Abstract
While great strides have been made in the improvement of outcome for newly diagnosed pediatric acute lymphoblastic leukemia (ALL) due to refinements in risk stratification and selective intensification of therapy, the prognosis for relapsed leukemia has lagged behind significantly. Understanding the underlying biological pathways responsible for drug resistance is essential to develop novel approaches for the prevention of recurrence and treatment of relapsed disease. High throughput genomic technologies have the potential to revolutionize cancer care in this era of personalized medicine. Using such advanced technologies, we and others have shown that a diverse assortment of cooperative genetic and epigenetic events drive the resistant phenotype. Herein, we summarize results using a variety of genomic technologies to highlight the power of this methodology in providing insight into the biological mechanisms that impart resistant disease.
Keywords: Relapsed leukemia, drug resistance, childhood ALL
Introduction
Despite vast improvements in the treatment of childhood acute lymphoblastic leukemia (ALL) over past few decades, the outlook for relapsed leukemia remains poor, indicating a need for innovative treatment approaches. A characteristic feature of refractory and recurrent disease is intrinsic “drug resistance.” This feature is manifested by in vitro studies demonstrating high levels of resistance to the commonly used chemotherapy agents compared to blasts harvested at diagnosis. Clinical manifestations include lower remission reinduction rates and the persistence of minimal residual disease (MRD), despite more intensive treatment1,2.
We and others have relied on the power of studying matched diagnosis-relapse patient pairs to identify the key cellular pathways accounting for chemoresistance. These approaches have been applied to cohorts of ALL cases enrolled on therapeutic trials from various cooperative groups worldwide and have led to the discovery of new biomarkers that predict a higher likelihood of relapse and importantly, the discovery of pathways that can serve as a target for novel therapeutic interventions. In this review, we discuss the advancements in the current understanding of the genetic landscape of relapsed leukemia using high-resolution genome technologies. With these methodologies, investigators can discover DNA sequence alterations of specific genes in the tumor cells at a very low level, and can also study changes in the “epigenome” which influence the gene transcription without affecting the DNA sequence by itself. Together, these genetic and epigenetic modifications govern the transcriptional regulation of leukemic cells leading to highly aggressive and resistant disease.
Gene Expression Microarrays
Gene expression profiling of leukemic blasts in matched diagnosis-relapse patient pairs has revealed a common gene signature reflective of relapse, marked by genes involved in proliferation and cell cycle regulation (DUSP6, UBE2V1, F2R), apoptosis (BIRC5, HRK), DNA repair (PTTG1, UBE2V1), and drug resistance (TYMS, RAB5C)3,4. Furthermore relapsed blasts may be locked into a more stem-cell-like transcriptional program due to over expression of genes such as HMGA15,6. While relapsed blasts from patients who relapse early (e.g. < 36 months from initial diagnosis) may show changes in common biological pathways, the expression of distinct genes within these pathways are altered3.
Importantly, information gained from gene expression microarrays can be used therapeutically. As an example, survivin (BIRC5), which is a member of the inhibitor of apoptosis (IAP) family regulating cell division and inhibiting caspase function7,8, has been found to be consistently upregulated in the majority of relapse samples, and is an attractive therapeutic target. Studies in ALL cell lines and xenograft models have shown its role in chemoresistance9. Based on these preclinical studies and preliminary results from a phase 1 trial in adults with relapsed or refractory solid tumors and lymphoma10, a clinical trial was designed in children with second or greater bone marrow relapses using a survivin anti-sense oligonucleotide in combination with conventional chemotherapy. Unfortunately patients experienced toxicity, which precluded further clinical development11. Nonetheless, survivin remains an attractive target.
Theoretically, the gene expression signature can be targeted by seeking agents that “reverse” the signature (e.g. relapse to diagnosis) to restore chemosensitivity. Recently, the connectivity map concept12 was applied to identify drugs, which could reverse the genetic signature suggestive of drug resistant relapse13. Of note, the histone deacetylase inhibitor, vorinostat, was the top agent in this search, indicating the potential to reverse the relapse-specific gene expression profile and potentially endow chemosensitivity13. Histone acetyltransferases acetylate lysines on the tails of histones thereby making DNA more accessible or “open” to transcription factors. Deacetylases thus result in closed chromatin structure. In this scenario vorinostat would maintain genes in an active chromatin state. In fact, vorinostat not only “flipped” the gene expression signature characteristic of relapse in cell lines and patient samples14, but also showed a synergistic effect when used as pretreatment followed by traditional chemotherapy13.
DNA Methylation arrays
Given that epigenetic mechanisms play an essential role in the emergence of relapsed disease, exploring the DNA methylation profiles of leukemic cells is a logical approach. Methylation of DNA usually at CpG islands can result in suppression of transcription. For example, methylation of tumor suppressor genes is involved in the genesis of many human cancers15. Genome wide DNA methylation profiling performed on 33 matched pairs demonstrated that the relapsed genome was distinctly more hypermethylated as compared to those at diagnosis14. In this study, overall 867 CpG sites were differentially methylated between diagnosis and relapse in more than 30% of cases14. Furthermore, concordant down-regulation of mRNA expression was observed for a subset of differentially methylated genes namely WT1, APC, CDKN2A, PTPRO, GATA4, HOXA9 among many others14. In a subsequent study, re-expression of these hypermethylated and down regulated genes was observed with the use of demethylating agent, decitabine, which is a DNA methyltransferase inhibitor (DNMTi)13. Moreover, enhanced chemosensitivity was observed when ALL cell lines and primary patient samples were pretreated with decitabine followed by conventional cytotoxic chemotherapy13. This work and changes in the gene expression signature with HDAC inhibitors described above highlights the role of cooperating epigenetic mechanisms in regulating the aberrant transcription responsible for relapsed disease and has led to a clinical trial in order to determine the therapeutic potential of this approach in relapsed ALL (http://Clinicaltrials.gov:NCT01483690)13,16. DNA methylation studies have also been used for subtype classification of newly diagnosed ALL17,18 and to predict outcome and risk of relapse18, however, further studies are needed in an expanded cohort of patients to accurately identify the biomarkers of relapse.
Copy Number Abnormalities
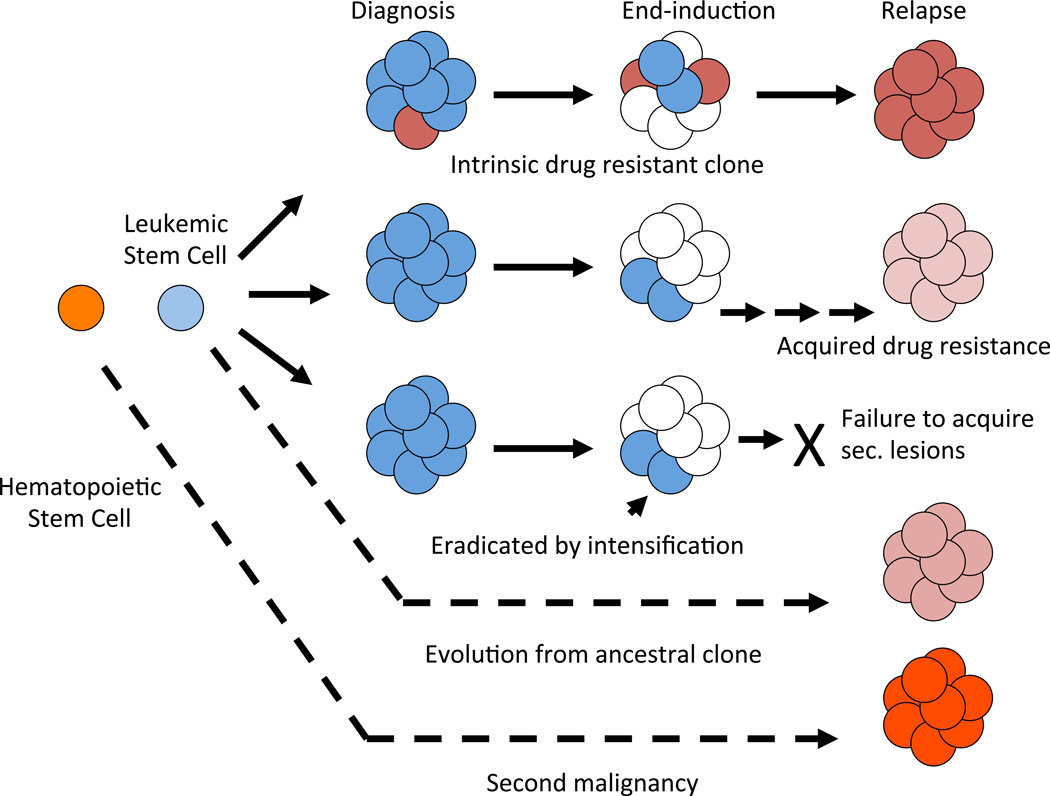
It has long been known that leukemic blasts undergo cytogenetic19 and immunophenotypic drift20,21 from diagnosis to relapse. However sentinel genetic lesions are preserved indicating that relapse cells are clonally related to the original leukemia with very rare exceptions. Over the past decade, genome wide DNA copy number profiling studies have been utilized to detect minor deletions and amplifications more precisely. Mullighan and colleagues studied 61 diagnosis relapse pairs, in which majority of samples were of B-lineage, and elegantly demonstrated the clonal relationship of diagnosis and relapse genome22. This study revealed that in about a third of cases, relapse blasts shared the majority of genetic lesions with leukemic population predominant at the initial disease presentation, while in another half of cases, back-tracking of the specific genomic alterations suggested that the relapse leukemia cells resembled “pre-leukemic” or an ancestral clone that was present as a minor subpopulation at the time of diagnosis and emerged during ALL therapy (Figure 1). In only a small percentage of cases relapse cells did not share any lesions with diagnostic blasts, suggesting a secondary leukemia22. Similar patterns of clonal evolution have been described in specific ALL subtypes such as ETV6-RUNX1 fusion positive leukemia23,24 and cases with high hyperdiploidy25.
Figure 1. Model of evolution of relapsed disease.
A preleukemic stem cell (LSC, light blue) gives rise to frank ALL (dark blue cells). Intrinsically drug resistant clones (red) may be present at diagnosis at low levels. However, LSCs and other subclones may survive initial treatment and acquire additional lesions that result in acquired drug resistance. Rarely, relapse clone is genetically distinct from that at diagnosis (second malignancy).
Likewise, a study of 20 trios (diagnosis, relapse and germline samples from individual patients) demonstrated somatic copy number alterations of CDKN2A/B, PAX5, IKZF1 and EBF1, genes involved in B-cell development, differentiation and lineage commitment26. Specific deletions were enriched in this cohort of relapse cases compared to studies performed in patients at diagnosis (Table 1). Such deletions may be predicted to be biomarkers of relapse and that is indeed the case, as deletions in IKZF1, a gene encoding the lymphoid transcription factor IKAROS has been associated with poor outcome and is a strong predictor of relapse27,28. Moreover, recurrent relapse-specific deletions were found in MSH6, a gene implicated in DNA mismatch repair, which has been linked with 6-mercaptopurine and steroid resistance14,26. Overall, these data suggest that recurring genomic alterations play an essential role in driving drug resistance at relapse. Future refinements in existing risk and treatment algorithms can be envisioned upon further investigation and validation of these potential targets.
Table 1.
Comparison of selected CNA events (copy number losses) from reported literature (adapted from Hogan et al, Blood 2011)14
| Gene | Yang et al, Blood 200826 and Hogan et al. Blood, 201114 n=76 |
Mullighan et al. Science, 200822 n=47 |
Mullighan et al. Nature, 200729 n=192 |
|---|---|---|---|
| CDKN2A/B | 42.1% | 36.20% | 33.90% |
| IKZF1 | 38.2% | 25.50% | 8.90% |
| VPREB1 | 35.5% | N/A | N/A |
| PAX5 | 28.9% | 25.50% | 29.70% |
| EBF1 | 15.8% | 4.20% | 4.20% |
| MSH6 | 6.6% | None | None |
| TBL1XR1 | 9.2% | 10.60% | 3.60% |
| BTG1 | 15.8% | 2.10% | 6.70% |
| NR3C1 | 3.9% | 8.50% | 4.70% |
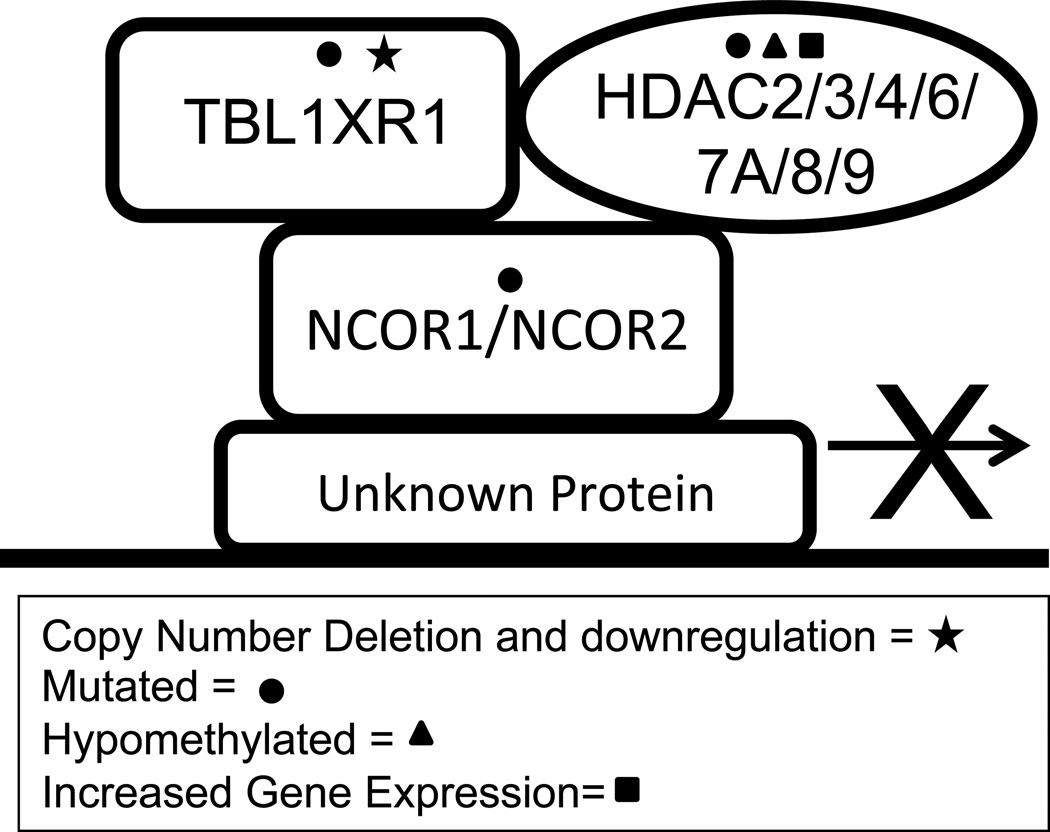
Glucocorticoid (GC) resistance is a hallmark of relapsed blasts. Enrichment of deletions in BTG1/BTG2, NR3C1 and TBL1XR1, genes involved in glucocorticoid signaling, have been observed in relapse samples in various studies14,22,26,29. These deletions have all been shown to be functionally relevant in steroid resistance30–32. While deletions in NR3C1, the glucocorticoid receptor, are rare, deletions in BTG1 and TBL1XR1 have been observed in 10–14% of patients at relapse. BTG1 functions as a co-activator of the glucocorticoid receptor30 while TBL1XR1 operates to degrade the inhibitory nuclear co-repressor complex (NCoR)33,34 residing at GC responsive elements32. Importantly, BTG1 and TBL1XR1 deletions are mutually exclusive indicating a single deletion in a gene involved in GC signaling is enough to result in steroid resistance in patients14. In addition to TBL1XR1, proteins that make up the nuclear receptor co-repressor complex (NCoR) including NCoR1 and HDACs have also been found to be altered in ALL patients at relapse14,35–37. Thus one unifying model of GC resistance is that mutations and/or deletions converge on the transcriptional complex that operates to activate or repress GC target genes (Figure 2). We have shown that knockdown of TBL1XR1 results in steroid resistance in ALL cell lines by altering the activity of the NCoR complex32. This data suggests that altering many of the components of the NCoR complex may result in steroid resistance making this a potentially interesting complex to target therapeutically in recurrent disease.
Figure 2. NCoR complex mutations identified in relapsed ALL.
Schematic illustrating mutations, copy number abnormalities, differential methylation, and altered gene expression in protein components of the NCoR complex. Only those NCoR complex proteins that have been found to be altered at relapse are shown here.
The role of host (patient) in response to treatment due to inherited genetic variations is now known to play an integral role in assessing the risk of relapse38. In an extensive cohort of 2535 children with ALL, 14 of the 134 relapse SNPs were associated with unfavorable pharmacokinetics of dexamethasone, methotrexate and asparaginase, suggesting that host disposition of antileukemic drugs affects the risk of relapse39. More recently Perez-Andreu and colleagues have discovered that inherited genetic variation at the GATA3 locus is associated with an increased risk of developing a high risk subtype of ALL with an unfavorable response to therapy40. Early identification of such patients and allocating them into intensified regimen may improve their outcome.
Next Generation Sequencing
While the earlier generation genomic technologies using array based strategies to determine changes in gene expression, copy number and methylation have provided helpful insight into molecular evolution of relapse, next generation sequencing techniques provide precision in mapping small deletions, insertions and single base changes.
Targeted sequencing performed by PCR and capillary resequencing of 300 genes in 23 matched diagnosis-relapse patient pairs with B-lymphoblastic leukemia identified novel mutations in CREBBP, a gene encoding CREB binding protein with histone acetyltransferase (HAT) activity. In an extended cohort of samples 18.3% of relapse cases had sequence or deletion mutations of CREBBP. Less commonly mutations in other important epigenetic regulators were seen such as NCoR1, Nuclear corepressor complex, EP300, a paralog of CREBBP, EZH2, the histone methyltransferase gene, and CTCF, a zinc finger protein involved in histone modification35. Additionally, transcriptome sequencing has identified relapse specific mutations in other epigenetic regulatory genes such as CBX3, a gene encoding heterochromatin protein, PRMT2, a gene encoding protein arginine methyltransferase 2 and MIER3, a gene involved in chromatin binding; providing further evidence of aberrant epigenetic mechanisms that play a role at relapse41.
Furthermore, recurrent somatic mutations were identified in the Ras pathway genes (NRAS, KRAS, NF1 and PTPN11), transcription factor genes (ETV6, PAX5), the tumor suppressor gene TP53, the ubiquitin ligase gene FBXW7 and the Janus Kinase genes JAK135. The discovery of JAK mutations has led to an increased interest in targeting the JAK-STAT pathway in ALL as well as other cooperating pathways42,43.
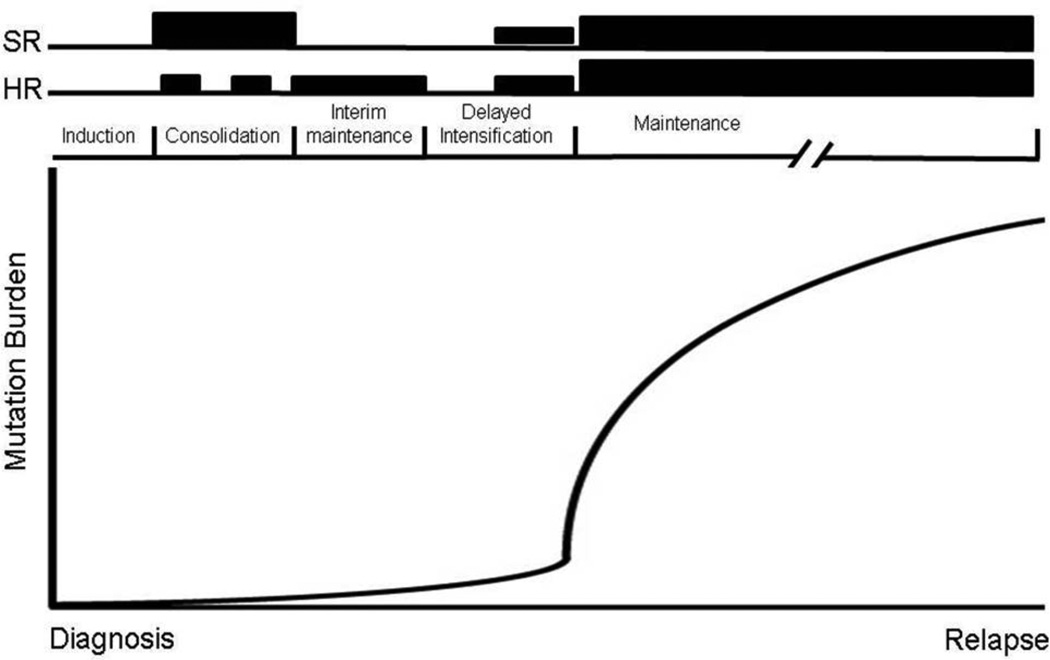
Recently, two independent groups41,44 have discovered relapse-specific mutations in NT5C2, a gene encoding 5’ nucleotidase enzyme, which is responsible for maintaining intracellular nucleotide pools. These mutations are gain-of-function mutations resulting in enhanced enzymatic activity of this enzyme, leading to inactivation of cytotoxic metabolites of the purine analogs 6-thioguanine and 6-mercaptopurine. Of note, in vitro cytotoxic assays demonstrated remarkable resistance limited to purine nucleoside analogs, while there was no effect on other commonly used chemotherapy agents41,44. NT5C2 mutations were identified exclusively in patients who relapsed early leading to the hypothesis that such mutations may be present in a very small subclone at diagnosis and that outgrowth occurs when 6-MP becomes the major component of treatment during maintenance therapy (Figure 3). Indeed targeted, high-density sequencing indicates that at least in some patients these clones are present in 0.01% of cells from the time of diagnosis. Lastly, the finding that NT5C2 mutations were discovered in both B and T ALL, two distinctly different molecular subtypes of ALL, mimics convergent evolution that occurs in the natural world. Different species can develop identical phenotypic traits if similar selective pressures (e.g. chemotherapy) are applied.
Figure 3. Schematic representation of purine nucleoside analogue therapy during ALL treatment.
SR (standard risk) and HR (high risk) back bones are shown above with height of the bars representing relative amounts of 6MP/6TG used during treatment cycles. The solid line represents theoretical outgrowth of NT5C2 mutant clones during maintenance therapy leading to relapse disease.
Furthermore, whole exome sequencing in 20 pairs from patients with high risk B-ALL has confirmed the high prevalence of relapse specific mutations in NT5C2 (35%) but also the histone acetyltransferase CREBBP (10%)45. Interestingly mutations in WHSC1, USH2A and NT5C1B (also involved in purine metabolism) were also enriched at relapse. The NT5C2 and WHSC1 mutations were mutually exclusive with a combined frequency of 55%. Most of the WHSC1 mutations occurred in a single amino acid (E1099K) that results in increased activity of this histone methyltransferase46. In addition, examining an extended cohort of diagnosis-relapse pairs from patients with both standard risk and high risk ALL also confirmed the prevalence of NT5C2 and WHSC1 E1099K mutations and mutual exclusivity (10 and 7% respectively)41 (Meyer, unpublished). Overall mutations in five pathways are enriched at relapse: Ras signaling, histone modification, purine metabolism, tyrosine kinase signaling and B cell development45.
Integrated Genomic Analysis
It is clear that relapsed leukemia is a heterogeneous disease and that distinct genetic/epigenetic alterations may be unique to small subgroups of patients. However, information gathered from the various genomic platforms can provide valuable insights into the unifying pathways, which are implicated in driving relapse. Hogan and colleagues have identified WNT/β-catenin and mitogen activated protein kinase (MAPK) pathways to be upregulated at relapse by integrating the data obtained from gene expression, SNP and methylation arrays14. In this study, several negative regulators of the WNT pathway were down-regulated and/or hypermethylated and/or had a copy number loss, while the downstream WNT target genes such as BIRC5 and CCND1 were upregulated at relapse14. Preclinical studies using specific inhibitors to interrupt the WNT pathway in ALL cell lines, patient samples and xenografts are underway and may guide therapeutic options for the treatment of resistant disease47.
Conclusions and Future Directions
In summary, studies to date indicate that: 1) relapsed blasts utilize a variety of biological pathways to evade therapy, 2) individual alterations in different patients may converge on similar pathways (e.g. the glucocorticoid receptor transcriptional complex, Wnt pathway, MAPK pathway etc.), 3) in many cases mutations exist in a small subclone at diagnosis and such clones become dominant under the selective pressure of chemotherapy, 4) different biological subtypes of ALL (and possibly other hematological malignancies) may utilize common pathways to evade therapy if similar chemotherapeutic agents are part of the treatment, 5) inherited genetic susceptibility influence the treatment outcome by affecting host metabolism of chemotherapeutic drugs and susceptibility to high risk forms of ALL and, 6) relapse specific pathways can be targeted to reverse the intrinsic drug resistance characteristic of relapse.
While many efforts have been made to characterize the genomic landscape of pediatric relapsed ALL there is still a lot left to be learned. Further integration of genomic and epigenetic platforms will be essential to continue to identify genetic alterations that could be potential drug targets or biomarkers. While the genomic studies on patient samples accurately assess the dynamic molecular events, it still remains difficult to discern “drivers” from those of “passenger” lesions. Recent advances in genomic technologies have progressed at a rapid pace, accompanied by plunging costs, allowing investigators to address wide range of biologically important questions. Currently, a genome wide short hairpin RNA (shRNA) screening approach is being deployed to determine various genetic pathways that could sensitize leukemic blasts to conventional cytotoxic chemotherapeutic agents. By performing functional studies utilizing the strengths of unbiased shRNA screens in conjunction with data from genomic platforms, our ability to identify potential targets for drug development will be greatly enhanced. Finally exploration of epigenetic mechanisms using genome-wide mapping of the key histones marks essential in regulating transcription by chromatin immunoprecipitation sequencing would allow us the discovery of key genes and pathways that are primarily epigenetically driven in relapsed disease. All of this information now provides a framework for the development of biologically-driven clinical trials to prevent and treat relapsed disease.
Acknowledgments
Acknowledgement of sources of support: This work was supported by the National Institute of Health (NIH) 5 R01 CA 140729-04 (WLC), the Ira Sohn Conference Foundation grant (TB), NIH Training in Pharmacological Sciences T32 GM066704 (CLJ), the American Society of Hematology Research Training Awards for fellows (NAV), the Silber Pediatric Leukemia Fund and “Jackson Takes Action” Fund.
Footnotes
The authors declare no conflict of interest.
References
- 1.Klumper E, et al. In vitro cellular drug resistance in children with relapsed/refractory acute lymphoblastic leukemia. Blood. 1995;86:3861–3868. [PubMed] [Google Scholar]
- 2.Hongo T, Fujii Y. In vitro chemosensitivity of lymphoblasts at relapse in childhood leukemia using the MTT assay. International journal of hematology. 1991;54:219–230. [PubMed] [Google Scholar]
- 3.Bhojwani D, et al. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children's Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beesley AH, et al. The gene expression signature of relapse in paediatric acute lymphoblastic leukaemia: implications for mechanisms of therapy failure. British journal of haematology. 2005;131:447–456. doi: 10.1111/j.1365-2141.2005.05785.x. [DOI] [PubMed] [Google Scholar]
- 5.Staal FJ, et al. DNA microarrays for comparison of gene expression profiles between diagnosis and relapse in precursor-B acute lymphoblastic leukemia: choice of technique and purification influence the identification of potential diagnostic markers. Leukemia. 2003;17:1324–1332. doi: 10.1038/sj.leu.2402974. [DOI] [PubMed] [Google Scholar]
- 6.Roy S, et al. HMGA1 overexpression correlates with relapse in childhood B-lineage acute lymphoblastic leukemia. Leukemia & lymphoma. 2013;54:2565–2567. doi: 10.3109/10428194.2013.782610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knauer SK, et al. The survivin isoform surviving-3B is cytoprotective and can function as a chromosomal passenger complex protein. Cell Cycle. 2007;6:1502–1509. [PubMed] [Google Scholar]
- 8.Pennati M, Folini M, Zaffaroni N. Targeting survivin in cancer therapy: fulfilled promises and open questions. Carcinogenesis. 2007;28:1133–1139. doi: 10.1093/carcin/bgm047. [DOI] [PubMed] [Google Scholar]
- 9.Morrison DJ, et al. Endogenous knockdown of survivin improves chemotherapeutic response in ALL models. Leukemia. 2012;26:271–279. doi: 10.1038/leu.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolcher AW, et al. Results of a phase 1, open-label, dose-escalation study evaluating the safety and tolerability of EZN-3042, a survivin mRNA antagonist, administered with or without docetaxel in adult patietns with advanced solid tumors or lymphoma; Proceedings of the 102nd Annual Meeting of AACR, LB-409 (abstract); 2011. p. 409. [Google Scholar]
- 11.Raetz EA, et al. A Phase I Study of EZN-3042, a Novel Survivin Messenger Ribonucleic Acid (mRNA) Antagonist, Administered in Combination With Chemotherapy in Children With Relapsed Acute Lymphoblastic Leukemia (ALL): A Report From the Therapeutic Advances in Childhood Leukemia and Lymphoma (TACL) Consortium. Journal of Pediatric Hematology/Oncology. doi: 10.1097/MPH.0b013e3182a8f58f. Publish Ahead of Print, 10.1097/MPH.1090b1013e3182a1098f1058f (9000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 13.Bhatla T, et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood. 2012;119:5201–5210. doi: 10.1182/blood-2012-01-401687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan LE, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118:5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 16.Bachmann PS, et al. Epigenetic silencing of BIM in glucocorticoid poor-responsive pediatric acute lymphoblastic leukemia, and its reversal by histone deacetylase inhibition. Blood. 2010;116:3013–3022. doi: 10.1182/blood-2010-05-284968. [DOI] [PubMed] [Google Scholar]
- 17.Figueroa ME, et al. Integrated genetic and epigenetic analysis of childhood acute lymphoblastic leukemia. The Journal of clinical investigation. 2013;123:3099–3111. doi: 10.1172/JCI66203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milani L, et al. DNA methylation for subtype classification and prediction of treatment outcome in patients with childhood acute lymphoblastic leukemia. Blood. 2010;115:1214–1225. doi: 10.1182/blood-2009-04-214668. [DOI] [PubMed] [Google Scholar]
- 19.Raimondi SC, Pui CH, Head DR, Rivera GK, Behm FG. Cytogenetically different leukemic clones at relapse of childhood acute lymphoblastic leukemia. Blood. 1993;82:576–580. [PubMed] [Google Scholar]
- 20.van Wering ER, et al. Immunophenotypic changes between diagnosis and relapse in childhood acute lymphoblastic leukemia. Leukemia. 1995;9:1523–1533. [PubMed] [Google Scholar]
- 21.Pui CH, et al. Shifts in blast cell phenotype and karyotype at relapse of childhood lymphoblastic leukemia. Blood. 1986;68:1306–1310. [PubMed] [Google Scholar]
- 22.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuster L, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood. 2011;117:2658–2667. doi: 10.1182/blood-2010-03-275347. [DOI] [PubMed] [Google Scholar]
- 24.van Delft FW, et al. Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood. 2011;117:6247–6254. doi: 10.1182/blood-2010-10-314674. [DOI] [PubMed] [Google Scholar]
- 25.Davidsson J, et al. Relapsed childhood high hyperdiploid acute lymphoblastic leukemia: presence of preleukemic ancestral clones and the secondary nature of microdeletions and RTK-RAS mutations. Leukemia. 2010;24:924–931. doi: 10.1038/leu.2010.39. [DOI] [PubMed] [Google Scholar]
- 26.Yang JJ, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112:4178–4183. doi: 10.1182/blood-2008-06-165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. The New England journal of medicine. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuiper RP, et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24:1258–1264. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- 29.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 30.van Galen JC, et al. BTG1 regulates glucocorticoid receptor autoinduction in acute lymphoblastic leukemia. Blood. 2010;115:4810–4819. doi: 10.1182/blood-2009-05-223081. [DOI] [PubMed] [Google Scholar]
- 31.Irving JA, Minto L, Bailey S, Hall AG. Loss of heterozygosity and somatic mutations of the glucocorticoid receptor gene are rarely found at relapse in pediatric acute lymphoblastic leukemia but may occur in a subpopulation early in the disease course. Cancer research. 2005;65:9712–9718. doi: 10.1158/0008-5472.CAN-05-1227. [DOI] [PubMed] [Google Scholar]
- 32.Jones C. Deletions In TBL1XR1 Results In Glucocorticoid Resistance By Decreasing Glucocorticoid Signaling In Childhood B-Lymphoblastic Leukemia. American Society of Hematology Annual meeting abstract; New Orleans, LA. 2013. [Google Scholar]
- 33.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 34.Perissi V, et al. TBL1 and TBLR1 phosphorylation on regulated gene promoters overcomes dual CtBP and NCoR/SMRT transcriptional repression checkpoints. Molecular cell. 2008;29:755–766. doi: 10.1016/j.molcel.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruhn B, et al. The expression of histone deacetylase 4 is associated with prednisone poor-response in childhood acute lymphoblastic leukemia. Leukemia research. 2013;37:1200–1207. doi: 10.1016/j.leukres.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 37.Moreno DA, et al. Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. British journal of haematology. 2010;150:665–673. doi: 10.1111/j.1365-2141.2010.08301.x. [DOI] [PubMed] [Google Scholar]
- 38.Yang JJ, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA : the journal of the American Medical Association. 2009;301:393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang JJ, et al. Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood. 2012;120:4197–4204. doi: 10.1182/blood-2012-07-440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Andreu V, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nature genetics. 2013;45:1494–1498. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer JA, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nature genetics. 2013;45:290–294. doi: 10.1038/ng.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maude SL, et al. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaFave LM, Levine RL. JAK2 the future: therapeutic strategies for JAK-dependent malignancies. Trends in Pharmacological Sciences. 2012;33:574–582. doi: 10.1016/j.tips.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Tzoneva G, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nature medicine. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J. Comparison Of Mutational Profiles Of Diagnosis and Relapsed Pediatric B-Acute Lymphoblastic Leukemia: A Report From The COG ALL Target Project. in. American Soceity of Hematology meeting abstract; New Orleans, LA. 2013. [Google Scholar]
- 46.Oyer JA, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2013 doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dandekar S. Inhibition Of The Wnt Pathway Leads To Improved Chemosensitivity In Pediatric Acute Lymphoblastic Leukemia. American Soceity of Hematology annual meeting abstract; New Orleans, LA. 2013. [Google Scholar]