Abstract
How cells regulate the bioavailability of utilizable sulfur while mitigating the effects of hydrogen sulfide toxicity is poorly understood. CstR (Copper-sensing operon repressor (CsoR)-like sulfurtransferase repressor) represses the expression of the cst operon encoding a putative sulfide oxidation system in Staphylococcus aureus. Here, we show that the cst operon is strongly and transiently induced by cellular sulfide stress in an acute phase and specific response and that cst-encoded genes are necessary to mitigate the effects of sulfide toxicity. Growth defects are most pronounced when S. aureus is cultured in chemically defined media with thiosulfate (TS) as a sole sulfur source, but are also apparent when cystine is used or in rich media. Under TS growth conditions, cells fail to grow as a result of either unregulated expression of the cst operon in a ΔcstR strain or transformation with a non-inducible C31A/C60A CstR that blocks cst induction. This suggests that the cst operon contributes to cellular sulfide homeostasis. Tandem high resolution mass spectrometry reveals derivatization of CstR by both inorganic tetrasulfide and an organic persulfide, glutathione persulfide, to yield a mixture of Cys31-Cys60’ interprotomer crosslinks, including di-, tri- and tetrasulfide bonds, which allosterically inhibit cst operator DNA binding by CstR.
Keywords: hydrogen sulfide, sulfide sensing, repressor, tandem mass spectrometry, sulfur oxidation
Introduction
Hydrogen sulfide (H2S) is a recently classified “gasotransmitter” or signaling molecule that plays important roles in a number of (patho) physiological processes, including vasorelaxation, cardioprotection, and neurotransmission in mammals (Kabil & Banerjee, 2010, Kolluru et al., 2013, Paul & Snyder, 2012). H2S is produced endogenously by the action of cystathionine γ-lyase (CSE) (Chiku et al., 2009, Singh et al., 2009), cystathionine β-synthase (CBS) (Singh et al., 2009) and cysteine aminotransferease (CAT) in combination with 3-mercaptopyruvate sulfurtransferase (3MST) (Kimura, 2010). Hydrogen sulfide is a weak acid and exists primarily as soluble hydrogen sulfide, HS−. H2S freely diffuses across membranes and once inside the cell, HS− predominates where it is either assimilated, or in some organisms, effluxed via active transport (Czyzewski & Wang, 2012). H2S is toxic at high concentrations due to its ability to inhibit cytochrome c oxidase.
Cells must be capable of regulating and assimilating bioavailable sulfur, a process that is poorly understood beyond studies of the master regulator of cysteine metabolism CymR, in Bacillus subtilis and Staphylococcus aureus (Tanous et al., 2008, Soutourina et al., 2009). Sulfane sulfur (S0) is two e− oxidized relative to H2S (S2−) and is generally accepted as the major form of bioavailable sulfur that is trafficked in the cell (Mueller, 2006). This “reductant-labile” form of sulfur is found in low-molecular weight (LMW) hydrodisulfides or persulfides, RSSH, hydropolysulfides, RS(Sn)SH, and polysulfides, RS(Sn)SR which are derived from LMW thiols, RSH, e.g., glutathione (Bailey et al., 2014). Recent studies in mammalian cells suggest that these forms of sulfur may accumulate to millimolar levels and thus may represent a highly dynamic source of sulfane sulfur (Ida et al., 2014). Proteins can also undergo reversible S-sulfhydration at cysteine residues (Pan & Carroll, 2013, Zhang et al., 2014), notable examples of which include cysteine desulfurases, rhodaneses and sulfide:quinone oxidoreductases, but may include many additional targets (Ida et al., 2014). Indeed, S-sulfhydration may well represent an important oxidative posttranslational modification, much like S-nitrosation (RSNO) mediated by nitric oxide (NO•) donors (Derakhshan et al., 2007). Recent evidence suggests an interplay between NO• and H2S signaling pathways in mammals (Filipovic et al., 2012, Filipovic et al., 2013, Eberhardt et al., 2014).
Staphylococcus aureus is a medically relevant, opportunistic Gram-positive pathogen that is the causative agent of numerous illnesses ranging from minor skin infections to life-threatening diseases and can colonize virtually any tissue in the body. Bacteria, including S. aureus, that harbor deletions of the genes encoding CBS and CSE produce significantly less HS−; this in turn leads to an increased susceptibility to oxidative stress and general microbial stress induced by antibiotics (Shatalin et al., 2011). This protective effect might be traced to LMW hydropolysulfides and related polysulfide species (Greiner et al., 2013, Ida et al., 2014). Interestingly, bacterial cells in culture are known to produce increased amounts of hydrogen sulfide under conditions of oxidative stress (Shatalin et al., 2011).
Staphylococcus aureus encodes what appears to be a complete HS− oxidation system in a single operon, termed cst (Grossoehme et al., 2011) and this operon is duplicated in highly pathogenic S. aureus strains. The physiological function of the cst operon is unknown, but is proposed to oxidize S2− to thiosulfate, S2O32−, which, unlike the preferred bacterial sulfur source, sulfate (SO42−), can be assimilated by this organism. The cst operon is under the control of the transcriptional repressor CstR (CsoR-like sulfurtransferase repressor) in S. aureus (Fig. 2A, below) (Grossoehme et al., 2011). CstR reacts with chalcogen oxyanions and the oxidant tetrathionate to negatively regulate DNA binding in vitro; this in turn, regulates the transcription of tauE, cstR, cstA, cstB, and sqr (Grossoehme et al., 2011, Luebke et al., 2013). CstA is a three-domain sulfurtransferase or rhodanese, enzymes historically characterized as thiosulfate sulfurtransferases (TSTs) that cleave the sulfur-sulfur bond of thiosulfate to form an enzyme cysteine-bound persulfide, RSSH, and sulfite (SO32−) (Cipollone et al., 2007). More recently rhodaneses have been shown to shuttle persulfides in molybdopterin biosynthesis (Dahl et al., 2011, Dahl et al., 2013), detoxify hydrogen sulfide (Hildebrandt & Grieshaber, 2008) and biosynthesize 2-thiouridine (Ikeuchi, 2006). CstB contains a sulfur dioxygenase-like (SDO) domain that is similar to other identified SDOs, including and human ETHE1 (Holdorf, 2008) and Blh (beta-lactamase-like hydrolase) from the plant pathogen Agrobacterium tumefaciens (Guimaraes et al., 2011). Chromosomal deletion of the mammalian SDO, ETHE1, leads to embryonic lethality in mice as a result of sulfide toxicity-induced ethylmalonic encephalopathy (Tiranti et al., 2009). A. tumefacians SDO is also required for growth and sulfide detoxification under hypoxic conditions that might be found in a biofilm, hypothesized to be required to clear highly toxic HS− under conditions of low oxygen availability (Guimaraes et al., 2011).
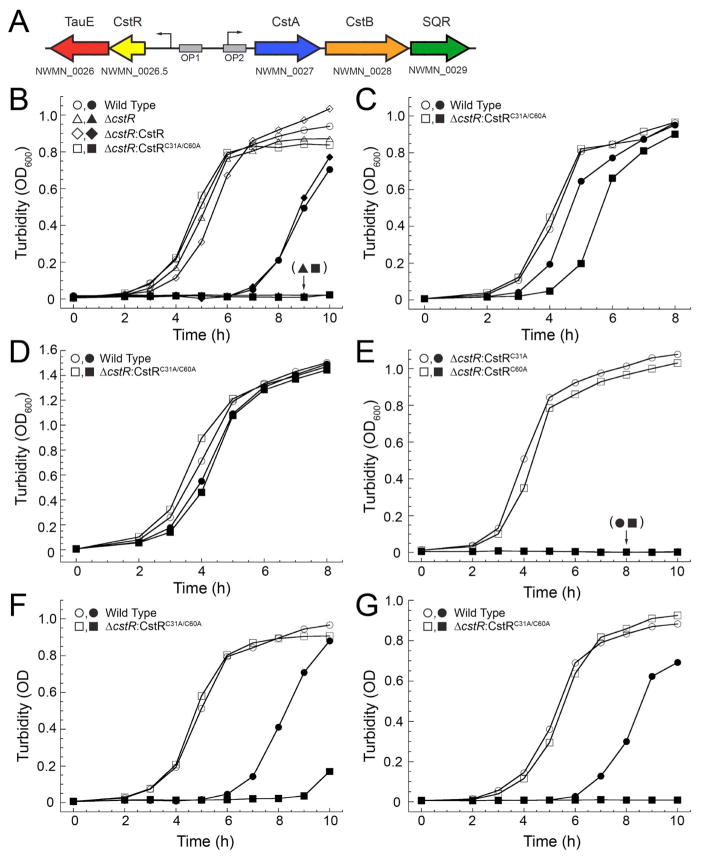
Figure 2. CstR is required for S. aureus defense against sulfide stress.
(A) Schematic representation of the cst operon, NWMN_0026-NWMN_0029, and operator binding sites OP1 and OP2 (Grossoehme et al., 2011). Arrows indicate promoter regions. (B–E) Growth curves of wild-type (WT) and ΔcstR S. aureus mutant strains transformed with the indicated CstR allele carried on pOS1 complementation vector under aerobic conditions at 37 °C with shaking in the absence (open symbols) or presence (filled symbols) of 0.2 mM NaHS. (B) WT (circles), ΔcstR:pOS1 (triangles), ΔcstR:CstR (diamonds), and ΔcstR:CstRC31A/C60A (squares) S. aureus grown in HHWm minimal media supplemented with 0.5 mM thiosulfate (HHWm+TS). (C) WT and ΔcstR:CstRC31A/C60A S. aureus strains grown in HHWm supplemented with 0.25 mM cystine. (D) WT and ΔcstR:CstRC31A/C60A grown on rich TSB media. (E) Growth of ΔcstR:CstRC31A (circles) and ΔcstR:CstRC60A (squares) in HHWm+TS. (F) WT and ΔcstR:CstRC31A/C60A S. aureus grown in the absence (open symbols) or presence (filled symbols) of 25 μM sodium tetrasulfide (Na2S4). (G) WT and ΔcstR:CstRC31A/C60A S. aureus grown in the absence (open symbols) or presence (filled symbols) of 0.2 mM sodium sulfide (Na2S). The data points represent a single representative growth curve.
In a microarray study reported prior to the discovery of CstR (Grossoehme et al., 2011), S. aureus N315 strain grown anaerobically in a biofilm in the presence vs. absence of sodium nitrite (NO2−) was found to induce adaptation to general oxidative and nitrosative stress, as well as high iron, that leads to the impairment of polysaccharide intercellular adhesion (PIA) synthesis and inhibition or dispersal of existing biofilms. Furthermore, these effects were promoted by slightly acidic pH and were found to be quenched by the addition of nitric oxide (NO•) scavengers. Strikingly, all genes of the cst operon from tauE to sqr, were among the most highly induced genes in the cell under these conditions (Schlag et al., 2007). NO• has also been shown to play a role in biofilm dispersal of Pseudomonas aureginosa (Barraud et al., 2006) and in many other bacteria (Barraud et al., 2014), while anaerobically grown E. coli respiring on nitrate induces protein S-nitrosation that seems to be controlled by an anaerobic “moonlighting” function of the peroxide sensor, OxyR (Seth et al., 2012).
In this work, we significantly extend these earlier studies and establish that CstR is a sulfane sulfur-sensing transcriptional repressor in Staphylococcus aureus. Acute sodium hydrogen sulfide (NaHS), disodium sulfide (Na2S) or sodium tetrasulfide (Na2S4) stress introduced into mid-log cells in a variety of growth media induces the cst operon in a manner that requires both cysteine residues in CstR, Cys31 and Cys60. Neither nitrite or nitric oxide induce the operon under these aerobic conditions. Although Na2S is an inducer of the cst operon, this induction is indirect since cysteine thiols on purified CstR do not react directly with Na2S. In contrast, CstR readily reacts with a sulfane sulfur (S0) donor under anaerobic conditions to form a mixture of di-, tri-, and tetrasulfide crosslinks as determined by high resolution tandem mass spectrometry, modifications that lead to dissociation of CstR from the DNA operator in vitro. These results are consistent with a model in which CstR governs sulfide homeostasis in S. aureus by functioning as a polysulfide or persulfide-sensing repressor that mediates transcriptional depression of the cst-encoded genes, the products of which allow assimilation of reductant-labile cellular sulfides. The potential connection between these findings and those previously reported for the cst operon and nitrite stress in biofilms is discussed.
Results
CstR binds Cu(I) but Cu(I) binding does not negatively regulate cst operator binding
CstR and the bona fide copper sensor CsoR from S. aureus strain Newman are 31% identical but function as paralogous repressors in the same cytoplasm (Grossoehme et al., 2011) and partition into separate clades on the basis of a global sequence alignment (Chang et al., 2014). The cst operon encodes what appears to be a sulfur oxidation system under the transcriptional control of CstR, while CsoR regulates the transcription of the Cu(I)-specific P1B-type ATPase efflux pump under conditions of Cu(I) stress (Grossoehme et al., 2011). Addition of copper salts to the growth medium does not induce the expression of cstA or tauE relative to untreated cells thus revealing that CstR is incapable of sensing Cu toxicity (Grossoehme et al., 2011). These and other experiments establish that CsoR and CstR function independently in the S. aureus cytoplasm.
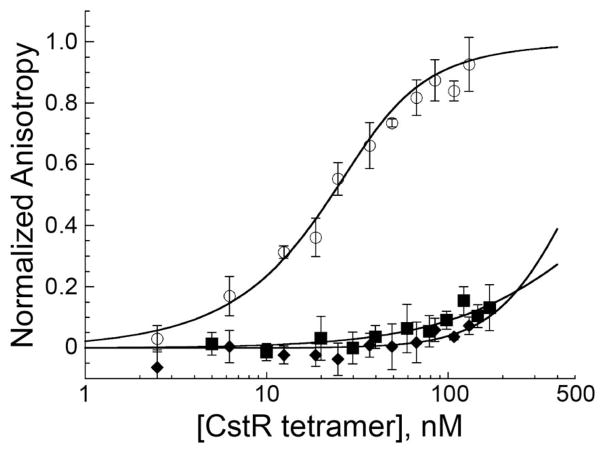
To further probe this dichotomy of function, a series of anaerobic Cu(I) binding experiments were carried out by titrating reduced CstR into a solution of Cu(I):BCA2 (bicinchoninic acid; log KCu=17.2) and observing the change in absorption of the Cu(I):BCA2 complex (A562) as a result of Cu(I) binding by CstA (Fig. 1A). Global analysis of three independent experiments carried out at different Cu(I):BCA2 concentrations reveals log KCu=14.0 (±0.3). This value is four orders of magnitude weaker than S. aureus CsoR (18.0 ±0.1) and is in fact, within a factor of ≈5 of KCu for to H70A S. aureus CsoR (Grossoehme et al., 2011) and the analogous H64A Mtb CsoR (Ma et al., 2009b), the latter which is known to form a bis-thiolate coordination complex (Liu et al., 2007) similar to what is likely formed by CstR on the basis of an absorption spectrum dominated by Cu(I)-S bonds (Fig. S1). Cu(I)-bound CstR binds to the cst OP1 operator with an affinity that is similar to that of apo-reduced CstR (Fig. 1B; Table S1). Thus, despite the fact that CstR binds Cu(I), Cu(I) binding is poorly allosterically competent to drive dissociation of CstR from the operator in vitro or in vivo (Grossoehme et al., 2011) and the affinity is such that the intracellular Cu(I) concentration is unlikely to rise to a level required to be bound by CstR, due to regulation by CsoR (Reyes-Caballero et al., 2011).
Figure 1. CstR forms a modest affinity complex with Cu(I) and metal binding does not negatively regulate cst DNA binding.
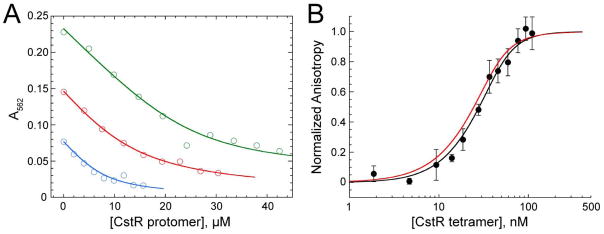
(A) Representative Cu(I)-bicinchoninic acid competition assays with apo CstR. Binding curves were obtained under anaerobic conditions. Open symbols represent the A562 of the Cu(I):BCA2 complex and the solid lines represent the global fitting of three individual experiments to a single-site binding, direct competition model. The global Cu(I):CstR binding constant was calculated to 1.0±0.4×1014 M−1. Green: 29.6 μM Cu(I), 70 μM BCA. Red: 18.9 μM Cu(I), 50 μM BCA. Blue: 10 μM Cu(I), 30 μM BCA. (B) Representative fluorescence anisotropy titration of Cu(I)-bound CstR to a fluorescently labeled cst OP1 DNA fragment. The macroscopic binding constant, Ktet, was determined to be 4.3±1.7×107 M−1. The red line is a simulated curve defined by the binding parameters for apo-reduced CstR (see Fig. 6) under the same solution conditions (Ktet,= 6.3±0.5×107 M−1).
The cst operon is essential for normal growth under sulfide stress
The next series of experiments were carried out in an effort to define the inducer(s) of cst operon. Given the presumed connection to sulfur oxidation (Fig. 2A), wild-type (WT) S. aureus was first grown in HHWm chemically defined media (Hussain et al., 1991) supplemented with either 0.5 mM thiosulfate (TS, HHWm+TS) or 0.25 mM cystine (Cys, HHWm+Cys) as the sole sulfur source (Grossoehme et al., 2011) or on a rich growth medium (TSB) in the absence or presence of 0.2 mM NaHS (Fig. 2B–D). Growth in HHWm+TS in the presence of NaHS results in a significant growth defect while cultures grown in HHWm+Cys or TSB are comparatively less negatively impacted by sulfide stress (Fig. 2B–D).
Wild-type S. aureus was next compared to a ΔcstR deletion strain (Grossoehme et al., 2011). In the ΔcstR strain, expression of the cst operon is derepressed leading to massive overexpression of those genes regulated by OP1 (tauE) or OP2 (cstA, cstB, sqr) (Fig. 2A). When this strain is grown in the presence of NaHS, it fails to recover (Fig. 2B). The ΔcstR strain was then transformed with the S. aureus complementation vector pOS1-Plgt (Bubeck Wardenburg et al., 2006) encoding a wild-type cstR allele (ΔcstR:CstR). The ΔcstR:CstR strain was grown in the presence of 0.2 mM NaHS and yielded a growth phenotype identical to that of the WT S. aureus strain (Fig. 2B). In contrast, a ΔcstR strain transformed with a double cysteine mutant allele of cstR (ΔcstR:CstRC31A/C60A) exhibits a severe growth defect and therefore fails to complement the ΔcstR strain. These results reveal that the two cysteine residues in CstR are essential to mitigate the effects of sulfide toxicity on cell viability.
Wild-type and ΔcstR:CstRC31A/C60A S. aureus strains were next grown in HHWm+Cys (Fig. 2C) or in rich TSB media (Fig. 2D). When grown in HHWm+Cys under NaHS stress, WT S. aureus does not exhibit a significant growth defect while the ΔcstR:CstRC31A/C60A strain is clearly impaired (Fig. 2C). When grown in TSB medium, there is a small but measurable growth defect in the presence of HS− stress for both strains (Fig. 2D). These results collectively suggest that limitation of nutrients or sulfur availability enhances the susceptibility of S. aureus to sulfide stress. The requirement to synthesize cysteine from TS may significantly impair the ability of the organism to acclimate to sulfide toxicity in the minimal HHWm+TS medium; however, an inducible cst operon (in either the WT or ΔcstR:CstR strain) is clearly required for assimilation of organic sulfur (cystine) in the presence of NaHS as well. High cellular TS may also inhibit cst enzymes as TS is a common byproduct of HS− detoxification (Kabil et al., 2014).
The ΔcstR strain was next transformed with plasmids encoding individual cysteine mutants of CstR, ΔcstR:CstRC31A and ΔcstR:CstRC60A, as well as that expressing the paralogous copper sensor, CsoR (ΔcstR:CsoR) (Grossoehme et al., 2011). Previous work indicated that chemical modification of Cys31 in CstR was both necessary and sufficient to negatively regulate DNA binding in vitro while modification of Cys60 alone had little affect on DNA binding (Luebke et al., 2013). Both ΔcstR:CstRC31A and ΔcstR:CstRC60A strains grown in HHWm+TS under 0.2 mM NaHS stress fail to grow (Fig. 1E) revealing that although Cys31 alone is sufficient in vitro (Luebke et al., 2013), both Cys31 and Cys60 of CstR are essential for wild-type-like growth under sulfide stress in vivo. Constitutively expressed CsoR also fails to complement the growth phenotype of the ΔcstR strain grown on HHWm+TS with 0.2 mM NaHS (Fig. S2), further confirming the physiological specificity of the biological response to sulfide stress by CstR (Grossoehme et al., 2011).
Recent studies reveal that commercial sources of NaHS are contaminated with polysulfides of the general formula, −S-Sx-S− (where x=1–6) that form spontaneously from NaHS in the presence of molecular oxygen, and can be catalyzed by contaminating divalent metal ions (Greiner et al., 2013). The NaHS stock used here was found to possess low but detectable levels (0.3%) of polysulfide by absorbance spectroscopy (Fig. S3) consistent with previous work (Greiner et al., 2013). This prompted us to assess the growth phenotype of wild-type and ΔcstR:CstRC31A/C60A S. aureus strains in HHWm+TS media (cf. Fig. 2B) when challenged with 25 μM sodium tetrasulfide (Na2S4) (Fig. 1F) or 0.15 mM sodium sulfide (Na2S) that is devoid of polysulfide contamination (Fig. S3) (Fig. 2G). Virtually identical growth phenotypes are observed when the WT strain is stressed with each stressor vs. NaHS; in both cases as well, the ΔcstR:CstRC31A/C60A fails to complement the ΔcstR strain. Thus, both reduced S2− or partially oxidized forms of sulfur (S−, S0) present in polysulfides are deleterious to the viability of S. aureus, with a wild-type CstR mediating a protective effect against this stress.
CstA, CstB, and SQR are each essential for mitigating sulfide stress
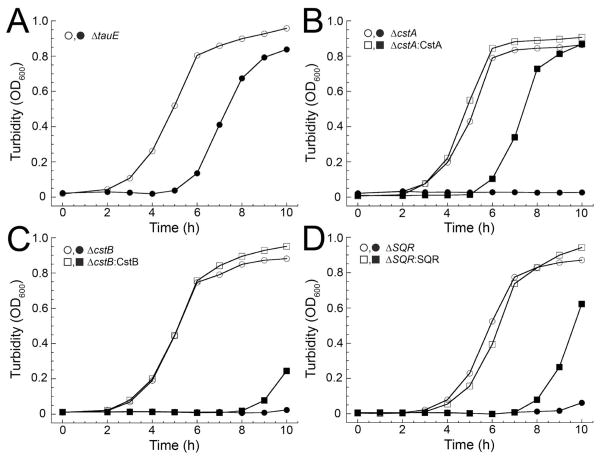
Individual S. aureus deletion strains of the remaining genes in the cst operon, ΔtauE, ΔcstA, ΔcstB, and Δsqr were next generated and cultured in the presence or absence of sulfide stress in HHWm+TS medium. We observe no growth defect for the ΔtauE strain under sulfide stress and thus the physiological function of TauE remains undefined by these experiments (Fig. 3A). In strong contrast, the ΔcstA, ΔcstB, and Δsqr strains each exhibit a severe growth phenotype when grown in the presence of sulfide stress (Fig. 3B–D). Complementation with the corresponding wild-type allele constitutively expressed under control of the pOS1-Plgt promoter and comparison to the wild-type and cstR-complemented ΔcstR growth curves (Fig. 2B) reveals that while wild-type cstA provides full complementation (Fig. 3B), the wild-type cstB and sqr complemented strains appear to give an intermediate growth phenotype (Fig. 3C, D). These data provide strong evidence in support of the proposal that in order for S. aureus to mitigate the cellular effects of (poly)sulfide toxicity, the CstR-dependent transcriptional derepression of the downstream half of the cst operon requires all three genes for survival under sulfide stress.
Figure 3. cst genes are required for cellular sulfide resistance.
Representative growth curves of individual cst operon gene deletion strains (circles) and corresponding complementation strain (squares) grown in the absence (open symbols) or presence of 0.2 mM NaHS (closed symbols) in HHWm+TS. (A) ΔtauE (no complementation strain shown) (B) Δ3cstA and ΔcstA:CstA. (C) ΔcstB and ΔcstB:CstB. (D) Δsqr and Δsqr:SQR.
Acute exogenous (poly)sulfide stress induces the cst operon
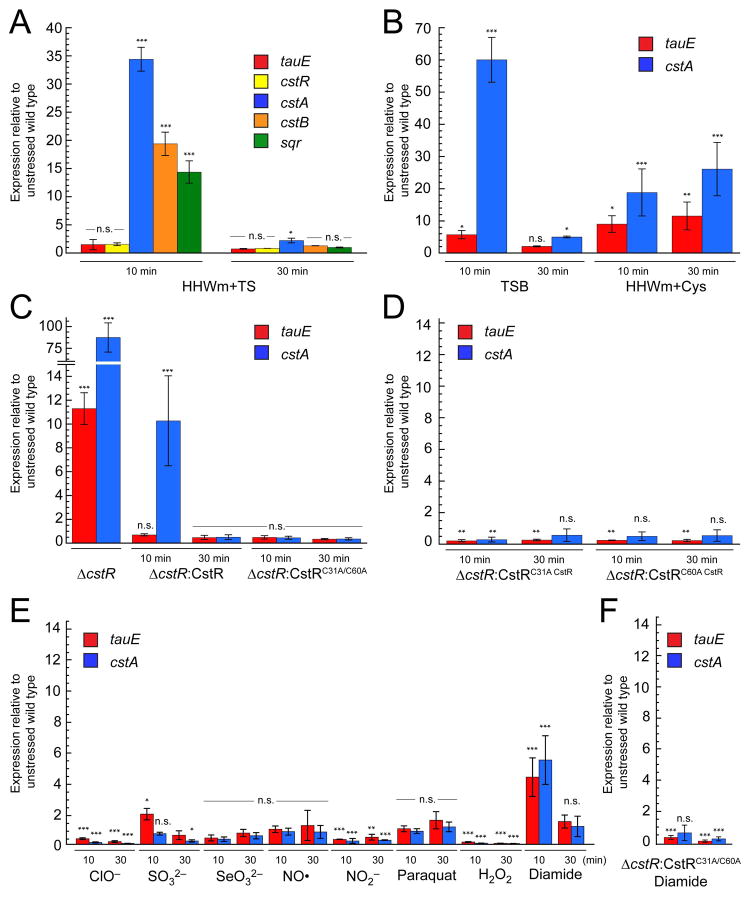
Expression levels of the tauE, cstR, cstA, cstB, and sqr genes were monitored following acute NaHS toxicity in HHWm+TS at OD600 of ≈0.2 by quantitative real-time PCR (qRT-PCR). A 15–30-fold induction of the OP2-regulated downstream cst genes, cstA, cstB, and sqr, was observed at 10 min post stress, with comparatively little induction of the cstR and tauE genes in this divergently transcribed operon (Fig. 4A). This relative induction of each side of the operon qualitatively matches that previously observed in a ΔcstR strain (Grossoehme et al., 2011) (see also Fig. 4C). Strikingly, by 30 min, the mRNA levels of all induced genes return nearly to pre-induction levels, consistent with an acute phase response to sulfide toxicity (Fuchs et al., 2013). The experiment was repeated for cells grown in TSB and HHWm+Cys for tauE and cstA as they report on the induction of the upstream and downstream regions of this divergently transcribed operon, respectively. In TSB, an induction profile similar to that obtained for cells grown in HHWm+TS was observed where there is stronger upregulation for the downstream cstA than the upstream tauE gene. Additionally, mRNA levels return to baseline by 30 min (Fig. 4B). When the experiment is performed for cultures grown in HHWm+Cys, both tauE and cstA are induced at 10 min and both genes remain de-repressed at 30 min post-addition of NaHS (Fig. 4B).
Figure 4. The cst operon is regulated by hydrogen sulfide in vivo.
Quantitative RT-PCR experiments for WT and mutant S. aureus cultures grown to an OD600 of 0.2 and challenged with 0.2 mM NaHS added to the growth medium at t=0. Aliquots for analysis were collected at 10 and 30 min post addition. All cultures were grown in HHWm+TS unless otherwise noted. (A) Relative expression levels for individual cst operon genes post addition of NaHS. (B) Levels of tauE and cstA expression in TSB (left) or HHWm+Cys (right). (C) ΔcstR (left), ΔcstR:CstR (middle), and ΔcstR:CstRC31A/C60A (right). (D) ΔcstR:CstRC31A (left) and ΔcstR:CstRC60A (right) individual cysteine mutants of CstR. (E) RT-PCR analysis for WT S. aureus exposed to acute toxicity of 2.4 mM hypochlorite (ClO−), 10 mM sulfite (SO32−), 0.2 mM selenite (SeO32−), 0.5 mM nitric oxide (NO) as MAHMA NONOate, 5 mM nitrite (NO2−), 25 nM paraquat, 10 mM hydrogen peroxide (H2O2), or 1 mM diamide. (F) ΔcstR:CstRC31A/C60A S. aureus exposed to 1 mM diamide stress. N = 3 error bars represent one s.d. from the mean, with fold-expression relative to wild-type, unstressed cells. Two-way ANOVA analysis was performed relative to 16S RNA at the indicated time point (*** = p < 0.001, ** = p < 0.005, * = p < 0.050, and n.s. = not statistically significant).
The experiments were repeated for the ΔcstR strains transformed with pOS1 vectors constitutively expressing wild-type or mutant CstRs. The cst operon is massively upregulated in the ΔcstR strain as expected (Fig. 4C) (Grossoehme et al., 2011) but when complemented with either wild-type cstR (ΔcstR:CstR) or C31A/C60A cstR (ΔcstR:CstRC31A/C60A) alleles, repression of the operon is restored prior to addition of NaHS to these cultures. Upon addition of NaHS to the ΔcstR:CstR culture, the cst operon is induced at 10 min post addition and returns to baseline by 30 min (Fig. 4C), albeit to a lesser extent than with the WT strain. The induction level may be lower here as a result of the constitutive expression of CstR, which might function as a partial sink for (poly)sulfide toxicity leading to an attenuated induction upon sulfide stress. In the case of the WT strain under the same conditions, cstR is not significantly upregulated (Fig. 4A). In any case, this finding recapitulates the wild-type strain, and is consistent with complementation of the growth curve (Fig. 2B). When the same experiment is performed with the ΔcstR:CstRC31A/C60A strain, no induction of the operon is observed in the presence of NaHS stress (Fig. 4C). This experiment was repeated for the individual CstR cysteine mutants, ΔcstR:CstRC31A and ΔcstR:CstRC60A strains. Here, both mutant strains fail to respond to NaHS stress in vivo and the operon remains repressed through the duration of the experiment (Fig. 4D). Identical findings characterize mid-log wild-type cells stressed with 25 μM sodium tetrasulfide or 150 μM disodium sulfide (Fig. S4).
It was next of interest to assess the specificity of the induction of the cst operon through CstR by (poly)sulfide. To do this, transcriptional depression of the tauE and cstA genes was determined upon addition of a range of biologically relevant oxidative and nitrosative stressors. These include hypochlorite (HOCl), sulfite (SO32−), selenite (SeO32−), nitric oxide (NO•), nitrite (NO2−), paraquat (1,1′-dimethyl-4,4′-bipyridinium dichloride), hydrogen peroxide (H2O2) and diamide (3-(dimethylcarbamoylimino)-1,1-dimethylurea). Of these, only diamide strongly induces both tauE and cstA at 10 min post addition of the reagent, with the expression returning to baseline by 30 min (Fig. 4E). In the ΔcstR:CstRC31A/C60A strain, diamide fails to induce the operon revealing that the induction observed is CstR-dependent and not through some alternative pathway (Fig. 4F). These experiments were carried out such that there was no noticeable growth phenotype upon addition of the inducer to the growth medium (Fig. S5). Sulfite was capable of inducing tauE but not cstA at 10 min post addition (Fig. 4E), consistent with the previous findings that sulfite reacts with CstR in vitro (Grossoehme et al., 2011, Luebke et al., 2013). However, sulfite is clearly not a primary inducer in vivo. Finally, SeO32−, NO•, H2O2 and paraquat stress do not significantly induce cst operon expression; however, the relative expression of tauE and cstA appears to decrease with ClO−, NO2− and H2O2 stress (Fig. 4E).
Sulfur metabolite profiling of acute NaHS- and tetrasulfide-stressed cells
Wild-type S. aureus was conditioned in HHWm+TS growth medium to an OD600=0.2 and acutely stressed with 0.2 mM NaHS or 25 μM Na2S4 with aliquots removed at t=0, 10 and 30 min post-cst induction, as described above for the qRT-PCR analysis, and subjected to sulfur metabolite profiling using a standard fluorescence-based monobromobimane (mBBr) labeling assay (Fahey & Newton, 1987, Newton et al., 1996) (Fig. S6). As expected, the major change in these profiles is a dramatic increase of intracellular sulfide in the NaHS-stressed cells, going from ≈25 nmol mg−1 protein at t=0 to ≈460 nmol mg−1 protein at 10 min and ≈300 nmol mg−1 protein 30 min post-induction of the cst operon (Fig. 5A). Concomitant with this increase and subsequent decrease in total sulfide, the TS concentration, while not detectable before the addition of sulfide to the growth medium (TS must be efficiently assimilated as the sole sulfur source), rises to ≈13 nmol mg−1 protein which persists for 30 min (Fig. 5B); in contrast, total reduced cysteine drops by ≈40% to ≈4 nmol mg−1 protein from pre-induction levels (Fig. 5C). These profiling experiments are consistent with the hypothesis that sulfide stress is sensed by CstR which upregulates the synthesis of CstA, CstB and SQR and may direct cellular sulfide to the cst-encoded sulfur oxidation system at the expense of cysteine biosynthesis. A major product of sulfur oxidation is TS which may then be assimilated by the organism (see Discussion).
Figure 5. Extracellular sodium hydrogen sulfide (NaHS) enters the cytoplasm of S. aureus resulting in a concomitant increase in thiosulfate (TS) and decrease in cysteine.
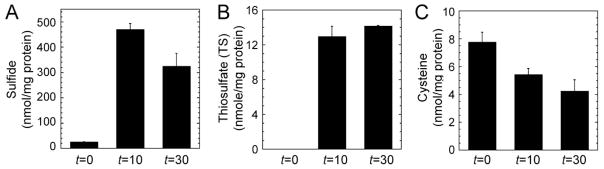
Cellular LMW sulfur metabolites were derivatized with mBBr were detected using a fluorescence-detected profiling method (Fahey & Newton, 1987) before (t=0) and after (t=10 min and t=30 min) the addition of 0.2 mM NaHS to the culture medium (see Fig. S6 for representative liquid chromatograms). (A) Sulfide; (B) thiosulfate; and (C) cysteine, each expressed in nmol mg−1 protein.
CstR reacts with a sulfane sulfur donor to negatively regulate DNA binding through the formation of di-, tri-, and tetrasulfides in vitro
The experiments outlined above suggest that CstR cysteine residue(s) react directly with sulfide and/or polysulfide, which in turn, drives DNA operator dissociation and transcriptional derepression of the cst operon. As pointed out by others (Greiner et al., 2013, Zhang et al., 2014), the reaction of a protein thiolate in CstR with S2− is chemically not possible in the absence of contaminating, partially oxidized forms of sulfur. To further define this relationship between CstR, Na2S, NaHS or Na2S4, and DNA binding in vitro, we reacted fully reduced recombinant CstR with a 5-fold thiol molar excess of Na2S, NaHS or Na2S4 under strictly anaerobic conditions in the presence of a divalent cation chelator, and measured the DNA binding to the operator binding site, cst OP1 (see Fig. 2A), by fluorescence anisotropy (Fig. 6), and characterized the reaction products by LC-ESI-MS (Fig. 7) and high resolution tandem mass spectrometry (Fig. 8). Fully reduced CstR binds to OP1 and OP2 operators with similar affinities, of ≈107 M−1 when analyzed using a two-tetramer binding model (Grossoehme et al., 2011). When the titration is performed with NaHS- or tetrasulfide-reacted CstR, the binding affinities decrease to by ≈100–200 fold, consistent with strong negative regulation of DNA operator binding by the inducers (Fig. 6, Table S1).
Figure 6. Reaction of CstR with polysulfide negatively regulates DNA operator binding.
Fluorescence anisotropy titrations of fully reduced CstR (open circles) and NaHS- (closed squares) or Na2S4-reacted CstR (closed diamonds) with fluorescently-labeled cst OP1-containing DNA. Data were fit to a sequential tetramer binding model where two non-dissociable tetramers bind stepwise to one operator DNA binding site. Stepwise binding constants, K1 and K2, were used to determine the average macroscopic binding constant, Ktet (Ktet = (K1•K2)½). WT CstR-OP1 affinity is 7.4 (±1.8)×107 M−1 (see Table S1 for all previously determined Ktet values) (Luebke et al., 2013, Grossoehme et al., 2011), while Ktet for NaHS- and Na2S4-reacted CstRs have upper limits of 0.06±0.05×107 M−1 and 0.03±0.03×107 M−1, respectively. Binding curves represent a single representative titration. Conditions: 10 nM cst OP1 DNA, 10 mM HEPES, 0.2 M NaCl, pH 7.5, 25 °C.
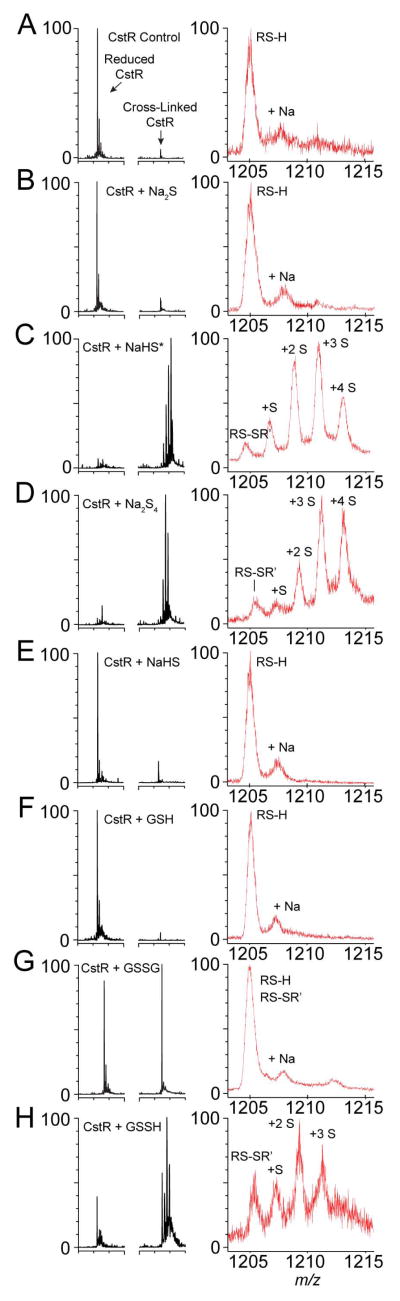
Figure 7. CstR reacts with NaHS, Na2S4, and GSSH to form a series of mixed di-, tri-, and tetrasulfide crosslinks.
LC-ESI-MS spectra of intact reduced CstR (A), CstR following reaction with a 5-fold S:Cys molar excess of (B) Na2S, (C) NaHS* (D) Na2S4, (E) NaHS, (F) GSH, (G) GSSG, and (H) GSSH. Black traces represent the ratio of reduced (left) or cross-linked (right) CstR in the deconvoluted mass spectra. Red traces represent m/z ratios of the +8 or +16 charge states of reduced or cross-linked dimeric CstR species, respectively, with corresponding post-translational modification assignments shown. RS-H indicates reduced CstR and RS-SR’ represents an interprotomer disulfide bond between Cys31 and Cys60’. Each ‘S’ represents a mass shift of +32 Da relative to the RS-SR’ disulfide in the deconvoluted spectra. For a sample like GSSG (panel G), the +8 and +16 m/z overlap but can be deconvoluted based on the m/z distribution of the reduced vs. oxidized forms (Luebke et al., 2013). For a summary of the observed masses, refer to Supplemental Table S2. (*) indicates a reaction performed with NaHS in 10 mM HEPES, 200 mM NaCl, pH 7.0. All other reactions were performed in 10 mM PO43−, 200 mM NaCl, 1 mM EDTA, pH 7.0.
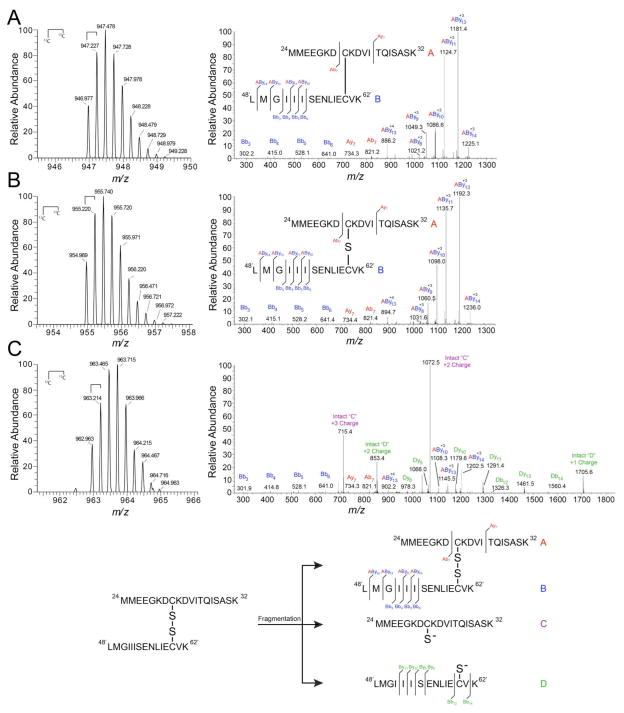
Figure 8. High-resolution tandem mass spectrometry confirms di-, tri-, and tetrasulfide mass shift assignments.
High-resolution LTQ-orbitrap tandem mass spectra of NaHS-reacted CstR tryptic peptides in the +4 charge state (left) and corresponding fragmentation patterns (right). Peptide A (red), 24MMEEGKDCKVITQISASK42, includes Cys31 and Peptide B (blue), 48′LMGIIISENLIECVK62′, includes Cys60’. Peptide fragments were assigned relative to either peptide “A” or “B” where Bb3 corresponds to the peptide fragment b ion 48′LMG50′ with a mass of 302 Da. Cross-linked peptides are denoted as “AByn” where peptide “A” remained intact and fragmentation occurred on peptide “B”. Inset: map of fragmentation pattern. (A) Disulfide. (B) Trisulfide. (C) Tetrasulfide.
To define the nature of the chemical adducts in NaHS-reacted and Na2S4-reacted CstR, CstRs were subjected to LC-ESI-MS analysis. Unreacted CstR gives no disulfide crosslinks and is nearly quantitatively reduced (Luebke et al., 2013) (Fig. 7A). As expected (Greiner et al., 2013), CstR mixed with authentic Na2S gives rise to no reaction products under these conditions (Fig. 7B). In contrast, both NaHS- and sodium tetrasulfide-reacted CstRs form a series of mixed disulfides, including an interprotomer disulfide bond and a number of mass shifted species corresponding to the incorporation of 0–4 sulfur atoms (Fig. 7C–D). Given that authentic Na2S gives no reaction, the reaction products observed in the presence of NaHS must derive from contaminating polysulfide that is found or formed in situ in this sample; this is confirmed by observing no reaction when the experiment is repeated in a phosphate-containing buffer in the presence of EDTA (Fig. 7E) used to suppress production of metal-catalyzed HS• radicals and polysulfides (Zhang et al., 2014). In the NaHS-and Na2S4- reacted sample, these masses correspond to +32.0, +63.1, +92.8, and +124.0 Da associated with each crosslinked CstR dimer (CstR is a dimer of dimers; the noncovalent tetramer interface of which is disrupted upon MS analysis). These mass shifts were assigned as di- (0 S), tri- (1 S, +32.0), and tetrasulfide (2 S, +63.1) cross-links (Tables S2–S3); the +3 S and +4 S species are predicted to correspond to CstR dimers that possess mixed interprotomor tri- and tetrasulfide linkages (+3 S) and a pair of tetrasulfide linkages (+4 S), respectively.
The NaHS-reacted CstR was then digested with trypsin and analyzed by high-resolution LTQ-orbitrap tandem mass spectrometry (Fig. 8A–C). The highest intensity peaks containing CstR cross-links were in the +4 charge state as 24MMEEGK|DCK|DVITQISASK42 (Peptide A, where “|” indicates a missed tryptic cleavage site) and 48′LMGIIISENLIECVK62′ (Peptide B) for the Cys31 and Cys60’-containing peptides, respectively (see Table S4 for peptide mass assignments). Upon inspection of the high mass accuracy LTQ data, we identified a mass corresponding to the Cys31-Cys60’ disulfide cross-linked peptides at 946.977 Da (946.978 Da expected; Table S4. This assignment was confirmed by inspection of the fragmentation pattern (Fig. 8A) and previous data (Luebke et al., 2013). We next searched for masses corresponding to the addition of one or two sulfur atoms and identified peaks at 954.969 Da (954.972 Da expected) and 962.963 Da (962.965 Da expected), respectively. The mass shifts between the di- and trisulfide cross-linked peptides match the monoisotopic mass of 32S at +31.968 Da (31.970 Da expected) and +63.944 Da (63.941 Da expected) for the di- and tetrasulfide (Fig. 8A–C). The tri- and tetrasulfide assignment is further confirmed by inspection of the fragmentation pattern of the 954.969 and 962.965 Da peptides. This reveals a series of cross-linked A and B peptides that contain either a +32 of +64 Da mass shift relative the disulfide (Fig. 5A–C; Fig. S7).
Given the similar reaction profiles obtained with NaHS in HEPES buffer and Na2S4, we conclude that CstR preferentially reacts with the more oxidized, electrophilic, internal sulfur atoms within inorganic polysulfide rather than HS− directly (Zhang et al., 2014) and this reaction is sufficient to allosterically inhibit DNA binding in vitro. The chemical speciation of these more oxidized forms of sulfur, including sulfane S0, are not known but the LMW disulfide, GSSG, or glutahione persulfide, GSSH, are possible reactants. To test this, reactions were performed with GSH, GSSG and GSSH and the reaction products characterized by LC-ESI-MS (Fig. 7F–H). As expected reduced GSH does not react with CstR (Fig. 7F). In contrast, a cleanly disulfide-crosslinked CstR species is the only crosslinked product observed with GSSG, consistent with S-glutathionylation of Cys31, followed by resolution by Cys61’ and release of GSH (Fig. 7G). Strikingly, incubation with GSSH results in a mixture of crosslinked species that incorporate 0, 1, 2 or 3 S atoms (Fig. 7H), qualitatively similar to the products formed with polysulfides (Fig. 7C–D). The structurally and evolutionarily related copper sensor CsoR reacts with Na2S4 poorly under the same conditions (Fig. S8), again consistent with the inducer specificity of this sensing response (vide supra).
Discussion
In the work presented here, we further establish that S. aureus CstR and the Cu(I)-sensing CsoR are functionally independent paralogous repressors in the cell. The distinguishing feature is that although CstR binds Cu(I), it does so with weakly relative to bona fide CsoRs, and Cu(I)-binding fails to disassemble cst OP1-CstR complexes. Despite the fact that both the CstR and CsoR conserve the analogous Cys pair (Higgins & Giedroc, 2014), CstR thiolates are far more reactive toward oxidized sulfur sources and this is central to the functional distinction between the two repressors. Although perturbation of the LMW thiol pool under conditions of sulfide stress may well lead to changes in the cytoplasmic Cu(I)-buffering capacity (Tottey et al., 2012), there is insufficient free Cu(I) under these conditions to induce copA expression as sensed by CsoR (Fig. S2B). Structural studies of reduced vs. oxidized CstRs, relative to those already in hand for a number of CsoRs (Liu et al., 2007, Dwarakanath et al., 2012, Chang et al., 2014), will be required to understand inducer discrimination at the molecular level.
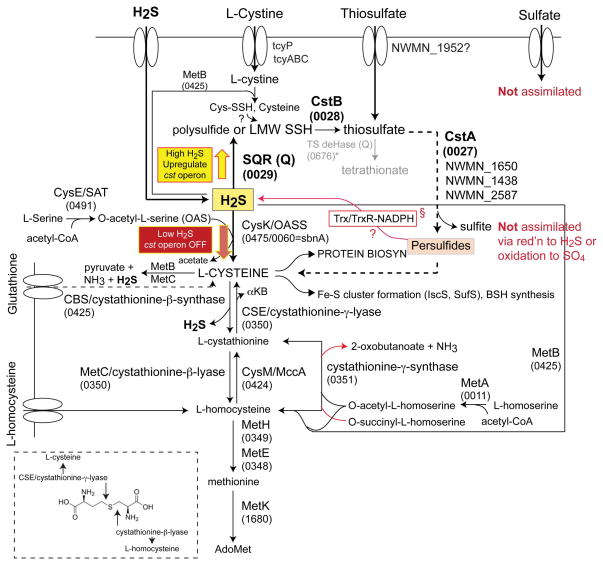
We establish that CstR is a persulfide- and polysulfide-sensing repressor (Figs. 2 and 4) under the aerobic culture conditions used here. Persulfide-sensing by CstR leads to derepression of the transcription of what is predicted to be a sulfide oxidation system, with all components required to be functional in order to mitigate the effects of sulfide toxicity, particularly when cells are grown in a chemically defined growth medium (HHWm) with inorganic TS added as sole sulfur source (Fig. 2). A relatively more modest, but detectable, growth phenotype is observed with cystine provided as the sole sulfur source. This is consistent with the proposal that sulfide toxicity interferes directly with cellular sulfur assimilation, with the effect more pronounced when S. aureus is forced to utilize an inorganic sulfur source (TS), but there are of course other possibilities. The mobilizable sulfane sulfur in TS is likely first “fixed” as a cysteine persulfide by one of five cellular sulfur transferases (rhodaneses) (Fig. 9), which is ultimately trafficked to the cysteine biosynthesis complex via the activity of O-acetyl-L-serine sulfhydratase (OASS; CysK), perhaps via thioredoxin/thioredoxin reductase system or to other cellular needs, e.g., Fe-S cluster biogenesis (Fig. 9). The identity of those rhodaneses that function in this process are not known, although it is interesting to note that two are found in the cst operon itself (cstA and cstB). One possible scenario is that high intracellular persulfide or polysulfides (Greiner et al., 2013) poison key sulfur shuttling steps and/or maintenance of cellular reduction potential, or leads to an increase in deleterious proteome S-sulfhydration (Hildebrandt et al., 2013) that induction of the cst operon serves to mitigate.
Figure 9. An abbreviated rendering of sulfur assimilation and hydrogen sulfide metabolism in S. aureus strain Newman.
The 4-number (wxyx) S. aureus strain Newman gene locus tags (NWMN_wxyz) are given for each enzyme that is annotated with a standard abbreviation based on recent work (Soutourina et al., 2009). OASS: O-acetyl-L-serine sulfhydrylase; SAT + OASS: cysteine synthase complex. The TS quinone-dependent dehydrogenase marked with an asterisk and shaded grey (NWMN_0676) is likely not functional given the absence of a gene encoding the small subunit. §, highlighted to implicate a reductive path to the generation of H2S as a substrate for OASS from cellular protein-bound persulfides (Ida et al., 2014). The large yellow and red arrows illustrate the concept of sulfide homeostasis, which is dictated by the coordinate action of H2S oxidation (by proteins encoded by the cst operon) and assimilatory pathways.
The OP2-regulated genes of the cst operon are strongly transcriptionally induced in a response that requires both sensing cysteines (Cys31 and Cys60) in CstR. This provides direct evidence that thiol chemistry involving both Cys31 and Cys60 is required for the biological function of CstR. Transcriptional depression is observed in mid-log cells in an acute phase response to (poly)sulfide toxicity. The induction is high 10 min post-addition with expression levels of the operon returning to near baseline by 30 min post-addition of sulfide stress. These kinetics of mRNA induction mirror the temporal aspects of the changes in the proteomics profiles in S. aureus COL strain by a number of inducers (Fuchs et al., 2013), but do not track with the cellular sulfide levels which remain elevated at 30 min post-induction (Fig. 4). The origin of this discrepancy is unknown, but suggests there may be a delay in synthesizing sufficient enzymes encoded by OP2-regulated cst genes (CstA, CstB and SQR) to fully metabolize cellular sulfide or polysulfides to less toxic species over this time. Consistent with this prediction, SQR and CstB working together are predicted to oxidize S2− to sulfite and/or TS (Fig. 9); significant TS is indeed detected in cell lysates following sulfide addition coupled with a corresponding reduction in cellular sulfide and free cysteine levels over this time frame (Fig. 4). CstA itself may also react with LMW persulfide and polysulfide sulfur donors as well, and this is the subject of ongoing studies.
The kinetics of the CstR-dependent induction response further suggest that the cst operon functions in (poly)sulfide homeostasis under normal housekeeping conditions, and in particular under conditions where the sulfur source is not optimal, i.e., with thiosulfate (TS) (Fig. 9). Additional evidence in support of a homeostatic control is the initially puzzling finding that both too much or too little of the expression of genes encoding CstR-regulated enzymes gives rise to the same strong growth defect on HHWm+TS medium in these aerobic conditions. This suggests that overexpression of cstA, cstB and sqr may well divert TS-derived sulfur from cellular rhodaneses dedicated to pushing sulfur into cysteine biosynthesis and other metabolite needs, e.g., to sulfur-containing metabolites of Fe-S biogenesis, to sulfide oxidation along with NaHS and thus depriving the cell of useable sulfur (Fig. 9). On the other hand, too little of the sulfide oxidation system may essentially overrun the ability of the organism to utilize sulfide, which itself is a poor sulfur source (Grossoehme et al., 2011). The observed increased cellular thiosulfate and decreased cysteine upon sulfide stress (Fig. 4) are consistent with this model, although other interpretations are possible. For example, the cysteine level could fall in response in an increase in export, or as a result of cysteine catabolism, which would also increase the cellular sulfide levels.
Sulfur adduction of purified, reduced CstR under anaerobic conditions as analyzed by high resolution tandem mass spectrometry reveals a number of interprotomor crosslinked species, including di-, tri- and tetrasulfide Cys31-Cys60’ crosslinked products (Fig. 7). Although additional kinetic and mechanistic studies are required, these crosslinked products clearly derive from a reaction of CstR thiolates with sulfane sulfur (S0) within tetrasulfide and the related organic hydrodisulfude, e.g., glutathione persulfide or from metal-catalyzed one-electron oxidized form of HS−, HS•, rather than HS−, S2− or an organic thiolate, glutathione (Zhang et al., 2014) (Fig. 7). Although the precise chemical inducer(s) of the cst operon in cells is unknown, LMW inorganic polysulfides or organic persulfides and polysulfides, including bacillithiol persulfide, BSSH, or cysteine persulfide, CysSSH, are reasonable candidates, since the model persulfide, GSSH, gives rise to the same product profile as polysulfide (Fig. 7H). Each contains internal, more electrophilic S0 atoms as part of a sulfur chain. Such hydrodisulfides (persulfides) are predicted to be formed from the reaction of oxidized bacillithiol (BSSB) and cystine with excess HS− (Ida et al., 2014), while the former is formed noncatalytically in solutions of sulfide itself (Greiner et al., 2013) or enzymatically via the action of SQR (Marcia et al., 2009) (Fig. 9).
In this context, it is interesting to note that diamide, as the only other strong inducer of the cst operon relative to polysulfide, is a disulfide-generating electrophile that strongly perturbs intracellular disulfide-dithiol balance and induces S-thiolation of proteins in the cytoplasmic proteome of S. aureus and B. subtilis (Pother et al., 2009). This suggests that CstR might also be sensitive to S-thiolation, which if formed on Cys31 (Luebke et al., 2013) would then be immediately converted to a disulfide bond via nucleophilic attack by the resolving Cys60’. This is exactly what is observed with GSSG (Fig. 7G). It is known that formation of a disulfide bond between Cys31 and Cys60’ across the tetramer interface is necessary and sufficient to weaken the affinity of the CstR for the operator (Grossoehme et al., 2011, Luebke et al., 2013).
We favor the hypothesis that under conditions of acute sulfide stress in S. aureus, cysteine persulfides, bacillithiol persulfides and/or polysulfides represent a highly dynamic source of sulfane sulfur (Ida et al., 2014). Further studies are required to identify the chemical nature of the cellular adducts of CstR in sulfide-stressed cells, as well as to more fully characterize the entire LMW and proteomic pools of S-sulfhydrated species in cells, and how these pools change upon sulfide stress. In this context, it is interesting to note that in ETHE1 (CstB)-deficient mice, significant protein S-sulfhydration is proposed to accumulate as a result of sulfide toxicity (Hildebrandt et al., 2013). It is also of interest to connect previous findings, which establish that nitrite stress in S. aureus biofilms induces the cst operon (Schlag et al., 2007), with the persulfide induction of the cst operon reported here. One exciting possibility is that elevated nitric oxide and increased endogenous H2S production required for molybdopterin and Fe-S cluster biogenesis needed for nitrate/nitrite reductases, for example (Schlag et al., 2007), converge to form thionitrous acid (HSNO), nitroxyl (HNO), and possibly polysulfides (Filipovic et al., 2012, Filipovic et al., 2013). Indeed, recent work suggests a significant NO•/H2S interplay in cardiovascular vasodilation which has been traced to HNO-mediated disulfide bond formation in a sensory chemoreceptor channel (Eberhardt et al., 2014). These studies make the prediction that CstR senses nitroxyl directly. Studies designed to test this hypothesis are underway in our laboratory.
Experimental Procedures
Chemicals and Reagents
Sodium hydrogen sulfide (NaHS·xH2O; Sigma-Aldrich, 161527; CAS# 207683-19-0), sodium thiosulfate (Na2S2O3; Sigma-Aldrich 217263; CAS# 7772-98-7; ≥99% reagent-plus grade), sodium sulfite (Na2SO3, Sigma-Aldrich S0505; CAS# 7757-83-7; ≥98% ACS-certified grade), sodium tetrasulfide (Na2S4; Alfa Aesar 88697; CAS# 12034-39-8; ≥90%), and disodium sulfide (Na2S, Sigma-Aldrich 407410; CAS# 1313-82-2; 97–103%) were obtained as crystalline solids, and used as freshly dissolved stock solutions in degassed, deionized metal-free water, without purification. Freshly prepared NaHS and Na2S solutions were compared with freshly prepared stock solutions of authentic Na2S4 and NaHS was estimated to contain 0.3% sulfur as polysulfide (expressed as tetrasulfide sulfur equivalents) by UV-Vis absorption spectroscopy quantified at 372 nm using a standard curve (Greiner et al., 2013, Debiemme-Chouvy et al., 2004) (Fig. S3). All reagents used in the preparation of the chemically defined growth medium (HHWm) were obtained from Fisher or Sigma-Aldrich as tissue culture grade and were used without purification.
Plasmid Construction and Protein Purification
Wild type CstR and CsoR and mutant CstRs were expressed and purified as previously described (Grossoehme et al., 2011, Luebke et al., 2013).
CstR Cu(I) Binding and Affinity Measurements
Bicinchoninic acid (BCA) was used to determine the Cu(I) binding affinity of CstR essentially as described (Xiao et al., 2011, Fu et al., 2013). A fresh Cu(I) stock was prepared anaerobically for each titration in 10 mM HEPES, 200 mM NaCl, pH 7.0 by dissolution of solid Cu(I) chloride, subsequent centrifugation, and collection of the supernatant. The Cu(I) stock concentration was determined by atomic absorption spectroscopy (PerkinElmer AAS-400). 120 μL aliquots containing 9.98, 18.9, or 29.6 μM Cu(I) and 30, 50, or 70 μM BCA, respectively, were prepared and titrated with increasing reduced CstR titrant. Prior to CstR addition, all Cu(I) is bound as Cu-BCA2 complex. An absorbance value of 562 nm (A562) and the extinction coefficient 7,700 M−1 cm−1 were used to determine Cu(I):BCA2 concentration. All titrations were globally fit to a 2direct competition model using Dynafit (Kuzmic, 1996) as described previously (Fu et al., 2013).
Fluorescence Anisotropy-based DNA Binding Titrations
Double-stranded fluorescein-labeled DNAs were prepared as previously described (Ma et al., 2009a, Luebke et al., 2013) with cst OP1 (Grossoehme et al., 2011). Each anisotropy experiment was performed under strictly anaerobic conditions with 10 nM fluorescently-labeled cst OP1 dsDNA in 10 mM HEPES, pH 7.0, 200 mM NaCl at 25 °C. Injections of the indicated CstR were equilibrated for 3 min prior to each anisotropy measurement. Fluorescein was excited at 490 nm and polarization monitored with a 515 nm cut-off filter in the L-format using an ISS PC1 Spectrofluorometer (Champaign, IL). Five measurements of each injection were collected and averaged. Normalized r values for the fractional saturation of cst OP1 were calculated as (robs−r0)/(rcomplex−r0) from 0 to 1 where rcomplex represents the maximum anisotropy obtained and r0 represents free cst OP1 DNA. For titrations not reaching saturation, rcomplex was calculated from the addition of the anisotropy change of reduced CstR to r0 of a given non-saturating CstR. Data were fit to a sequential non-dissociable tetramer (CstR4) binding model using Dynafit (Kuzmic, 1996) assuming a linear relationship between robs and vi and the binding density at the ith addition of the titrant (Grossoehme et al., 2011). The macroscopic binding constant, Ktet, is reported and was determined as Ktet = (K1•K2)½ due to high uncertainty in extracting unique Ki values, K1 and K2, as there is a strong inverse correlation and little sigmoidal behavior in the binding isotherms as previously described (Luebke et al., 2013, Grossoehme et al., 2011). Reported affinities (Ktet) are the average of three independent experiments (Table S1).
In vitro Reaction of NaHS, Na2S, and Na2S4 with reduced CstR
15 μM samples of CstR (protomer) were reacted anaerobically in a Vacuum Atmospheres (Amesbury, MA) glovebox (≤0.5% O2) in fully degassed 10 mM PO43−, 200 mM NaCl, 1 mM EDTA at pH 7.0 with a 5-fold thiol excess of NaHS, Na2S, or Na2S4 for 17 h at 22° C. NaHS reactions were also performed in fully degassed 10 mM HEPES, 200 mM NaCl at pH 7.0. After 17 h, samples were sealed in septa cap vials for immediate LC-ESI-MS analysis or prepared for tryptic digest. Samples for digest were precipitated with a 12.5% final concentration of trichloroacetic acid (TCA) and placed on ice for 1 h. Precipitated samples were sealed in septa cap vials and centrifuged at 4 °C for 20 min. The resulting pellets were washed twice with ice-cold acetone and resuspended in 10 mM ammonium bicarbonate at pH 8.2 and 10% acetonitrile. Proteomics-grade trypsin from porcine pancreas (Sigma) was added at a 1:50 ratio and digested for 1 h at 37 °C. A final concentration of 0.25% trifluoroacetic acid (TFA) was added to quench the digests. Finally, samples were desalted using a C18-packed ZipTip™ column (Millipore), dried, and resuspended in Milli-Q water. All preparative steps were performed anaerobically to avoid oxidation of cysteine and methionine residues. An additional CstR sample was digested and prepared aerobically following reaction with NaHS as a control.
Glutathione Persulfide (GSSH) Preparation and CstR reactions
Glutathione persulfide (GSSH) was prepared by mixing an excess of sodium sulfide (Na2S) with oxidized glutathione (GSSG) (Pan & Carroll, 2013) in a 5:1 ratio and reacted anaerobically at 30° C for 30 min in fully degassed 10 mM phosphate buffer, 200 mM NaCl, and 1 mM EDTA. To confirm and quantify the formation of persulfide, a cyanolysis assay (Kelly et al., 1969) was performed and it was determined that 98.9% of GSSG was converted to GSSH and GSH. This ≈1:1 mixture of GSH/GSSH was reacted anaerobically with reduced CstR exactly as described above for other sulfur compounds in fully degassed 10 mM PO43−, 200 mM NaCl, 1 mM EDTA at pH 7.0 with a 5-fold thiol excess. Control reactions were carried out with GSSG starting material and GSH (see text for details), allowing attribution of any reaction products observed with the GSH/GSSH mixture specifically to GSSH.
LC-ESI and LTQ-Orbitrap Mass Spectrometry Analysis of CstR
LC-ESI-MS was performed as described (Luebke et al., 2013) using a Water/Micromass LCT Classic time of flight (TOF, Waters) with a CapLC inlet at the Indiana University Mass Spectrometry Facility. All data were collected and analysed using the MassLynx software package (Waters). Tandem mass spectrometry analysis of CstR peptides was performed at the Laboratory for Biological Mass Spectrometry using a ThermoFinnigan LTQ-Orbitrap XL mass spectrometer equipped with an UltiMate 3000 nanoLC system (Dionex, Sunnyvale, CA) as described (Luebke et al., 2013, Ma et al., 2009b). Briefly, 5 μL of protein digest was loaded onto a 15 mm×100 mm i.d. C18 reversed-phase trapping column and eluted through a 150 mm×75 mm internal diameter analytical column packed with 5 μm, 100 Å Magic C18AQ packing material (Microm BioResourses Inc.), using a 30 min gradient from 97% to 60% solvent A, 97:3:0.1 water/acetonitrile/formic acid (Solvent B is 0.1% formic acid in acetonitrile) at 250 nL min−1. The column eluent was ionized and electrosprayed directly.
Construction of S. aureus Deletion Strains
S. aureus strain Newman and its derivatives were used for all experiments carried out in this study. To construct the ΔtauE (NWMN_0026), ΔcstA (NWMN_0027), ΔcstB (NWMN_0028), and Δsqr (NWMN_0029) the 5′ and 3′ flanking regions were amplified using the indicated primers (Table S5). These fragments were then combined and introduced into pKOR1 via recombination and in frame deletions were created via allelic replacement as previously described (Bae & Schneewind, 2006). For the ΔtauE and ΔcstA constructs the flanking regions were initially cloned into pCR2.1 before being introduced into pKOR1. For the ΔcstB and Δsqr constructs the 5′ and 3′ regions were joined by overlapping PCR and transferred to pKOR1. All constructs were sequenced prior to use and the resulting deletion strains were confirmed to be hemolytic.
Preparation of S. aureus Complementation Strains
Individual S. aureus genes were subcloned into the pOS1 vector harboring the constitutive Plgt promoter (Bubeck Wardenburg et al., 2006) between the NdeI and BamHI restriction sites and sequenced using appropriate primers (Table S5). For CstB, a short sequence was looped into the multiple cloning site of pOS1, GTCTAGA, to utilize the Xho1 restriction site of pOS1 because an NdeI site resides in the cstB gene sequence. Each construct was electroporated into S. aureus RN4220 to obtain a properly methylated plasmid prior to electroporation into S. aureus strain Newman. WT and deletion strains were also transformed with empty vector. Plasmid DNA was maintained by addition of 10 μg mL−1 chloramphenicol (Cm) to plates and growth media.
S. aureus Growth Curves
5 mL fresh TSB broth was inoculated with S. aureus from frozen glycerol cell stocks and grown to saturation overnight (~14 h). A 1 mL aliquot was pelleted by centrifugation and resuspended in an equal volume TSB or HHWm minimal media (Toledo-Arana et al., 2005, Hussain et al., 1991, Grossoehme et al., 2011) and then diluted in 15 mL TSB or HHWm supplemented with either 0.50 mM thiosulfate or 0.25 mM cystine and the indicated stressor. All strains carried the pOS1 empty vector (WT) or the indicated allele and were grown in the presence of 10 μg mL−1 Cm with the exception of ΔtauE strain, which did not carry pOS1 and was grown in the absence of Cm. All cultures were grown aerobically at 37 °C in duplicate with shaking (200 rpm) in loosely-capped 50 mL Falcon tubes unless otherwise noted. Cell density was recorded hourly for 10 h by removal of 0.5 mL and measuring cell density (OD600) with a Spectronic® 20 Genesys® spectrophotometer (Thermo Scientific). The starting OD600 of each culture was ≈0.007.
S. aureus qRT-PCR Experiments
RNA extraction
For preparation of samples for RNA extraction, 15 mL cultures were grown to an OD600 of 0.2 in HHWm with the indicated sulfur source or in TSB as described above at which point the indicated stressor was added to the growth media. The stressors used in these experiments were 0.2 mM NaHS, 2.4 mM sodium hypochlorite (Chang et al., 2007) (ClO−), 10 mM sodium sulfite (SO32−), 0.2 mM sodium selenite (SeO32−), 0.5 μM nitric oxide (Hochgräfe et al., 2008) (NO) presented as the NO donor MAHMA NONOate (Hochgräfe et al., 2008) ([6-(2-hydroxy-1-methyl-2-nitrosohydrazino)-N-methyl-1-hexanamine]; Sigma Aldrich), 5 mM sodium nitrite (Schlag et al., 2007) (NO2−), 0.025 μM paraquat (Wolf et al., 2008), or 1 mM diamide (Wolf et al., 2008). At 10 or 30 min post addition of stressor to the culture, a 5 mL aliquot was removed and placed on ice until centrifugation at 4 °C (~2 min). Following centrifugation, the cell pellet was washed with PBS, centrifuged, and stored at −80°C. An analogous protocol was performed at 30 min post-induction. Pellets were thawed on ice and resuspended in 1 mL TriReagent (Cat. #TR-118, Molecular Research Center, Inc.). Cells were placed in tubes containing 0.1 mm silica beads (Lysing matrix B tubes, Cat. #6911-100, MP Biomedicals) and lysed in a bead beater (FastPrep®-24, MP Biomedicals). RNA was extracted by adding 200 μL of chloroform, mixing and centrifuging for 15 minutes at 16,100 x g. The aqueous layer was extracted and added to one volume of 70% ethanol. RNA purification was completed using the RNeasy minikit (Cat. #74104, Qiagen), including the on-column DNase I treatment (Cat. #79254, Qiagen). 5 μg total RNA was subsequently digested with the DNA-free™ kit (Cat. #AM1906, Ambion) and diluted five-fold. First-strand cDNA was synthesized using random hexamers (Quanta Biosciences) and the qScript Flex cDNA synthesis kit (Cat. #95049-100, Quanta Biosciences). Reactions without reverse transcriptase were also prepared to check for possible DNA contamination.
Quantitative Real Time PCR (qRT-PCR)
Reactions contained 10 μL 2x Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Cat. #600882, Agilent), 2 μL each of 2 μM QPCR primers (Table S5), 0.3 μL of 2 μM ROX reference dye and 6 μL diluted cDNA. Relative transcript amounts were measured using the MX3000P thermocycler (Stratagene) running the SYBR Green with dissociation curve program, and normalized to the amount of 16S rRNA. The thermal profile contained 1 cycle at 95°C for 3 min, 40 cycles at 95°C for 20 s to 59°C for 20 s. Subsequently, a dissociation curve starting at 55°C going to 95°C in 0.5°C increments with a dwell time of 30 s was performed to assess the specificity of the reactions. At least two, and typically three biologically independent samples were measured for each treatment, and the mean (±SD) values are reported. Transcript amounts were compared using two-way ANOVA with Bonferroni tests (GraphPad Prism, ver 5.0).
Cellular sulfur metabolite profiling of S. aureus
Cell growth and sulfide stress conditions
WT S. aureus complemented with empty pOS1 was grown overnight in 10 mL TSB with 10 μg mL−1 Cm. Cells were pelleted, washed by PBS and cultures were initiated at OD600 of ≈0.02 in HHWm medium with 10 μg mL−1 Cm and 0.5 mM TS. NaHS was added to the cultures at a final concentration of 0.2 mM when the OD600 reached ≈0.2. All cultures were grown in loosely-capped 50 mL Falcon tubes at 37 °C with shaking (200 rpm). Aliquots were removed at 0, 10, and 30 min post-addition of NaHS (equivalent to 15 mL of OD600=0.2 cells) and harvested by centrifugation at 3,000 rpm for 10 min with culture medium supernatant removed. Cell pellets were then washed with PBS, pelleted again by centrifugation at 13,200 rpm for 5 min and kept frozen at −80 °C until analysis by monobromobimane (mBBr) labeling.
Cellular thiol extraction, mBBr labeling and fluorescence-detected RP-HPLC analysis of sulfur-mB derivatives
The extraction and labeling procedure was slightly modified from previous work (Fahey & Newton, 1987, Newton et al., 1996). The total cell pellet obtained above was thawed, resuspended in 100 μL mBBr labeling buffer (20 mM Tris·HBr, pH 8.0, 50% acetonitrile, 1 mM mBBr, and 2 μM N-acetyl-L-cysteine (NAC)) and incubated at 60 °C for 1 h in the dark in a screw capped Eppendorf tube to avoid liquid loss by evaporation. The cellular debris and proteins were pelleted by centrifugation at 13,200 rpm for 5 min and the supernatant transferred to an Eppendorf tube containing 300 μL 10 mM methanesulfonic acid to terminate the labeling reaction. These samples were centrifuged at 13,200 rpm for 5 min through 0.2 μm centrifugal filter unit (Millipore, UFC30GVNB) to remove particulates and 40 μL samples were injected onto a Kinetex C18 reversed-phase column (Phenomenex, P/No. 00F-4601-E0, 4.6 mm×150 mm, 5 μm, 100 Å) outfitted with a Zorbax Eclipse Plus C18 guard column (Agilent, P.N. 820950-936, 4.6 mm×12.5 mm, 5 μm) and chromatographed on a Waters 600 high-performance liquid chromatography system equipped with a Waters 717 plus autosampler, a Waters 474 scanning fluorescence detector (λex=384 nm and λem=478 nm) and Empower chromatography software installed on a standard PC running Windows XP. Duplicate samples were typically analyzed using a methanol-based gradient system (Solvent A: 10% methanol, 0.25% acetic acid, pH 3.9; Solvent B: 90% methanol, 0.25% acetic acid, pH 3.9) with the elution protocol at 25 °C and a flow-rate of 1.2 mL min−1 as follows: 0–10 min, 0% B isocratic; 10–22 min, 0–24% B, linear gradient; 24–32 min, 24% B isocratic; 32–45 min, 24–45% B, linear gradient; 45–50 min, 45–82% B, linear gradient; 50–52 min, 82–100% B, linear gradient, followed by re-equilibration to 0% B. Authentic standards including sodium thiosulfate (TS), sodium sulfite, NaHS, L-cysteine, and N-acetyl-L-cysteine, were subjected to the same mBBr labeling protocol at 60 °C and chromatographed on a C18 reverse-phase LC column as described above (Fig. S6).
Supplementary Material
Acknowledgments
This work was supported by a grant from the US National Institutes of Health to D.P.G. (R01 GM097225) and E.P.S. (R01 AI069233) and will be submitted to the Indiana University Graduate School in partial fulfillments of the Ph.D. degree by J.L.L. and J.S. T.K.F. kindly acknowledges support from NIH grants F32 AI100480 and K22 AI104805. The authors gratefully acknowledge Drs. Jonathan Karty and Randy Arnold and Mrs. Angela Hanson in the Laboratory for Biological Mass Spectrometry at Indiana University of their assistance in acquiring high resolution mass fragmentation data and help with the data analysis.
Footnotes
The authors declare no conflict of interest in this work.
References
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Bailey TS, Zakharov LN, Pluth MD. Understanding hydrogen sulfide storage: probing conditions for sulfide release from hydrodisulfides. J Am Chem Soc. 2014;136:10573–10576. doi: 10.1021/ja505371z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud N, Kelso MJ, Rice SA, Kjelleberg S. Nitric oxide: A key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des. 2014 doi: 10.2174/1381612820666140905112822. in press. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci U S A. 2006;103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang FM, Coyne HJ, Ramirez CA, Fleischmann PV, Fang X, Ma Z, et al. Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR) J Biol Chem. 2014;289:19204–19217. doi: 10.1074/jbc.M114.556704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MW, Toghrol F, Bentley WE. Toxicogenomic response to chlorination includes induction of major virulence genes in Staphylococcus aureus. Environ Sci Technol. 2007;41:7570–7575. doi: 10.1021/es070929k. [DOI] [PubMed] [Google Scholar]
- Chiku T, Padovani D, Zhu W, Singh S, Vitvitsky V, Banerjee R. H2S biogenesis by human cystathionine γ-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J Biol Chem. 2009;284:11601–11612. doi: 10.1074/jbc.M808026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollone R, Ascenzi P, Visca P. Common themes and variations in the rhodanese superfamily. IUBMB Life. 2007;59:51–59. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- Czyzewski BK, Wang DN. Identification and characterization of a bacterial hydrosulphide ion channel. Nature. 2012;483:494–497. doi: 10.1038/nature10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JU, Radon C, Bühning M, Nimtz M, Leichert LI, Denis Y, et al. The sulfur carrier protein TusA has a pleiotropic role in Escherichia coli that also affects molybdenum cofactor biosynthesis. J Biol Chem. 2013;288:5426–5442. doi: 10.1074/jbc.M112.431569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JU, Urban A, Bolte A, Sriyabhaya P, Donahue JL, Nimtz M, et al. The identification of a novel protein involved in molybdenum cofactor biosynthesis in Escherichia coli. J Biol Chem. 2011;286:35801–35812. doi: 10.1074/jbc.M111.282368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debiemme-Chouvy C, Wartelle C, Sauvage FX. First evidence of the oxidation and regeneration of polysulfides at a GaAs electrode, under anodic conditions. A study by in situ UV–visible spectroelectrochemistry. J Phys Chem B. 2004;108:18291–18296. [Google Scholar]
- Derakhshan B, Wille PC, Gross SS. Unbiased identification of cysteine S-nitrosylation sites on proteins. Nat Protoc. 2007;2:1685–1691. doi: 10.1038/nprot.2007.210. [DOI] [PubMed] [Google Scholar]
- Dwarakanath S, Chaplin AK, Hough MA, Rigali S, Vijgenboom E, Worrall JA. Response to copper stress in Streptomyces lividans extends beyond genes under direct control of a copper-sensitive operon repressor protein (CsoR) J Biol Chem. 2012;287:17833–17847. doi: 10.1074/jbc.M112.352740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt M, Dux M, Namer B, Miljkovic J, Cordasic N, Will C, et al. H2S and NO cooperatively regulate vascular tone by activating a neuroendocrine HNO-TRPA1-CGRP signalling pathway. Nat Comm. 2014;5:4381. doi: 10.1038/ncomms5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey RC, Newton GL. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- Filipovic MR, Eberhardt M, Prokopovic V, Mijuskovic A, Orescanin-Dusic Z, Reeh P, et al. Beyond H2S and NO interplay: hydrogen sulfide and nitroprusside react directly to give nitroxyl (HNO). A new pharmacological source of HNO. J Med Chem. 2013;56:1499–1508. doi: 10.1021/jm3012036. [DOI] [PubMed] [Google Scholar]
- Filipovic MR, Miljkovic J, Nauser T, Royzen M, Klos K, Shubina T, et al. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. J Am Chem Soc. 2012;134:12016–12027. doi: 10.1021/ja3009693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Tsui HC, Bruce KE, Sham LT, Higgins KA, Lisher JP, et al. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat Chem Biol. 2013;9:177–183. doi: 10.1038/nchembio.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs S, Zuhlke D, Pane-Farre J, Kusch H, Wolf C, Reiss S, et al. Aureolib - a proteome signature library: towards an understanding of Staphylococcus aureus pathophysiology. PLoS One. 2013;8:e70669. doi: 10.1371/journal.pone.0070669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner R, Palinkas Z, Basell K, Becher D, Antelmann H, Nagy P, et al. Polysulfides link H2S to protein thiol oxidation. Antioxid Redox Signal. 2013;19:1749–1765. doi: 10.1089/ars.2012.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N, Kehl-Fie TE, Ma Z, Adams KW, Cowart DM, Scott RA, et al. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J Biol Chem. 2011;286:13522–13531. doi: 10.1074/jbc.M111.220012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, et al. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J Biol Chem. 2011;286:26148–26157. doi: 10.1074/jbc.M111.234039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins KA, Giedroc D. Insights into protein allostery in the CsoR/RcnR Family of transcriptional repressors. Chem Lett. 2014;43:20–25. doi: 10.1246/cl.130965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM, Di Meo I, Zeviani M, Viscomi C, Braun HP. Proteome adaptations in Ethe1-deficient mice indicate a role in lipid catabolism and cytoskeleton organization via post-translational protein modifications. Bioscience Rep. 2013;33:e00052. doi: 10.1042/BSR20130051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008;275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- Hochgräfe F, Wolf C, Fuchs S, Liebeke M, Lalk M, Engelmann S, et al. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2008;190:4997–5008. doi: 10.1128/JB.01846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdorf MM, Bennett B, Crowder MW, Markaroff CA. Spectroscopic studies on Arabidopsis ETHE1, a glyoxalase II-like protein. J Inorg Biochem. 2008;102:1825–1830. doi: 10.1016/j.jinorgbio.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M, Hastings JGM, White PJ. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol. 1991;34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A. 2014;111:7606–7611. doi: 10.1073/pnas.1321232111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multible sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Molec Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Kabil O, Banerjee R. Redox biochemistry of hydrogen sulfide. J Biol Chem. 2010;285:21903–21907. doi: 10.1074/jbc.R110.128363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabil O, Motl N, Banerjee R. HS and its role in redox signaling. Biochim Biophys Acta. 2014;1844:1355–1366. doi: 10.1016/j.bbapap.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Chambers LA, Trudinger PA. Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal Chem. 1969;41:898–901. [Google Scholar]
- Kimura H. Hydrogen Sulfide: From Brain to Gut. Antioxid Redox Signal. 2010;12:1111–1123. doi: 10.1089/ars.2009.2919. [DOI] [PubMed] [Google Scholar]
- Kolluru GK, Shen X, Bir SC, Kevil CG. Hydrogen sulfide chemical biology: Pathophysiological roles and detection. Nitric Oxide. 2013;35:5–20. doi: 10.1016/j.niox.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- Luebke JL, Arnold RJ, Giedroc DP. Selenite and tellurite form mixed seleno- and tellurotrisulfides with CstR from Staphylococcus aureus. Metallomics. 2013;5:335–342. doi: 10.1039/c3mt20205d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Cowart DM, Scott RA, Giedroc DP. Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry. 2009a;48:3325–3334. doi: 10.1021/bi900115w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Cowart DM, Ward BP, Arnold RJ, DiMarchi RD, Zhang L, et al. Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J Am Chem Soc. 2009b;131:18044–18045. doi: 10.1021/ja908372b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci U S A. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EG. Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, Aharonowitz Y, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Carroll KS. Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem Biol. 2013;8:1110–1116. doi: 10.1021/cb4001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. H2S signalling through protein sulfhydration and beyond. Nat Rev Mol Cell Biol. 2012;13:499–507. doi: 10.1038/nrm3391. [DOI] [PubMed] [Google Scholar]
- Pother DC, Liebeke M, Hochgrafe F, Antelmann H, Becher D, Lalk M, et al. Diamide triggers mainly S-thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191:7520–7530. doi: 10.1128/JB.00937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Caballero H, Campanello GC, Giedroc DP. Metalloregulatory proteins: metal selectivity and allosteric switching. Biophys Chem. 2011;156:103–114. doi: 10.1016/j.bpc.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag S, Nerz C, Birkenstock TA, Altenberend F, Götz F. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol. 2007;189:7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth D, Hausladen A, Wang YJ, Stamler JS. Endogenous protein S-Nitrosylation in E. coli: regulation by OxyR. Science. 2012;336:470–473. doi: 10.1126/science.1215643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- Singh S, Padovani D, Leslie RA, Chiku T, Banerjee R. Relative contributions of cystathionine β-synthase and γ-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J Biol Chem. 2009;284:22457–22466. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina O, Poupel O, Coppee JY, Danchin A, Msadek T, Martin-Verstraete I. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulphur source utilization and plays a role in biofilm formation. Mol Microbiol. 2009;73:194–211. doi: 10.1111/j.1365-2958.2009.06760.x. [DOI] [PubMed] [Google Scholar]
- Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, Gilles AM, et al. The CymR regulator in complex with the enzyme CysK controls cysteine metabolism in Bacillus subtilis. J Biol Chem. 2008;283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- Tiranti V, Viscomi C, Hildebrandt T, Di Meo I, Mineri R, Tiveron C, et al. Loss of ETHE1, a mitochondrial dioxygenase, causes fatal sulfide toxicity in ethylmalonic encephalopathy. Nat Med. 2009;15:200–205. doi: 10.1038/nm.1907. [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, Lasa I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol. 2005;187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tottey S, Patterson CJ, Banci L, Bertini I, Felli IC, Pavelkova A, et al. Cyanobacterial metallochaperone inhibits deleterious side reactions of copper. Proc Natl Acad Sci U S A. 2012;109:95–100. doi: 10.1073/pnas.1117515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C, Hochgräfe F, Kusch H, Albrecht D, Hecker M, Engelmann S. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: Diverse responses to different oxidants. Proteomics. 2008;8:3139–3153. doi: 10.1002/pmic.200701062. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Brose J, Schimo S, Ackland SM, La Fontaine S, Wedd AG. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: detection probes and affinity standards. J Biol Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Macinkovic I, Devarie-Baez NO, Pan J, Park CM, Carroll KS, et al. Detection of protein S-sulfhydration by a tag-switch technique. Angew Chem Int Ed Engl. 2014;53:575–581. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.