-µg total protein aliquots) to 5 mM. Samples were then incubated for 1 h at room temperature with gentle shaking before SDS was added to 2% (w/v). The labeling reaction was allowed to proceed for a further 2 h after a second biotin hydrazide addition had been made. Parallel experiments where biotin hydrazide addition was omitted were performed to allow assessment of non-specific binding. Hydrazone-bond reduction (thus fixing the label) was achieved by adding sodium cyanoborohydride to 15 mM and then incubating at 4°C for 1 h. Samples were then desalted using PD midiTrap G-25 columns (GE Healthcare; Piscataway, NJ) and protein concentrations were determined using the bicinchoninic acid (BCA) assay (Pierce Biotechnology; Rockford, IL).
2.5. Western blotting for protein carbonyls
Beginning with unlabeled homogenates, OxyBlot kits (Millipore; Billerica, MA) were used as recommended by the manufacturer to visualize protein carbonyls through derivatization with 2,4-dinitrophenylhydrazine (DNPH) followed by western blotting with an antibody specific for the dinitrophenyl group. Enhanced chemiluminescence kits (GE Healthcare; Piscataway, NJ) were used for visualization. Alternatively, starting with homogenates labeled with biotin hydrazide as described above, western blotting with HRP-conjugated streptavidin (Pierce Biotechnology), with visualization achieved using TMB substrate (Promega), was investigated.
2.6. Peptide biotin quantitation assay
Urea and DTT were added to biotinylated protein samples (and parallel samples where biotin hydrazide addition had been omitted) to achieve final concentrations of 8 M and 10 mM, respectively. Denaturation and reduction was allowed to proceed for 1 h at 37°C. Cysteines were capped by adding iodoacetamide to 55 mM and then continuing incubation at room temperature in the dark for 1 h. Samples were prepared for digestion by diluting 10× in PBS and then adding calcium chloride to 1 mM and trypsin to achieve a circa 1:50, trypsin:protein ratio. Digestion was allowed to proceed for 16 h at 37°C and then terminated by acidification with 10% (v/v) TFA (aq). The resulting peptide solutions were desalted using RP, C18 SPE cartridges (Sep-Pak; Waters; Milford, MA) and then dried by centrifugal evaporation. To determine aggregate biotinylation levels, the amount of peptide-bound biotin was measured using a biotin quantitation kit (Pierce Biotechnology), based on the displacement of 4’-hydroxyazobenzene-2-carboxylic acid (HABA) from avidin, as recommended by the manufacturer.
2.7. Enrichment of biotinylated proteins and on-bead tryptic digestion
Biotinylated protein samples were added to streptavidin UltraLink resin (Pierce Biotechnology), which had previously been washed three times with PBS, and incubated at room temperature overnight. Unbound proteins were removed by washing the resin three times with PBS followed by washing once with 8 M urea and 10 mM DTT in PBS. Bound proteins were denatured and reduced by incubating with 8 M urea and 10 mM DTT in PBS for 1 h at 37°C. Cysteines were then alkylated by adding iodoacetamide to 55 mM and continuing the incubation for a further 1 h at room temperature in the dark. Excess reagents were removed prior to digestion by washing three times with 50 mM ammonium bicarbonate (aq). Digestion with trypsin (1:50, trypsin:protein ratio; calcium chloride added to 1 mM) was allowed to proceed for 16 h at 37°C before being terminated by acidification with 10% (v/v) TFA (aq). Peptides were recovered by extensive washing of the beads with 50 mM ammonium bicarbonate (aq) followed by SPE (same method as biotin quantitation assay) and centrifugal evaporation.
2.8. iTRAQ labeling of peptides
Prior to labeling, entire dried peptide samples were dissolved in 30 µL of iTRAQ dissolution buffer (0.5 M triethylammonium bicarbonate, pH 8.5). For the Paraquat-exposure study, where a single control was used, two iTRAQ 4-plex reagents were required, while for the Sod2 knockdown experiment, where two controls were necessary, three iTRAQ 4-plex reagents were used. Parallel experiments where duplicate aliquots not labeled with biotin hydrazide were prepared required all four iTRAQ 4-plex reagents for the Paraquat-exposure studies. After allowing the labeling reaction to proceed for 2 h at room temperature, samples to be compared were combined and then dried in a centrifugal evaporator. Prior to LC–MS/MS analysis, the combined peptide samples were suspended in 20 µL of 0.1% formic acid (aq).
2.9. LC–MS/MS
Combined, iTRAQ-labeled samples were analyzed using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific; Waltham, MA) interfaced with a Waters nanoACQUITY UPLC system. The injection volume in each case was 2 µL, with a nanoACQUITY trapping column being used to reduce loading time. Labeled peptides were separated with a RP, C18 nanoACQUITY capillary analytical column (1.7 µm particles, 100 µm × 100 mm). Elution was achieved using a gradient of 0.1% formic acid in acetonitrile (B) versus 0.1% formic acid in water (A) at a flow rate of 0.4 µL min−1. Beginning with an equilibration mobile phase of 1% B, the linear gradient initially ran to 15% B over 25 min, then to 25% B over 35 min, then to 35% B over 40 min, and finally to 85% B over 20 min. Eluting peptides flowed into a nano-scale ESI source that was operated at 3.5 kV. Full scan mass spectra (m/z = 400–2000), which were collected at 60,000 resolution in the Orbitrap, were followed by ten data-dependent MS/MS spectra, using higher-energy C-trap dissociation (HCD) [55] to access iTRAQ reporter ions. HCD parameters included normalized collision energy set to 40 and activation time at 0.1 ms. Ions selected twice for fragmentation within a 30 s window were excluded for 180 s.
2.10. Data processing and analysis
Peptides were identified using the X! Tandem code fork included with version 4.6.3 of the Trans-Proteomic Pipeline (TPP) software package (Institute for Systems Biology; Seattle, WA). The protein sequence database used was generated from the whole D. melanogaster proteome (FlyBase version R5.35, FB2011_03; 23361 proteins), 112 common contaminant proteins (Global Proteome Machine; Common Repository of Adventitious Proteins; downloaded on April 8, 2011), and reversed versions of these sequences included for false positive rate estimation. Search parameters included a precursor ion mass tolerance of ±15 ppm, a requirement for at least one trypsin-conforming peptide terminus, and a maximum of one missed cleavage. Fixed mass shifts were applied for alkylation with iodoacetamide at cysteine (+57.021464 Da) and addition of 4-plex iTRAQ labels at lysine and the peptide N-terminus (+144.102063 Da). Differential mass shifts were included for methionine sulfoxide (+15.994915 Da) and for the 4-plex iTRAQ reagents at tyrosine. PeptideProphet [56], a component of the TPP software package, was used to filter identifications proposed by X! Tandem, with peptide-spectrum matches (PSMs) scoring ≥ 0.9 being accepted. The PSM false positive rate was estimated as described by Elias and Gygi [57] (2 × decoy PSM count/total accepted PSM count). PSMs were organized into protein groups defined by parent gene, rejecting PSMs that indicated more than one gene. Relative abundance ratios were calculated for each PSM having no missing reporter ions and then combined to produce consensus values for each gene-based protein group by calculating the geometric mean. These ratios were corrected independently for each preparation using the ratio observed for trypsin autolysis peptides. Only groups supported by at least five measurements were accepted.
3. Results
Approximate times to a 50% death rate were five days for Sod2 knockdown and three days for Paraquat exposure. Control flies exhibited few (~1%) deaths through the duration of these experiments. Sod2 knockdown fly stocks and associated controls were maintained from an earlier study in which successful minimization of SOD2 activity was confirmed [38].
3.1. Visualization of carbonylated proteins by western blotting
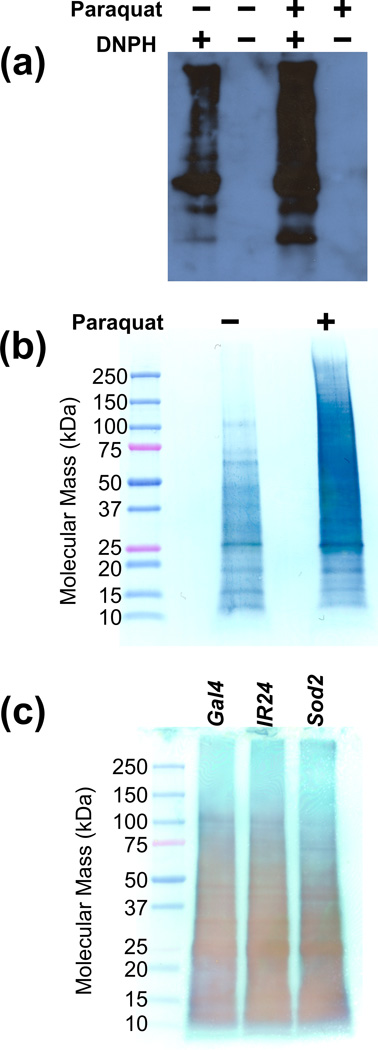
Protein carbonylation levels were initially assessed using two contrasting western blotting approaches. Loading controls could not reliably be used since widespread changes in protein carbonylation were expected in the Paraquat-exposure and possible in the Sod2-knockdown system. Instead, gel-loading amounts were equalized using spectrophotometric protein concentration assays. Approximately equal loading was confirmed by assessment of SDS-PAGE gels stained with Coomassie Brilliant Blue G-250 (a comparison of Paraquat-exposed with control whole-fly homogenates is provided as Supporting Information Figure S1). OxyBlot results for Paraquat-exposed and control flies are shown in Figure 1a. Paraquat exposure can clearly be seen to result in elevated levels of carbonylation when compared with control flies, which exhibit some level of background oxidation. Figure 1b gives the picture obtained when western blotting with HRP-conjugated streptavidin was applied to Paraquat-exposed and control samples that had been labeled with biotin hydrazide. These results also clearly indicate a higher degree of protein carbonylation in Paraquat-treated versus control flies. Figure 1c shows the result of applying the HRP-conjugated streptavidin method to the Sod2-knockdown system (a Coomassie-stained SDS-PAGE gel that was run in parallel with the gel that was transferred to create Figure 1c is provided as Supporting Information Figure S2). In this case, the difference between the Sod2 knockdown and controls (da-Gal4 driver alone and UAS-Sod2-IR24 transgene alone) was much less pronounced. From the western blotting, Paraquat exposure appears to be a more intense oxidative stress system than Sod2 knockdown.
Figure 1.
Western blotting was used to compare protein carbonylation levels between controls and oxidatively stressed D. melanogaster. In all cases, whole-fly homogenates were analyzed. First, the OxyBlot system was used to visualize the effects of Paraquat exposure, with lanes shown for Paraquat-exposed and control flies along with samples processed in parallel where the DNPH label was omitted (a). Second, the HRP-conjugated streptavidin approach was applied to Paraquat exposure (b) and Sod2 knockdown (c). For Sod2 knockdown, “Gal4” and “IR24” indicate the controls while “Sod2” indicates the knockdown flies. Elevated protein carbonylation levels are clearly visualized for Paraquat exposure, but not for Sod2 knockdown.
3.2. Biotin quantitation assay
Since the dynamic range of western blots can be narrow, judgments based solely on the darkness of the numerous observed bands have the potential to be misleading. Therefore, a spectrophotometric assay based on the displacement of HABA from avidin by biotin-labeled carbonylated proteins was used to compare protein carbonylation levels between Paraquat-exposed and control samples (reduction in absorbance of the HABA-avidin complex is measured). Using the third full repeat for the Paraquat-exposure system, peptide-bound biotin concentrations were determined. The assay was repeated three times and peptides originating from a control sample that had not been labelled with biotin hydrazide were also included. Expressed as average ± standard deviation, the results obtained were 2.6 ± 0.7, 32.1 ± 0.5, and 42.7 ± 3.7 pmol biotin per µg peptide for non-biotinylated control, biotinylated control, and biotinylated Paraquat-exposed samples, respectively. Subtracting the background amount from each, this implies a 36% increase in protein carbonylation on exposure to Paraquat.
3.3. Non-specific binding estimation by mass spectrometry
Retention of unlabeled proteins on streptavidin beads is a concern. Therefore, the degree of non-specific binding was estimated by creating duplicate samples that were not exposed to the labeling reagent, biotin hydrazide. This comparison was performed for the Paraquat-exposure system only, with three replicate LC–MS/MS runs performed for a single sample preparation. In the case of extensive non-specific binding, many proteins would be expected with non-biotinylated to biotinylated ratios of close to one for both the Paraquat-exposed and control pairs. After filtering, the dataset consisted of 6373 PSMs, four of which indicated decoy proteins (implying a PSM false positive rate of 0.13%). Processing into gene-based protein groups resulted in 101 groups. The distribution of observed ratios is provided as Supporting Information Figure S3, with ratios provided separately for each protein group in Supporting Information Table S1. Median non-biotinylated versus biotinylated ratios for control and Paraquat-exposed samples were 0.23 and 0.18, respectively, indicating some level of non-specific binding. For the control comparison, ratios greater than 0.5 were found for nine protein groups, while for the Paraquat comparison, three groups exceeded 0.5 (two of these overlapped with the nine from the controls). These ten protein groups were excluded from further analysis due their demonstrated propensity for non-specific binding. Interestingly, two protein groups had non-biotinylated to biotinylated ratios in excess of one for both the control and Paraquat-exposed samples. Acetyl-CoA carboxylase (FBgn0033246) gave non-biotinylated to biotinylated ratios of 3.36 for the control and 4.12 for the Paraquat-exposed comparison while CG1516 (FBgn0027580) gave 2.12 for the control and 2.00 for the Paraquat-exposed samples.
3.4. Mass spectrometry quantitation of Paraquat-induced changes in protein carbonylation
Paraquat exposure was expected to result in a significant increase in protein carbonylation levels over those found in the control. Three replicate LC–MS/MS runs were recorded for each sample preparation and the entire experiment was repeated three times, which resulted in 25621 PSMs after filtering. Only twelve of these identifications indicated decoy proteins, giving an estimated PSM false positive rate of 0.09%. Quantitative information was obtained for 249 protein groups after removal of non-specific binding proteins. The full list of quantified carbonylated proteins is provided with combined ratios for all repeats as Supporting Information Table S2 and with separate ratios for each repeat as Supporting Information Table S3. Those proteins found to exhibit at least a doubling of carbonylation levels over the control on exposure to Paraquat are shown in Table 1.
Table 1.
List of carbonylated proteins found in an MS-based proteomics study in D. melanogaster to be at least doubled in relative abundance on exposure to Paraquat.
| FlyBase gene ID used for protein-group construction |
Associated FlyBase gene symbol |
Associated FlyBase gene name | PSM count |
Paraquat-exposed versus control carbonylated protein relative abundance ratio |
|---|---|---|---|---|
| FBgn0010516 | wal | walrus | 16 | 2.75 |
| FBgn0010213 | Sod2 | superoxide dismutase 2 (Mn) | 25 | 2.73 |
| FBgn0031801 | CG9498 | 5 | 2.53 | |
| FBgn0024289 | Sodh-1 | sorbitol dehydrogenase 1 | 6 | 2.52 |
| FBgn0003067 | Pepck | phosphoenolpyruvate carboxykinase | 9 | 2.45 |
| FBgn0019957 | ND42 | NADH:ubiquinone reductase 42kD subunit precursor | 37 | 2.42 |
| FBgn0020653 | Trxr-1 | thioredoxin reductase-1 | 14 | 2.38 |
| FBgn0000556 | Ef1α48D | elongation factor 1α48D | 42 | 2.36 |
| FBgn0029823 | CG3011 | 23 | 2.30 | |
| FBgn0261014 | TER94 | TER94 | 76 | 2.27 |
| FBgn0020513 | ade5 | ade5 | 86 | 2.26 |
| FBgn0010482 | l(2)01289 | lethal (2) 01289 | 17 | 2.25 |
| FBgn0000052 | ade2 | adenosine 2 | 5 | 2.22 |
| FBgn0000319 | Chc | clathrin heavy chain | 17 | 2.21 |
| FBgn0036044 | CG14168 | 11 | 2.20 | |
| FBgn0260439 | Pp2A-29B | protein phosphatase 2A at 29B | 49 | 2.16 |
| FBgn0037607 | CG8036 | 48 | 2.16 | |
| FBgn0001219 | Hsc70-4 | heat shock protein cognate 4 | 31 | 2.14 |
| FBgn0040349 | CG3699 | 28 | 2.13 | |
| FBgn0003887 | βTub56D | β-tubulin at 56D | 15 | 2.09 |
| FBgn0004047 | Yp3 | yolk protein 3 | 842 | 2.09 |
| FBgn0004419 | me31B | maternal expression at 31B | 10 | 2.07 |
| FBgn0003676 | T-cp1 | Tcp1-like | 17 | 2.07 |
| FBgn0000261 | Cat | catalase | 82 | 2.06 |
| FBgn0039737 | CG7920 | 153 | 2.04 | |
| FBgn0012036 | Aldh | aldehyde dehydrogenase | 43 | 2.01 |
| FBgn0262559 | Mdh2 | malate dehydrogenase 2 | 637 | 2.01 |
| FBgn0019830 | colt | congested-like trachea | 9 | 2.01 |
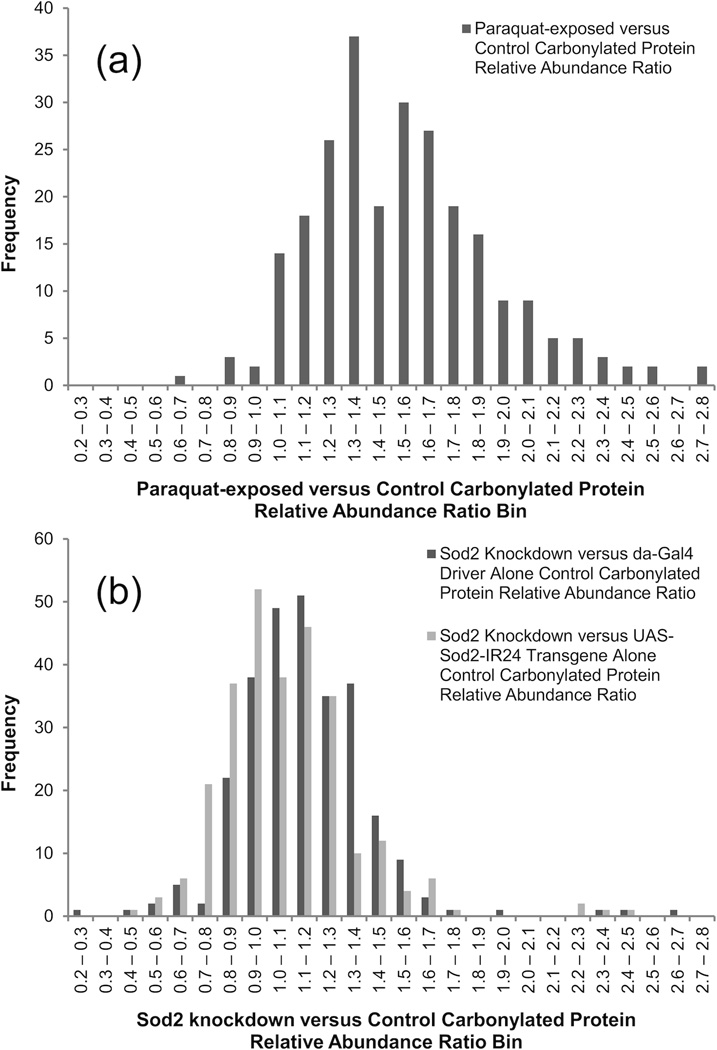
For the whole set, the median Paraquat-exposed to control ratio was 1.53. A histogram illustrating the ratio distribution is provided as Figure 2a. Of the 28 carbonylated proteins found to be at least doubled in relative abundance on exposure to Paraquat, SOD2, thioredoxin reductase-1, and catalase stand out as key antioxidant enzymes. Six proteins showed Paraquat-exposed to control ratios of less than one. While it would seem unlikely that the degree of carbonylation for any protein would fall on exposure to Paraquat, proteins falling in relative abundance in response to Paraquat is certainly possible. Functional analysis, using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) Bioinformatics Database (Release 6.7; http://david.abcc.ncifcrf.gov) [58,59], was used to search for connections between the whole set of 249 detected carbonylated proteins, which might best be regarded as a list of background carbonylated proteins. Since functional analysis tools usually manipulate gene lists only, the practice of defining protein groups by parent genes was beneficial. All 249 genes were recognized, although FlyBase secondary identifiers were required in some cases. Using the whole D. melanogaster proteome as background, strong associations were seen with the lipid particle (86 genes associated with GO:0005811; p = 1.5 × 10−61) and mitochondrion (81 genes associated with GO:0005739; p = 2.6 × 10−26) Gene Ontology cellular component classifications (37 proteins were annotated to both GO:0005811 and GO:0005739).
Figure 2.
Distribution of relative abundance ratios obtained when an MS-based proteomics approach was used to measure changes in carbonylated protein levels in response to Paraquat exposure (a) and Sod2 knockdown (b) in D. melanogaster. For the Sod2 knockdown case, separate distributions are provided versus the da-Gal4 driver alone and UAS-Sod2-IR24 transgene alone control. Paraquat exposure was found to result in a greater increase in protein carbonylation than Sod2 knockdown.
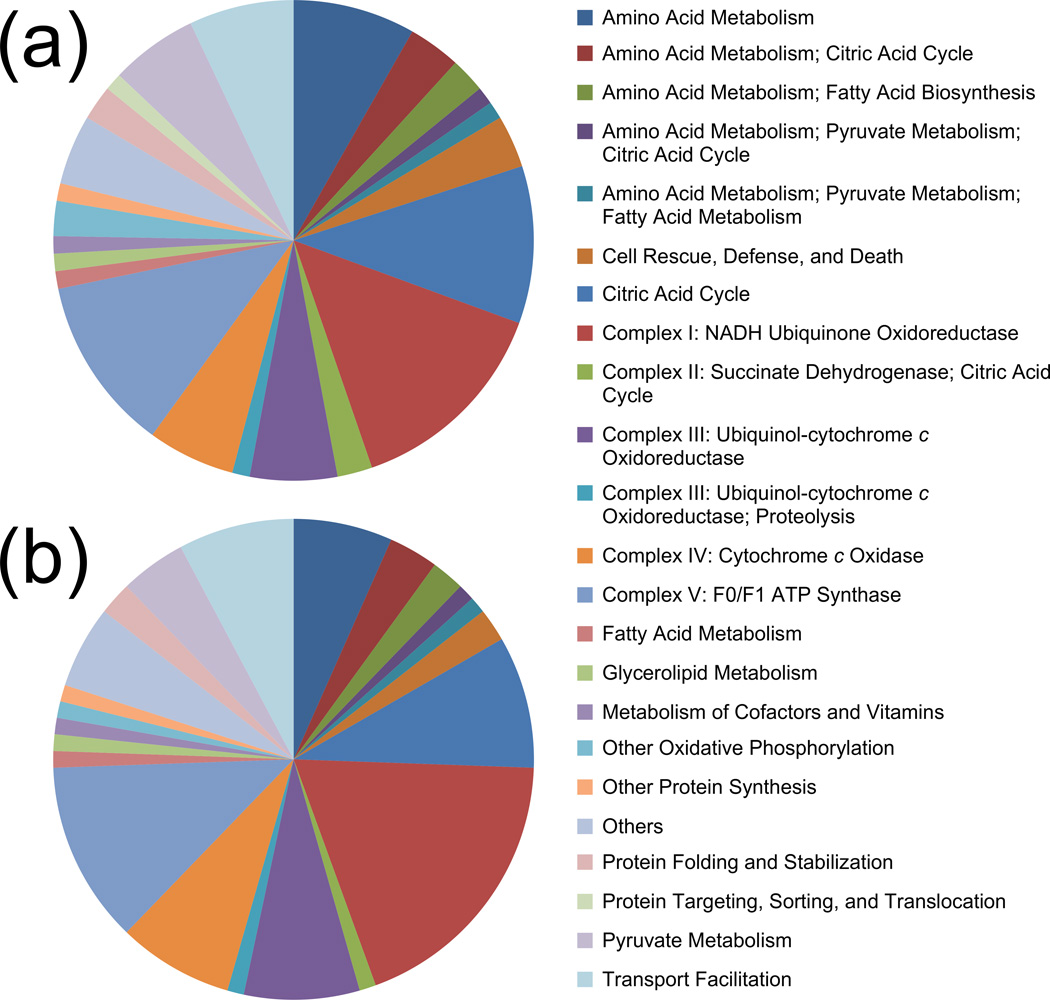
FlyBase (using the more recent FB2014_01 version) and DAVID annotations were combined with MitoDrome (http://mitodrome.ba.itb.cnr.it) [60] functional classifications to investigate the distribution of carbonylated mitochondrial proteins. Combining these information sources increased the total number of protein groups associated with mitochondria to 85. The distribution of these carbonylated mitochondrial proteins among MitoDrome classes is illustrated in Figure 3a. Briefly, twelve genes were associated with ETC Complex I (GO:0005747), two with Complex II (GO:0005749), six with Complex III (GO:0005750), five with Complex IV (GO:0005751), and ten with ATP synthase (GO:0005753). Similarly, fifteen genes were associated with the citric acid cycle (GO:0006099), although in this case there was significant overlap with other functional classifications. Of these gene-organized protein groups, three were found at double or greater relative abundance in the Paraquat-exposed versus control samples (all three repeats combined): NADH:ubiquinone reductase 42kD subunit precursor (FBgn0019957) at 2.42, Tcp1-like (ATP Synthase; FBgn0003676) at 2.07, and malate dehydrogenase 2 (FBgn0262559) at 2.01. Finally, three mitochondrial genes were associated with the MitoDrome cell rescue, defense, and death classification (this category includes antioxidant enzymes). Of these, the two that exceeded the significance threshold have been mentioned above: SOD2 (FBgn0010213) at 2.73 and thioredoxin reductase-1 (FBgn0020653) at 2.38.
Figure 3.
Distribution of MitoDrome functional classifications for the entire sets of identified carbonylated mitochondrial proteins from the Paraquat-exposure (a) and Sod2-knockdown (b) studies. Since numerous proteins were included in more than one functional class, some pie chart slices are annotated with a list of classification groups separated by semicolons. Proteins encoded by mitochondrial DNA and additional information from FlyBase and DAVID were also included. Carbonylated mitochondrial protein counts are 85 for Paraquat exposure (a) and 90 for Sod2 knockdown (b).
3.5. Mass spectrometry quantitation of Sod2 knockdown-induced changes in protein carbonylation
Higher protein carbonylation levels were also expected for the Sod2 knockdown system. As with Paraquat exposure, three replicate LC–MS/MS runs were collected for each of three full repeat experiments. Applying a PeptideProphet score acceptance threshold of 0.9, 27910 PSMs were obtained, of which 23 were hits to decoy proteins (implying a PSM false positive rate of 0.16%). Rejecting the non-specifically bound proteins, 276 gene-based protein groups were obtained. The whole set is presented with repeat runs combined as Supporting Information Table S4 and with repeat preparations separated as Supporting Information Table S5. The Sod2 knockdown results differ from the Paraquat-exposure results in having two controls: the da-Gal4 driver alone and the UAS-Sod2-IR24 transgene alone. Comparing reporter ion ratios between these controls provides a measure of experimental precision. Over 24473 measurements (PSMs with a complete set of reporter ions that indicate D. melanogaster proteins), the average (geometric mean) da-Gal4 driver alone to UAS-Sod2-IR24 transgene alone control ratio was 0.92, with a two standard deviation range of 0.61 to 1.40. Thus, adding uncertainty due to non-specific binding, it seems sensible to only consider a doubling or halving of relative abundance as significant in this work. Using DAVID on the whole set of 276 carbonylated proteins, results are similar to Paraquat exposure (lipid particle remains the most significant term) with the interesting exception of an increase in significance for the ribosome Gene Ontology cellular component classification (GO:0005840; from 31 proteins/p = 3.66 × 10−11 in Paraquat exposure to 51 proteins/p = 5.73 × 10−27 in Sod2 knockdown).
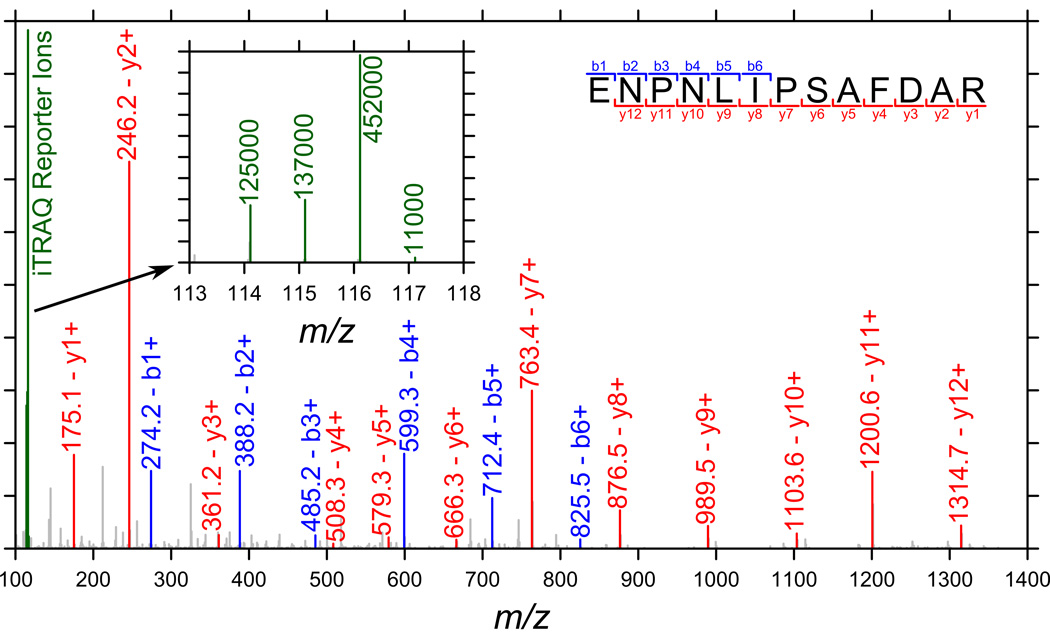
Figure 2b displays the distribution of Sod2 knockdown to da-Gal4 driver alone and UAS-Sod2-IR24 transgene alone control ratios. When compared with exposure to Paraquat (Figure 2a), it is clear that Sod2 knockdown produces a much smaller increase in global protein carbonylation levels. The median relative abundance ratios are 1.13 (versus da-Gal4 driver alone control) and 1.05 (versus UAS-Sod2-IR24 transgene alone control) and both distributions are similar. Against the da-Gal4 driver alone control, three proteins are observed to exceed a doubling in relative abundance on Sod2 knockdown; for the UAS-Sod2-IR24 transgene alone control, four proteins exceed this threshold. Two proteins exceed doubling in abundance in both Sod2 knockdown versus control comparisons: FBgn0036766 (CG5506) and FBgn0031830 (CG11015). While little information is available for FBgn0036766 (protein BLAST can provide no functional annotation), FBgn0031830 has been annotated as cytochrome c oxidase subunit Vb. The observed relative abundance ratio for cytochrome c oxidase subunit Vb on Sod2 knockdown is 2.39 versus the da-Gal4 driver alone and 2.26 versus the UAS-Sod2-IR24 transgene alone control. The protein group defined by FBgn0031830 contains a single protein, FBpp0078918, which is supported by a single peptide (35 detections) with sequence, ENPNLIPSAFDAR. Since the significance of cytochrome c oxidase subunit Vb rests on a single peptide, the MS/MS spectrum is provided as Figure 4. Identification is supported by clear, unbroken runs of singly charged b- and y-ions (six consecutive b- and twelve consecutive y-ions) in combination with few prominent, unexplained peaks.
Figure 4.
MS/MS spectrum associated with the identification of cytochrome c oxidase subunit Vb peptide ENPNLIPSAFDAR. The inset shows the region of the spectrum containing the iTRAQ reporter ions: the peak at 114 represents the da-Gal4 driver-alone control, 115 the UAS-Sod2-IR24 transgene-alone control, and 116 the Sod2 knockdown sample (117 was not used but some intensity is observed due to impurities in the other reporter ions).
Considering the complete set of 279 gene-based carbonylated protein groups identified in the three Sod2 knockdown repeats, a broadly similar picture is expected to that for Paraquat exposure (since the whole set effectively provides a list of background carbonylated proteins). As expected, there was extensive overlap between the systems (190 genes were indicated by both). Using MitoDrome, FlyBase, and DAVID annotations, 90 of 279 genes were associated with mitochondria. Figure 3b provides details of the distribution of these mitochondrial proteins using MitoDrome functional classifications. Clearly, the pattern for Sod2 knockdown is very similar to that for Paraquat exposure. Considering the mitochondrial ETC complexes, seventeen, one, eight, seven, and eleven genes associated with Complex I, II, III, IV, and ATP synthase, respectively. Similarly, thirteen genes were associated with the citric acid cycle and two with cell rescue, defense, and death (both antioxidant enzymes). Given the similarity of the functional classification distributions illustrated in Figure 3, it is clear that differences between Paraquat exposure and Sod2 knockdown lie mostly in the magnitude of changes in carbonylated protein relative abundances.
4. Discussion
Paraquat exposure and Sod2 knockdown both result in significant lifespan reduction, locomotor dysfunction, and mitochondrial degeneration in D. melanogaster. Paraquat exposure is expected to result in increased O2•− concentrations at numerous locations throughout the cell while the effects of Sod2 knockdown are expected to be localized to the mitochondrial matrix. In yeast, Sod2 knockout has been shown to result in increased carbonylation for specific proteins but not to lead to a significant increase in aggregate carbonylation [40]. Therefore, it was not surprising to observe similar levels of protein carbonylation for Sod2 knockdown and associated control samples using the HRP-conjugated streptavidin western blotting approach (Figure 1c) and median carbonylated protein relative abundance ratios close to one for the iTRAQ approach. In contrast, elevated protein carbonylation was observed in the Paraquat-exposure system as expected. Western blot bands (Figure 1a and Figure 1b) were noticeably darker when comparing Paraquat-exposed with control samples and the median carbonylated protein iTRAQ relative abundance (1.53) and biotin quantitation assay (1.36) ratios, which were broadly in agreement, indicated a substantial increase in aggregate carbonylation on Paraquat exposure. This would seem to indicate that while the impacts of Sod2 knockdown and Paraquat exposure are comparable, the mechanisms through which they act are dissimilar. The Sod2-knockdown phenotype could well be generated through an oxidative stress-related process that generates few carbonylated proteins. For example, the destruction of catalytic protein [4Fe-4S] clusters has been proposed as a mechanism for Sod2 knockout-associated O2•− toxicity in yeast [11].
Applying data mining software (DAVID) to the whole set of identified carbonylated proteins for both Paraquat exposure and Sod2 knockdown highlighted the Gene Ontology cellular component term, lipid particle (GO:0005811). Lipid particles, or lipid droplets, consist of a neutral lipid core surrounded by a phospholipid monolayer with numerous associated proteins [61,62]. Cermelli et al. [63] found the lipid droplet proteome in D. melanogaster to be unexpectedly complicated and proposed a role for the droplets in storage or sequestration of excess or incorrectly folded proteins. In this work, the carbonylated proteins associated with the lipid particle annotation clearly include both mitochondrial and ribosomal proteins. Rather than actually being in storage, mitochondrial proteins might be present in lipid droplet preparations as impurities [64]; however, association between lipid droplets and RNA has been observed [65], thus indicating ribosomal localization is conceivable. In contrast to the uncertain association with lipid particles, the connection to mitochondria seems obvious. Many ETC and citric acid cycle proteins were found to be carbonylated, with SOD2 found to be the second most increased in abundance with respect to the control in the Paraquat-exposure study.
While many carbonylated proteins become more abundant in D. melanogaster exposed to Paraquat, none stand out. However, for Sod2 knockdown, two gene-defined protein groups are prominent: FBgn0036766 (from protein FBpp0074923) and FBgn0031830 (from protein FBpp0078918; FBgn0031830 is associated with two distinct transcripts, but only one protein is found in the database because the resulting sequences are identical). Since both stand-outs are increased in abundance on Sod2 knockdown, it seems reasonable to rule out off-target RNA interference effects [66]. Unfortunately, little information is available for FBpp0074923, but FBpp0078918 is cytochrome c oxidase (ETC Complex IV) subunit Vb. Complex IV catalyzes electron transfer from cytochrome c to molecular oxygen. The mammalian form of Complex IV is composed of fourteen subunits [67], three of which are encoded by mitochondrial DNA. Subunit Vb is nuclear-encoded and necessary for proper regulation and assembly of the complex; knockouts have been shown to lead to reduced Complex IV activity and increased ROS production [68]. Furthermore, age-related declines in mitochondrial respiration in D. melanogaster have been associated in particular with decreases in cytochrome c oxidase activity [69,70]. Moreover, cytochrome c oxidase subunit Vb concentrations were found to be reduced with respect to the mitochondria-encoded subunits in prostate cancer [71]. Thus, the association between subunit Vb and Sod2 knockdown is clearly worthy of further investigation.
Immobilization of carbonylated proteins on streptavidin beads is central to this work. However, while the beads offered an effective means to sequester carbonylated proteins, since tight binding allowed extensive washing for minimization of non-specific binding, efficient elution could not be achieved. Therefore, on-bead tryptic digestion was performed, with the iTRAQ approach applied to the subsequently-eluted peptides for determination of relative abundance ratios. The chief disadvantage of this approach is that, since peptides containing carbonylation sites remain bound to the beads, site localization is not possible. Efficient elution of carbonylated proteins (or carbonylated peptides after digestion) from the beads was difficult because the harsh conditions required typically necessitated extensive clean-up that resulted in significant sample losses. Conversely, the on-bead denaturation, reduction, and alkylation procedure performed before buffer exchange for digestion could have resulted in some protein loss from the beads; however, losses are expected to be small since streptavidin has been reported to be structurally stable in high-concentration urea solutions [50] and contains no disulfide bridges. Alternative approaches that promised carbonylated protein elution under mild conditions, such as monomeric avidin, were found incapable of sufficient enrichment of carbonylated proteins to be effective. However, if enrichment is not required, elegant alternative approaches such as the carbonyl-reactive iTRAQ reagents reported by Palmese et al. [72], which have the advantage of reducing the two labeling steps to one, become attractive. Finally, the HRP-conjugated streptavidin western blotting provides some indication of discrimination against high-mass proteins, possibly resulting from sample losses occurring during the biotin hydrazide-labeling procedure.
In conclusion, while the association of carbonylated proteins with lipid particles in conjunction with a proposed role for the lipid particle in sequestration of damaged proteins is interesting, it would indicate that many of the identified carbonylated proteins are merely awaiting destruction. In contrast, the association of carbonylated proteins with the mitochondria is important and anticipated. Paraquat was expected to result in widespread O2•− generation and the general increase observed in carbonylated protein relative abundance is consistent with this expectation. Antioxidant enzymes were prominent in the set of carbonylated proteins most increased in abundance (Paraquat-exposed to control ratio > 2), but there were no outliers. Conversely, despite producing a similarly acute phenotype, Sod2 knockdown resulted in a much reduced global increase in carbonylated protein relative abundance. However, the two proteins that stood out clearly have the potential to elucidate aspects of the O2•− generation process or Sod2 knockdown phenotype. Unfortunately, limited information is available for one of these outliers, but for the other, cytochrome c oxidase subunit Vb, data exists supporting a role in aging and age-related disease. Further research targeting these proteins could advance our understanding of the role of SOD2 and the mechanistic underpinnings of age-related disorders associated with mitochondrial dysfunction.
Supplementary Material
Acknowledgments
Fruit flies were prepared by Lara Lewellyn and mass spectrometric data was collected by Dr. Kristina T. Nelson at the Virginia Commonwealth University Chemical and Proteomic Mass Spectrometry Core Facility. The support of the National Institutes of Health, National Institute on Aging is acknowledged: R01AG034167.
Abbreviations
- BCA
bicinchoninic acid
- DNPH
2,4-dinitrophenylhydrazine
- ETC
electron transport chain
- HABA
4’-hydroxyazobenzene-2-carboxylic acid
- HCD
higher-energy C-trap dissociation
- PSM
peptide-spectrum match
- SOD2/Sod2
Mn or mitochondrial superoxide dismutase (protein/gene)
- TPP
trans-proteomic pipeline
Footnotes
The authors have declared no conflict of interest.
References
- 1.Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech. Ageing Dev. 2004;125:811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Pelicano H, Carney D, Huang P. ROS stress in cancer cells and therapeutic implications. Drug Resist. Updat. 2004;7:97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Sugamura K, Keaney JF. Reactive oxygen species in cardiovascular disease. Free Radic. Biol. Med. 2011;51:978–992. doi: 10.1016/j.freeradbiomed.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishikawa T, Araki E. Impact of mitochondrial ROS production in the pathogenesis of diabetes mellitus and its complications. Antioxid. Redox Signal. 2007;9:343–353. doi: 10.1089/ars.2006.1458. [DOI] [PubMed] [Google Scholar]
- 5.Patten DA, Germain M, Kelly MA, Slack RS. Reactive oxygen species: stuck in the middle of neurodegeneration. J. Alzheimers. Dis. 2010;20:S357–S367. doi: 10.3233/JAD-2010-100498. [DOI] [PubMed] [Google Scholar]
- 6.Brown GC, Borutaite V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion. 2012;12:1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Brand MD. The sites and topology of mitochondrial superoxide production. Exp. Geront. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan CL, Treberg JR, Perevoshchikova IV, Orr AL, Brand MD. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radic. Biol. Med. 2012;53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller F. The nature and mechanism of superoxide production by the electron transpot chain: its relevance to aging. J. Amer. Aging Assoc. 2000;23:227–253. doi: 10.1007/s11357-000-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridovich I. Superoxide radical: an endogenous toxicant. Ann. Rev. Pharmacol. Toxicol. 1983;23:239–257. doi: 10.1146/annurev.pa.23.040183.001323. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan C, Liba A, Imlay JA, Selverstone Valentine J, Butler Gralla E. Yeast lacking superoxide dismutase(s) show elevated levels of “free iron” as measured by whole cell electron paramagnetic resonance. J. Biol. Chem. 2000;275:29187–29192. doi: 10.1074/jbc.M004239200. [DOI] [PubMed] [Google Scholar]
- 12.McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968–1988) Free Radic. Biol. Med. 1988;5:363–369. doi: 10.1016/0891-5849(88)90109-8. [DOI] [PubMed] [Google Scholar]
- 13.Kono Y, Fridovich I. Superoxide radical inhibits catalase. J. Biol. Chem. 1982;257:5751–5754. [PubMed] [Google Scholar]
- 14.Stadtman ER. Metal ion-catalyzed oxidation of proteins: biochemical mechanism and biological consequences. Free Radic. Biol. Med. 1990;9:315–325. doi: 10.1016/0891-5849(90)90006-5. [DOI] [PubMed] [Google Scholar]
- 15.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 16.Levine RL. Carbonyl modified proteins in cellular regulation, aging, and disease. Free Radic. Biol. Med. 2002;32:790–796. doi: 10.1016/s0891-5849(02)00765-7. [DOI] [PubMed] [Google Scholar]
- 17.Butterfield DA. Proteomics: A new approach to investigate oxidative stress in Alzheimer’s disease brain. Brain Res. 2004;1000:1–7. doi: 10.1016/j.brainres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 18.De Cristofaro R, Rocca B, Vitacolonna E, Falco A, et al. Lipid and protein oxidation contribute to a prothrombotic state in patients with type 2 diabetes mellitus. J. Thromb. Haemost. 2003;1:250–256. doi: 10.1046/j.1538-7836.2003.00072.x. [DOI] [PubMed] [Google Scholar]
- 19.Uchida K, Fukuda A, Kawakishi S, Hiai H, Toyokuni S. A renal carcinogen ferric nitrilotriacetate mediates a temporary accumulation of aldehyde-modified proteins within cytosolic compartment of rat kidney. Arch. Biochem. Biophys. 1995;317:405–411. doi: 10.1006/abbi.1995.1181. [DOI] [PubMed] [Google Scholar]
- 20.Yoritaka A, Hattori N, Uchida K, Tanaka M, et al. Immunohistochemical detection of 4-hydroxynonenal protein adducts in Parkinson disease. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2696–2701. doi: 10.1073/pnas.93.7.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 22.Dalle-Donne I, Rossi R, Giustarini D, Milzani A, Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 2003;329:23–38. doi: 10.1016/s0009-8981(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 23.Dalle-Donne I, Aldini G, Carini M, Colombo R, et al. Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madian AG, Regnier FE. Proteomic identification of carbonylated proteins and their oxidation sites. J. Proteome Res. 2010;9:3766–3780. doi: 10.1021/pr1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cabiscol E, Tamarit J, Ros J. Protein carbonylation: proteomics, specificity and relevance to aging. Mass Spectrom. Rev. 2014;33:21–48. doi: 10.1002/mas.21375. [DOI] [PubMed] [Google Scholar]
- 26.Drechsel DA, Patel M. Role of reactive oxygen species in the neurotoxicity of environmental agents implicated in Parkinson’s disease. Free Radic. Biol. Med. 2008;44:1873–1886. doi: 10.1016/j.freeradbiomed.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krall J, Bagley AC, Mullenbach GT, Hallewell RA, Lynch RE. Superoxide mediates the toxicity of Paraquat for cultured mammalian cells. J. Biol. Chem. 1988;263:1910–1914. [PubMed] [Google Scholar]
- 28.James AM, Cochemé HM, Smith RAJ, Murphy MP. Interactions of mitochondria-targeted and untargeted ubiquinones with the mitochondrial respiratory chain and reactive oxygen species. Implications for the use of exogenous ubiquinones as therapies and experimental tools. J. Biol. Chem. 2005;280:21295–21312. doi: 10.1074/jbc.M501527200. [DOI] [PubMed] [Google Scholar]
- 29.Mockett RJ, Bayne A-C, Kwong LK, Orr WC, Sohal RS. Ectopic expression of catalase in Drosophila mitochondria increases stress resistance but not longevity. Free Radic. Biol. Med. 2003;34:207–217. doi: 10.1016/s0891-5849(02)01190-5. [DOI] [PubMed] [Google Scholar]
- 30.Sampayo JN, Olsen A, Lithgow GJ. Oxidative stress in Caenorhabditis elegans: protective effects of superoxide dismutase/catalase mimetics. Aging Cell. 2003;2:319–326. doi: 10.1046/j.1474-9728.2003.00063.x. [DOI] [PubMed] [Google Scholar]
- 31.De Haan JB, Bladier C, Griffiths P, Kelner M, et al. Mice with a homozygous null mutation for the most abundant glutathione peroxidase, Gpx1, show increased susceptibility to the oxidative stress-inducing agents Paraquat and hydrogen peroxide. J. Biol. Chem. 1998;273:22528–22536. doi: 10.1074/jbc.273.35.22528. [DOI] [PubMed] [Google Scholar]
- 32.Jimenez-Del-Rio M, Guzman-Martinez C, Velez-Pardo C. The effects of polyphenols on survival and locomotor activity in Drosophila melanogaster exposed to iron and Paraquat. Neurochem. Res. 2010;35:227–238. doi: 10.1007/s11064-009-0046-1. [DOI] [PubMed] [Google Scholar]
- 33.Cochemé HM, Murphy MP. Complex I is the major site of mitochondrial superoxide production by Paraquat. J. Biol. Chem. 2008;283:1786–1798. doi: 10.1074/jbc.M708597200. [DOI] [PubMed] [Google Scholar]
- 34.Bové J, Prou D, Perier C, Przedborski S. Toxin-induced models of Parkinson’s disease. NeuroRx. 2005;2:484–494. doi: 10.1602/neurorx.2.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drechsel DA, Patel M. Paraquat-induced production of reactive oxygen species in brain mitochondria. Methods Enzymol. 2009;456:381–393. doi: 10.1016/S0076-6879(08)04421-2. [DOI] [PubMed] [Google Scholar]
- 36.Cochemé HM, Murphy MP. The uptake and interactions of the redox cycler Paraquat with mitochondria. Methods Enzymol. 2009;456:395–417. doi: 10.1016/S0076-6879(08)04422-4. [DOI] [PubMed] [Google Scholar]
- 37.Kirby K, Hu J, Hilliker AJ, Phillips JP. RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16162–16167. doi: 10.1073/pnas.252342899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin I, Jones MA, Rhodenizer D, Zheng J, et al. Sod2 knockdown in the musculature has whole-organism consequences in Drosophila. Free Radic. Biol. Med. 2009;47:803–813. doi: 10.1016/j.freeradbiomed.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wicks S, Bain N, Duttaroy A, Hilliker AJ, Phillips JP. Hypoxia rescues early mortality conferred by superoxide dismutase deficiency. Free Radic. Biol. Med. 2009;46:176–181. doi: 10.1016/j.freeradbiomed.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 40.O’Brien KM, Dirmeier R, Engle M, Poyton RO. Mitochondrial protein oxidation in yeast mutants lacking manganese- (MnSOD) or copper- and zinc-containing superoxide dismutase (CuZnSOD) J. Biol. Chem. 2004;279:51817–51827. doi: 10.1074/jbc.M405958200. [DOI] [PubMed] [Google Scholar]
- 41.Hensley K. Detection of protein carbonyls by means of biotin hydrazide-streptavidin affinity methods. Methods Mol. Biol. 2009;536:457–462. doi: 10.1007/978-1-59745-542-8_46. [DOI] [PubMed] [Google Scholar]
- 42.Anderson LB, Ouellette AJA, Eaton-Rye J, Maderia M, et al. Evidence for a post-translational modification, aspartyl aldehyde, in a photosynthetic membrane protein. J. Am. Chem. Soc. 2004;126:8399–8405. doi: 10.1021/ja0478781. [DOI] [PubMed] [Google Scholar]
- 43.Temple A, Yen T-Y, Gronert S. Identification of specific protein carbonylation sites in model oxidations of human serum albumin. J. Am. Soc. Mass Spectrom. 2006;17:1172–1180. doi: 10.1016/j.jasms.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 44.Mirzaei H, Regnier F. Identification of yeast oxidized proteins: chromatographic top-down approach for identification of carbonylated, fragmented and cross-linked proteins in yeast. J. Chromatogr. A. 2007;1141:22–31. doi: 10.1016/j.chroma.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 45.Meany DL, Xie H, Thompson LV, Griffin TJ, Arriaga EA. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 46.Grimsrud PA, Picklo MJ, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance: identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol. Cell. Proteomics. 2007;6:624–637. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Feng J, Xie H, Meany DL, Thompson LV, et al. Quantitative proteomic profiling of muscle type-dependent and age-dependent protein carbonylation in rat skeletal muscle mitochondria. J. Gerontol. 2008;63A:1137–1152. doi: 10.1093/gerona/63.11.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ugur Z, Coffey CM, Gronert S. Comparing the efficiencies of hydrazide labels in the study of protein carbonylation in human serum albumin. Anal. Bioanal. Chem. 2012;404:1399–1411. doi: 10.1007/s00216-012-6235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green NM. Avidin and streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 50.Kurzban GP, Bayer Ea, Wilchek M, Horowitz PM. The quaternary structure of streptavidin in urea. J. Biol. Chem. 1991;266:14470–14477. [PubMed] [Google Scholar]
- 51.Soreghan BA, Yang F, Thomas SN, Hsu J, Yang AJ. High-throughput proteomic-based identification of oxidatively induced protein carbonylation in mouse brain. Pharm. Res. 2003;20:1713–1720. doi: 10.1023/b:pham.0000003366.25263.78. [DOI] [PubMed] [Google Scholar]
- 52.Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 53.Bhandari P, Hill JS, Farris SP, Costin B, et al. Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes. Brain. Behav. 2012;11:387–397. doi: 10.1111/j.1601-183X.2012.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goddeeris MM, Cook-Wiens E, Horton WJ, Wolf H, et al. Delayed behavioural aging and altered mortality in Drosophila β integrin mutants. Aging Cell. 2003;2:257–264. doi: 10.1046/j.1474-9728.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 55.Olsen JV, Macek B, Lange O, Makarov A, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 56.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 57.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 58.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Sardiello M, Licciulli F, Catalano D, Attimonelli M, Caggese C. MitoDrome: a database of Drosophila melanogaster nuclear genes encoding proteins targeted to the mitochondrion. Nucleic Acids Res. 2003;31:322–324. doi: 10.1093/nar/gkg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- 62.Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006;7:373–378. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- 63.Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 64.Beller M, Riedel D, Jänsch L, Dieterich G, et al. Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Dvorak AM, Morgan ES, Weller PF. RNA is closely associated with human mast cell lipid bodies. Histol. Histopathol. 2003;18:943–968. doi: 10.14670/HH-18.943. [DOI] [PubMed] [Google Scholar]
- 66.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 67.Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378–386. doi: 10.1016/j.cmet.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 68.Galati D, Srinivasan S, Raza H, Prabu SK, et al. Role of nuclear-encoded subunit Vb in the assembly and stability of cytochrome c oxidase complex: implications in mitochondrial dysfunction and ROS production. Biochem. J. 2009;420:439–449. doi: 10.1042/BJ20090214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferguson M, Mockett RJ, Shen Y, Orr WC, Sohal RS. Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem. J. 2005;390:501–511. doi: 10.1042/BJ20042130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren J-C, Rebrin I, Klichko V, Orr WC, Sohal RS. Cytochrome c oxidase loses catalytic activity and structural integrity during the aging process in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2010;401:64–68. doi: 10.1016/j.bbrc.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herrmann PC, Gillespie JW, Charboneau L, Bichsel VE, et al. Mitochondrial proteome: altered cytochrome c oxidase subunit levels in prostate cancer. Proteomics. 2003;3:1801–1810. doi: 10.1002/pmic.200300461. [DOI] [PubMed] [Google Scholar]
- 72.Palmese A, De Rosa C, Chiappetta G, Marino G, Amoresano A. Novel method to investigate protein carbonylation by iTRAQ strategy. Anal. Bioanal. Chem. 2012;404:1631–1635. doi: 10.1007/s00216-012-6324-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.