Abstract
Expression of the nuclear receptor interacting factor 3 (NRIF3) coregulator in a wide variety of breast cancer cells selectively leads to rapid caspase-2–dependent apoptotic cell death. A novel death domain (DD1) was mapped to a 30– amino acid region of NRIF3. Because the cytotoxicity of NRIF3 and DD1 seems to be cell type–specific, these studies suggest that breast cancer cells contain a novel “death switch” that can be specifically modulated by NRIF3 or DD1. Using an MCF-7 cell cDNA library in a yeast two-hybrid screen, we cloned a factor that mediates apoptosis by DD1 and refer to this factor as DD1-interacting factor-1 (DIF-1). DIF-1 is a transcriptional repressor that mediates its effect through SirT1, and this repression is attenuated by the binding of NRIF3/DD1. DIF-1 expression rescues breast cancer cells from NRIF3/DD1-induced apoptosis. Small interfering RNA (siRNA) knockdown of DIF-1 selectively leads to apoptosis of breast cancer cells, further suggesting that DIF-1 plays a key role in NRIF3/DD1-mediated apoptosis. A protein kinase A inhibitor (H89) also elicits apoptosis of breast cancer cells but not of the other cell types examined, and DIF-1 also protects these cells from H89-mediated apoptosis. In addition, H89 incubation results in a rapid increase in NRIF3 levels and siRNA knockdown of NRIF3 protects breast cancer cells from H89-mediated apoptosis. Our results indicate that DIF-1 plays a key role in breast cancer cell survival and further characterizing this pathway may provide important insights into developing novel therapies to selec tively target breast cancer cells for apoptosis.
Introduction
Programmed cell death or apoptosis is a fundamental process in growth and development and is targeted in the treatment of various tumors. Apoptosis is mediated through activation of “initiator” caspases (e.g., caspase-2, caspase-8, caspase-9, and caspase-10), which then cleave and activate “effector” caspases (e.g., caspase-3, caspase-6, and caspase-7; refs. 1, 2), leading to cleavage of a wide variety of protein components in the cell. We have been interested in understanding the mechanism of retinoid-mediated inhibition of breast cancer cell growth (3, 4). We previously examined the role of a nuclear hormone receptor coactivator we cloned from HeLa cells [nuclear receptor interacting factor 3 (NRIF3); Fig. 1A] on the retinoid inhibition of growth (3, 4) of several breast cancer cell lines (5). Unlike HeLa cells, we found that NRIF3 expression in breast cancer cell lines rapidly leads to an apoptosis (within 5 hours of expression) independent of retinoid incubation (5, 6). The region of NRIF3-mediating apoptosis was mapped to amino acids 20 to 50, and we refer to this region as death domain-1 (DD1; Fig. 1A; refs. 5, 6). Caspase inhibitor and small interfering RNA (siRNA) studies indicated that DD1-mediated apoptosis resulted from activation of caspase-2 (5, 6). Ser28 is important for apoptosis because change of Ser28 to Ala28 abrogates the ability of DD1 to mediate apoptosis (5, 6). This effect of NRIF3 or DD1 seems selective for breast cancer cells because expression of DD1 in a wide variety of cell lines derived from other cell types, in addition to HeLa cells, did not lead to cell death (5, 6).
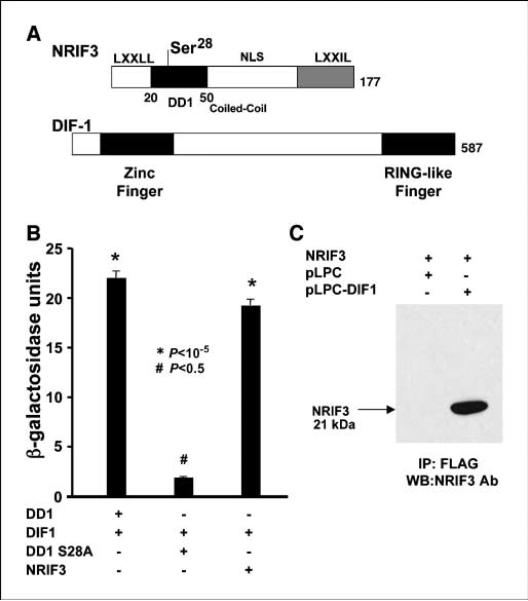
Figure 1.
Structure, domains, and interaction of NRIF3 and DIF-1. A, the domain structure of NRIF3 has been described previously (8). The DD1 region sufficient to mediate apoptosis (amino acids 20–50) is shown along with Ser28, which is essential to mediate apoptosis. DIF-1 contains an NH2 terminal zinc finger and a COOH terminal RING-like finger. DD1 interacts with DIF-1 through the RING-like finger. B and C, NRIF3/DD1 interacts with DIF-1 in yeast and mammalian cells. B, LexA-NRIF3, LexA-DD1, and LexA-DD1(S28A) were expressed in yeast along with a LexA-driven LacZ reporter (pSH18-34) and pJG4-5 conditionally expressing DIF-1 with a B42 activation domain. Shown are LacZ results where B42-DIF-1 was conditionally expressed by galactose. LexA-NRIF3 and LexA-DD1 interact with DIF-1, whereas LexA-DD1(S28A) exhibited little or no interaction. B, HeLa cells were transfected with pLPC-DIF-1 or the control pLPC vector along with pcDNA3.1 expressing NRIF3. Twenty-four hours later, the cells were lysed and incubated overnight with FLAG-antibody (M2) agarose beads. The protein bound to the beads was analyzed by Western blotting with NRIF3 antibody.
These findings suggest that breast cancer cells contain a novel “death switch” that is specifically triggered by NRIF3/DD1. To understand how NRIF3/DD1 expression leads to apoptosis in breast cancer cells, we screened an MCF-7 breast cancer cell cDNA library using a yeast two-hybrid screen and LexA-DD1 as bait. We identified a previously described factor (IRF-2BP2A) that had been suggested to mediate the repressive effect of IRF-2 on IRF-2–modulated genes (7). In this study, we show that IRF-2BP2A is an important target for DD1-mediated apoptosis, and thus, we refer to this factor in this article as DD1-interacting factor-1 (DIF-1; Fig. 1A). DIF-1 harbors an NH2 terminal zinc finger and COOH terminal RING-like finger, suggesting that it might function as an E3 ubiquitin or SUMO ligase, although in vivo and in vitro studies have not provided evidence for this. We found, however, that DIF-1 functions as a transcriptional repressor that mediates its effect through SirT1 and the repressive activity of DIF-1 is blocked by NRIF3. Additional evidence that DIF-1 plays a central role in NRIF3/DD1-mediated apoptosis are as follows: (a) change of Ser28 in DD1 to Ala28 abrogates the apoptogenic effect of DD1 and this mutation eliminates the binding of DD1 to DIF-1; (b) expression of DIF-1 blocks DD1-mediated apoptosis in breast cancer cells; and (c) RNA interference (RNAi) knockdown of DIF-1 in a wide variety of breast cancer cells (but not other cell types) leads to apoptosis.
These observations suggest that DIF-1 functions as an “anti-apoptotic” factor and “inhibition” of its activity through NRIF3/ DD1 binding or RNAi knockdown leads to apoptosis. Because NRIF3/DD1 expression or RNAi knockdown of DIF-1 seems to specifically lead to apoptosis of breast cancer cells, this pathway may provide important insights to develop novel therapies to selectively target breast cancer cells for apoptosis.
Materials and Methods
Yeast two-hybrid screen with an MCF-7 cell cDNA library
pEG202 expressing LexA-NRIF3 has been described previously (8). LexA-DD1 and LexA-DD1(S28A) were constructed by PCR of DD1 and DD1(S28A) from GFP-DD1 and GFP-DD1(S28A) vectors (5) and cloning the PCR-generated fragments into pEG202. An MCF-7 cDNA library (estrogen treated) in pJG4-5 was obtained from Origene. All methods and transformation procedures have been described previously (9). Because DD1(S28A) does not lead to apoptosis of breast cancer cells, an MCF-7 cell expressed protein from pJG4-5, which interacts with LexA-DD1 but not LexA-DD1(S28A), would be considered a candidate target of DD1 in breast cancer cells. One such clone was identified, which expressed 102 amino acids of the COOH terminal region, including the RING-like finger of a previously identified clone, IRF-2BP2A (7). In this study, we refer to IRF-2BP2A as DIF-1.
Yeast β-galactosidase assays
Details are given in Supplementary Materials and Methods.
Plasmids
Details on the construction of DIF-1 and other expression vectors are given in Supplementary Materials and Methods.
Cell culture and DNA transfection
HeLa cells and other cells, which were not derived from mammary epithelium, were routinely maintained in DMEM containing 10% bovine calf serum supplemented with glutamine and antibiotics. The various breast cancer cell lines (MCF-7, T-47D, SKBR3, MDA-435, and MDA-231) were maintained in DMEM containing 10% fetal bovine serum supplemented with glutamine and antibiotics. For the reporter gene studies, cells were seeded in 12-well plates at 100,000 to 150,000 cells per well, and the medium was replaced with DMEM containing resin-charcoal–treated bovine calf serum (10) before transfection. Transfections were performed with the amount of Gal4-chimeric vectors indicated in the figure legends using calcium phosphate coprecipitation. Typically, 100 ng of the chloramphenicol acetyltransferase (CAT) reporter plasmid pG5-SV-BCAT was used. This reporter contains five GAL4-responsive binding sites cloned upstream of the SV40 early promoter linked to the CAT gene (11). All transfections were performed in duplicate or triplicate. CAT activity was determined as previously described (12). In certain experiments, 150 ng/mL Trichostatin A (TSA; classes I and II HDAC inhibitor) or 50 nmol/L nicotimamide (a class III HDAC inhibitor) or 1 μmol/L resveratrol (a SirT1 agonist) was added 1 h before transfection.
TUNEL assay
Breast cancer cell lines were plated at a density of 3 × 104 cells per well (250 μL medium) on glass A coverslips in 48-well tissue culture plates. About 20 h later, the cells were transfected with indicated plasmid(s) using Lipofectamine 2000 (Invitrogen). Generally, the amount of plasmid used in transfections was 25 to 50 ng for GFP-DD1. In “rescue” experiments, cells were transfected with 100 ng of pLPC or pLPC-DIF-1. Twenty-four hours later, the cells were transfected with 50 ng of GFP-DD1 and then harvested 5 h later for TUNEL assay (In situ Cell Death Detection kit, TMR red; Roche Diagnostics GmbH). Cells were also stained with 4′,6-diamidino-2-phenylindole to visualize nuclei, mounted on slides, examined by fluorescent microscopy, and digitally imaged.
siRNA transfection
siRNA sequences and transfection conditions are given in Supplementary Materials and Methods.
Immunoprecipitation
HeLa cells were transfected with pLPC-DIF1 or the pLPC control vector along with a pcDNA3.1 vector-expressing untagged NRIF3. Twenty-four hours later, the cells were washed with isotonic saline and lysed in lysis buffer [50 mmol/L Tris-HCl (pH 7.4), 150 mmol/L KCl, 0.25% NP40, and 1 mmol/L DTT; Roche mini-complete protease inhibitor]. After a freeze-thaw cycle, the samples were centrifuged and the cell extracts were incubated with FLAG-M2-agarose beads (Sigma) at 4°C for 15 h. The beads were then washed, and NRIF3 bound to FLAG-HA-DIF-1 immobilized on the FLAG-M2-agarose beads was identified by Western blotting with NRIF3 antibody.
DNA-cellulose binding
Conditions for the binding of cell extracted DIF-1 are given in Supplementary Materials and Methods.
Results
Identification of DIF-1 as an interactor of DD1 and NRIF3 through a yeast two-hybrid screen
We used a yeast two-hybrid screen to identify candidate factors that might interact with NRIF3/DD1 to mediate apoptosis in breast cancer cells. Because DD1 of NRIF3 is sufficient to mediate apoptosis, we expressed DD1 as a LexA fusion in yeast and screened a pJG4-5 MCF-7 cell cDNA library. DD1(S28A) does not lead to apoptosis and any candidate factors identified with LexA-DD1 were rescreened with LexADD1(S28A). One of the clones that interacted with DD1, but not DD1(S28A), was IRF-2BP2A, which we refer to as DIF-1. The clone contained only the COOH terminal RING-like finger region (Fig. 1A) indicating that DD1 associates with the RING-like finger of DIF-1. A full-length clone of DIF-1 was generated and then cloned into (a) pJG4-5 for yeast interaction studies, (b) pLPC to generate NH2 terminal FLAG and HA tags for expression in mammalian cells, and (c) pEGFP-C3 for cell fluorescent studies. Full-length DIF-1 contains a predicted NH2 terminal zinc finger and a COOH terminal RING-like finger (Fig. 1A).
DIF-1 interacts with NRIF3/DD1 in yeast and mammalian cells
Figure 1B compares the yeast interaction (β-galactosidase activity) of LexA-DD1, LexA-DD1(S28A), and LexA-NRIF3 with full-length DIF-1 conditionally expressed with galactose from pJG4-5. DIF-1 did not interact with LexA-DD1(S28A), whereas DIF-1 interacted strongly with LexA-DD1 or LexA-NRIF3. These interactions parallel the apoptogenic potential of these peptides. To provide evidence that DIF-1 and NRIF3 interact in mammalian cells, HeLa cells were transfected with a vector expressing NRIF3 and with a pLPC vector expressing FLAG-HA–tagged DIF-1 or a control pLPC FLAG-HA vector. Twenty-four hours later, the cells were lysed, the extracts were incubated with FLAG-antibody(M2) immobilized on agarose beads, and the protein associated with the beads was analyzed by Western blotting (Fig. 1C). The Western blot from cells expressing FLAG-HA–tagged DIF-1 detected NRIF3 (~21 kDa) whereas no immunoreactive NRIF3 was found with the pLPC vector control.
DIF-1 is a nuclear protein
NRIF3 is a nuclear protein, and expression of GFP-DIF-1 in T-47D breast cancer cells indicates that DIF-1 is also a nuclear protein (Fig. 2A). Similar results were found with GFP-DIF-1 in HeLa cells (not shown). In addition, DIF-1 or components of a DIF-1 complex can bind to DNA-cellulose. FLAG-HA–tagged DIF-1 was expressed in T-47D cells and HeLa cells. Nuclear extracts (0.3 mol/L KCl) were bound to FLAG-antibody(M2) beads, washed, and eluted with FLAG peptide. The eluted fraction was incubated with DNA-cellulose or with just cellulose. The samples were washed and DIF-1 bound analyzed by Western blotting using FLAG-M2 antibody. Figure 2B shows that DIF-1, or a complex containing DIF-1, bound to DNA-cellulose but not the cellulose control.
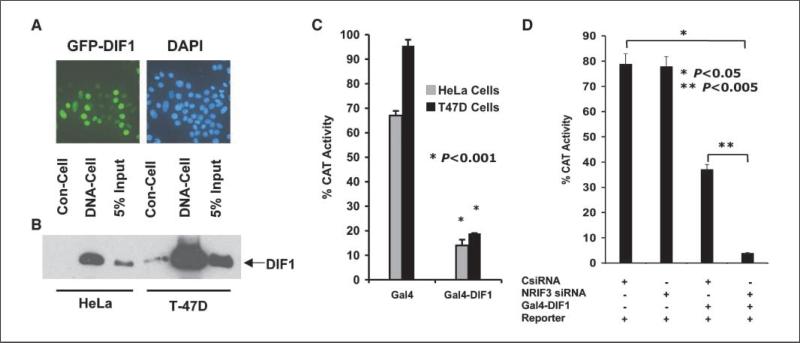
Figure 2.
DIF-1 is a nuclear protein that binds to DNA-cellulose and mediates transcriptional repression. A, GFP-DIF-1 expressed in T-47D cells localizes to the nucleus. B, HeLa and T-47D cells expressing FLAG-HA tagged DIF-1 were lysed in 0.3 mol/L KCl buffer. The supernatants were bound to FLAG antibody beads and then eluted with 3× FLAG peptide. Samples were incubated with DNA-cellulose (DNA-Cell) or with cellulose control (Con-Cell), and after washing, the cellulose beads were analyzed for DIF-1 by Western blotting with FLAG antibody. The input represents 5% of the sample before the binding assay. C, HeLa or T-47D cells were transfected with 100 ng of the pG5-SV-BCAT reporter along with 200 ng of vector expressing Gal4-DIF-1 or 150 ng (equal molar amount of plasmid) expressing only the Gal4 DBD. CAT activity was analyzed 30 h later. Gal4-DIF-1 represses the Gal4-CAT reporter in both cell types. D, HeLa cells were transfected as indicated with an NRIF3 siRNA (25 nmol/L) or a control-siRNA (25 nmol/L). Thirty hours later, the cells were transfected with 100 ng of pG5-SV-BCAT alone and with 50 ng of the Gal4-DIF-1 vector. CAT activity was determined 30 h later.
DIF-1 is a transcriptional repressor whose activity is blocked by NRIF3
IRF-2BP2A linked to the yeast Gal4 DNA-binding domain (DBD) was reported to show repression of a Gal4 reporter gene (7). To further examine this, we constructed a vector expressing a chimera of the Gal4-DBD and DIF-1 (Gal4-DIF-1). Expression of Gal4-DIF-1 in either HeLa or breast cancer cells leads to the marked repression of a Gal4-CAT reporter gene (Fig. 2C). To assess whether this repression is modulated by NRIF3, we expressed Gal4-DIF-1 at low levels in HeLa cells (which endogenously express NRIF3) and asked if repression by Gal4-DIF-1 was enhanced when NRIF3 was knocked down by NRIF3 siRNA. Figure 2D shows that low-level expression of Gal4-DIF-1 leads to incomplete transcriptional repression whereas knockdown of NRIF3 markedly enhances the level of repression by Gal4-DIF-1.
Repression mediated by DIF-1 seems to involve class III histone deacetylases
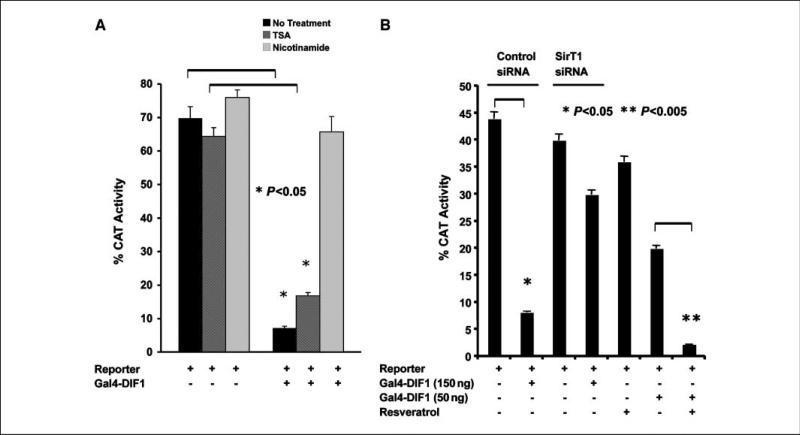
To assess the role of histone deacetylases (HDAC) in mediating repression by DIF-1, we examined the effect of TSA, a class I and class II HDAC inhibitor, and nicotinamide, a reversible inhibitor of the sirtuins, which are class III NAD+-dependent protein HDACs (13–15). Figure 3A shows that TSA minimally reverses Gal4-DIF-1–mediated repression whereas this repression is fully reversed by nicotinamide. Because both SirT1 and DIF-1 are nuclear proteins, we considered SirT1 as a potential candidate to mediate repression by DIF-1. To further explore this, we examined the effect of knockdown of SirT1 by siRNA on repression by DIF-1. Knockdown of SirT1 (~60% to 70% estimated by Western blotting) reversed repression mediated by DIF-1 (Fig. 3B). Further evidence that DIF-1–mediated repression occurs through SirT1 is supported by the finding that DIF-1–mediated repression is enhanced by resveratrol, a SirT1 agonist (Fig. 3B; ref. 16). Although resveratrol may have other targets, such as AMP kinase (17), enhancement of repression by resveratrol along with reversal of repression by SirT1 knockdown supports the notion that SirT1 mediates the repressive effects of DIF-1.
Figure 3.
Repression by DIF-1 is mediated by SirT1. A, T-47D cells were transfected with 100 ng of pG5-SV-BCAT and 200 ng of vector expressing Gal4-DIF-1. One hour before transfection, cells received either TSA (150 ng/mL), nicotinamide (50 nmol/L), or no treatment. Thirty hours later, the cells were harvested for CAT activity. B, T-47D cells were transfected with a SirT1 siRNA (25 nmol/L) or a control-siRNA (25 nmol/L). Before introduction of the siRNAs, cells were incubated with 20 μmol/L zVDVAD-fmk, a caspase-2 inhibitor, to prevent apoptosis that might occur as a result of knockdown of SirT1. Thirty hours after introduction of the siRNAs, the cells were transfected with 100 ng of the pG5-SV-BCAT reporter alone and with 150 ng of the Gal4-DIF-1 expression vector. Another set of cells did not receive siRNA but received 1 μmol/L resveratrol before transfection with 50 ng of the Gal4-DIF-1 vector. Thirty hours after transfection, the cells were analyzed for CAT activity.
DD1-mediated apoptosis in breast cancer cells is blocked by α-amanitin
The finding that DIF-1 is a transcriptional repressor, which is inactivated by NRIF3 or DD1, suggests that DIF-1 might repress one or more proapoptotic genes in breast cancer cells. To provide further support for this notion, we examined whether inhibition of RNA polymerase II by α-amanitin blocks DD1-mediated apoptosis. α-Amanitin does not rapidly enter cells and, depending on the concentration and cell type, has its effect after 5 to 6 hours of incubation. We took advantage of this by first incubating half of the cultured wells of T-47D cells with 2.5 umol/L α-amanitin 3 hours before transfection with a GFP-DD1 vector. This allowed for synthesis of these proteins, which seem stable as assessed by GFP fluorescence. Twenty hours later, cells were analyzed for GFP expression and for apoptosis by TUNEL assay (Fig. 4A). Incubation with α-amanitin does not lead to apoptosis, but it inhibits the ability of DD1 to mediate apoptosis (Fig. 4A, bottom left). Although there are likely a number of explanations for this finding, the results are consistent with DIF-1 acting to repress a proapoptotic gene(s) in breast cancer cells, which when expressed leads to apoptosis.
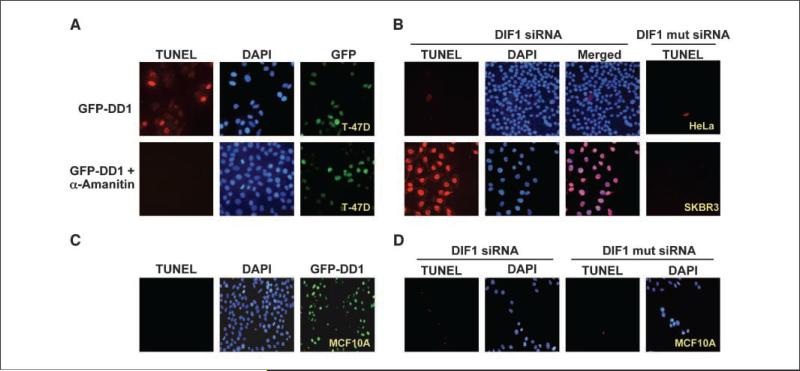
Figure 4.
Effect of α-amanitin on DD1-mediated apoptosis and knockdown of DIF-1 by siRNA leads to apoptosis of breast cancer cells. A, T-47D cells were incubated with or without α-amanitin (2.5 μmol/L) for 3 h and then transfected with vector (100 ng) to express GFP-DD1. Twenty hours later, the cells were analyzed for GFP-DD1 expression and apoptosis by TUNEL assay. Shown are representative fields. No apoptosis was identified in cells preincubated with α-amanitin and the levels of GFPDD1 were similar under both conditions. B, siRNA (25 nmol/L) was used to knockdown DIF-1 expression in five different breast cancer cell lines (SKBR3, MCF-7, T-47D, MDA-435, and MDA-231) or cells of other origin (U2OS, human osteosarcoma; 293, human kidney epithelium; UOK-145, kidney carcinoma; HepG2, human hepatoma; HeLa, human cervical epithelium). A control siRNA (25 nmol/L) contained four base changes (mut siRNA). Thirty hours later, cells were examined for apoptosis by TUNEL assay. All the breast cancer cell lines exhibited apoptosis, whereas the other cells did not. Shown are representative results from the study with SKBR3 breast cancer cells and HeLa cells. Supplementary Fig. S1 shows results with the other cell lines. Similar results were found with four different siRNAs that targeted DIF-1 mRNA. C, expression of DD1 (GFP-DD1) does not lead to apoptosis of MCF10A cells, a cell line derived from normal breast epithelial cells. D, DIF-1 siRNA does not lead to apoptosis of MCF10A cells.
siRNA knockdown of DIF-1 leads to apoptosis of breast cancer cells but not cells of other origin
NRIF3/DD1 might lead to apoptosis of breast cancer cells via DIF-1 by two general mechanisms. One proposes that DIF-1 is activated by NRIF3/DD1 to initiate an apoptotic pathway whereas the second is that DIF-1 acts as an antiapoptotic factor whose activity is interfered with by NRIF3/DD1. The findings in Fig. 2D support the second hypothesis. To further distinguish between these two general potential mechanisms, five different breast cancer cell lines were studied (SKBR3, MCF-7, T-47D, MDA-435, and MDA-231). We also examined MCF10A cells, which exhibit characteristics of normal breast epithelium, do not form tumors in nude mice, and lack anchorage-independent growth (18). The breast cancer cell lines and MCF10A cells were transfected with an siRNA that targets DIF-1 mRNA, whereas a control group received an siRNA containing four base changes. MCF10A cells showed no evidence for apoptosis, whereas all the breast cancer cell lines exhibited apoptosis (Fig. 4B and D). Because of space considerations, the results are only shown for SKBR3 in Fig. 4B. Supplementary Fig. S1 shows the results for MCF-7, T-47D, MDA-435, and MDA-231 cells. Parallel studies were carried out with other cell types (U2OS, human osteosarcoma; 293, human kidney epithelium; UOK-145, kidney carcinoma; HepG2, human hepatoma; HeLa, human cervical epithelium). These cells did not undergo apoptosis and, for space consideration, are only shown for HeLa in Fig. 4B. Supplementary Fig. S1 shows the results for U2OS, 293, UOK-145, and HepG2. Supplementary Fig. S2A shows that DIF-1 siRNA efficiently knocks down FLAG-HA–tagged DIF-1 (~90%) whereas the DIF-1 siRNA mutant (control) was without effect. Similar results were found for HeLa cells expressing FLAG-HA–tagged DIF-1 (not illustrated). The DIF-1 siRNA mutant did not lead to apoptosis in the breast cancer cell lines. Three other DIF-1 siRNAs (see Materials and Methods) also selectively led to apoptosis of the breast cancer cell lines. Interestingly, transfection with DIF-1 siRNA does not lead to apoptosis of MCF10A cells (Fig. 4D). Knockdown of DIF-1 in HeLa and the other nonbreast cancer lines does not lead to apoptosis (Supplementary Fig. S1), suggesting that breast cancer cells have the potential to selectively express a proapoptotic factor(s) that is(are) functionally inactivated by DIF-1 or whose expression is repressed by DIF-1.
That normal MCF10A cells do not undergo apoptosis when transfected with DIF-1 siRNA was an intriguing finding. Thus, we examined whether expression of GFP-DD1 leads to apoptosis of MCF10A cells (Fig. 4C). Interestingly, unlike the breast cancer cell lines (Supplementary Fig. S3), expression of DD1 does not lead to apoptosis of MCF10A cells (Fig. 4C), suggesting that there is something intrinsic to breast cancer cells and not just breast epithelial cells that sensitizes them to apoptosis. To further explore this possibility, we examined two mouse breast cancer cell lines (4T1 and 67NR; ref. 19) and a mouse mammary cell line (C57MG) derived from C57BL/6 mouse normal mammary epithelium (20). Expression of GFP-DD1 leads to apoptosis of 4T1 and 67NR cells but not C57MG cells (Supplementary Fig. S4), further suggesting that breast cancer cells are selectively sensitized to DD1-mediated apoptosis.
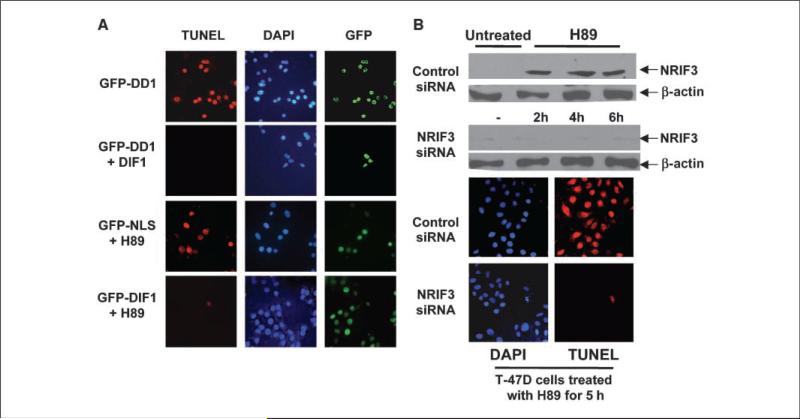
DIF-1 rescues breast cancer cells from DD1-mediated apoptosis and protects cells from H89-mediated apoptosis
Our results suggest that NRIF3/DD1 mediate their apoptotic effects by targeting DIF-1. A prediction of this conclusion is that further expression of DIF-1 might be expected to rescue breast cancer cells from DD1-mediated apoptosis. Figure 5A (rows 1 and 2) shows that DIF-1 expression rescues cells from DD1-mediated apoptosis. We explored whether DIF-1 might play a role in abrogating apoptosis of breast cancer cells initiated by other factors. Protein kinase A (PKA) plays an antiapoptotic role in breast cancer cells (21), and we found that the PKA inhibitor H89 rapidly leads to apoptosis (Fig. 5A, rows 3 and 4). GFP-DIF-1 was used to express DIF-1 in T-47D cells, whereas control cells received a control GFP-NLS vector. Twenty-four hours later, the cells were incubated with or without 200 nmol/L H89 for 5 hours followed by analysis by TUNEL assay. DIF-1 protected the T-47D cells from apoptosis within 5 hours of H89 incubation (Fig. 5A, rows 3 and 4), as well as after 24 hours of exposure (not illustrated). Similar findings with H89 were found with all breast cancer cell lines studied, whereas H89 did not mediate apoptosis in other cell types of different origin as described earlier (U2OS, 293, UOK-145, HepG2, and HeLa; not illustrated).
Figure 5.
DIF-1 rescues breast cancer cells from DD1-mediated and H89-mediated apoptosis. A, T-47D cells were transfected with 100 ng pLPC-DIF-1. Other cells were transfected with control pLPC vector. Twenty-four hours later, the cells were transfected with 50 ng of GFP-DD1 vector. Five hours later, cells were analyzed for apoptosis by TUNEL assay. Shown are represented fields. Expression of DIF-1 decreased DD1-mediated apoptosis by over 90%. For the H89 study, T-47D cells were transfected with GFP-DIF-1 vector (100 ng) or with a vector expressing GFP containing a nuclear localization signal (GFP-NLS; 75 ng). Twenty-four hours later, the cells received 200 nmol/L of H89, a PKA inhibitor. Five hours later, the cells were examined for apoptosis by TUNEL assay. Shown are representative fields. Expression of DIF-1 resulted in a >90% reduction in apoptosis mediated by H89. B, H89 mediates apoptosis in breast cancer cells through NRIF3. T-47D cells were treated with a NRIF3 siRNA (25 nmol/L) or with a control siRNA (25 nmol/L). Thirty hours later, the cells received 200 nmol/L H89. Cells were harvested for NRIF3 expression by Western blotting 2, 4, and 6 h after addition of H89. Cells were also examined for apoptosis by TUNEL assay 5 h after H89 incubation. H89 incubation results in a rapid increase in NRIF3. NRIF3 siRNA blocked the rapid H89-mediated increase in NRIF3 expression, as well as apoptosis. Supplementary Fig. S5 indicates that this rapid increase in NRIF3 occurs at the mRNA level.
Apoptosis mediated by H89 in breast cancer cells results from the rapid induction of NRIF3
The finding that DIF-1 rescues breast cancer cells from H89-mediated and NRIF3/DD1-mediated apoptosis suggests that H89 leads to apoptosis through a rapid increase in NRIF3, which targets DIF-1. Figure 5B shows that T-47D cells express very low levels of NRIF3 and that H89 incubation leads to a rapid increase in NRIF3 levels. Similar results were found for H89 and NRIF3 mRNA using quantitative reverse transcription–PCR (RT-PCR; Supplementary Fig. S5). Apoptosis is inhibited by siRNA knockdown of NRIF3 24 hours before addition of H89 (Fig. 5B), suggesting that NRIF3 mediates the rapid apoptotic response elicited by H89 in breast cancer cells. Supplementary Fig. S2B documents that the NRIF3 siRNA leads to knockdown of NRIF3. This finding raises the possibility that modulation of endogenous NRIF3 may play a role in the initiation of apoptosis in breast cancer cells by a variety of factors. Although HeLa cells express NRIF3, H89 incubation does not increase the levels of NRIF3 (Western blotting, not shown) or NRIF3 mRNA (Supplementary Fig. S5).
Discussion
In this study, we identified DIF-1 (IRF-2BP2A) as a potent antiapoptotic factor in a wide variety of breast cancer cell lines, but not in cell lines derived from other tissues. NRIF3/DD1 interacts with DIF-1 (Fig. 1B and C) and modulates DIF-1 activity (Fig. 2D), suggesting that DIF-1 is the target of NRIF3/DD1 that mediates apoptosis in breast cancer cells. Additional evidence that DIF-1 is the target of NRIF3/DD1 in mediating apoptosis comes from studies indicating that (a) DD1(S28A) does not interact with DIF-1 and does not lead to apoptosis, (b) expression of DIF-1 abrogates NRIF3/DD1-mediated apoptosis, and (c) siRNA knockdown of DIF-1 leads to apoptosis in breast cancer cell lines but not lines derived from other cell types.
IRF-2BP2A was initially cloned in a yeast two-hybrid screen as a factor that mediated the repressive effects of IRF-2 (7). The mechanism of this repression was not defined, although repression was not reversed by TSA, a class I and class II HDAC inhibitor. In this study, we found that DIF-1 is a potent transcriptional repressor, and this effect seems to be mediated by SirT1, a class III NAD+-dependent histone and protein deacetylase. Evidence supporting that DIF-1 acts through SirT1 comes from the findings that DIF-1 repression is reversed by siRNA knockdown of SirT1 and is enhanced by resveratrol, a SirT1 agonist (Fig. 3B). Although SirT1 has been identified with N-CoR/SMRT corepressors (22), which interact with class I or class II HDACs, our TSA findings do not support that class I or class II HDACs significantly mediate the repressive effects of DIF-1. The finding that DIF-1 or a component of the DIF-1 complex binds DNA (Fig. 2) suggests that DIF-1 via SirT1 may act to affect histone acetylation or other chromatin modifications (15) or deacetylation of other transcription factors (13–15).
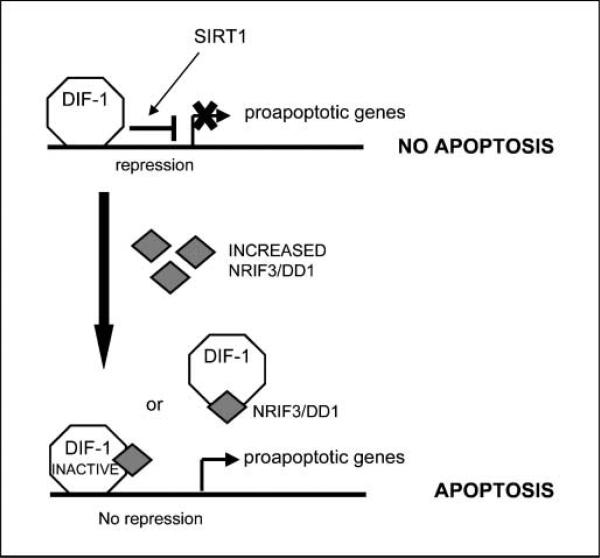
NRIF3 seems to inhibit repression mediated by DIF-1 because knockdown of endogenous NRIF3 in HeLa cells leads to enhanced DIF-1–mediated repression (Fig. 2D). In this regard, H89-mediated apoptosis in breast cancer cells seems to result from a rapid increase in NRIF3 levels, which would interact with and modify the activity of DIF-1. How H89 rapidly induces NRIF3 is unknown, but the rapidity of the response at both the mRNA (Supplementary Fig. S5) and protein level (Fig. 5B) suggests that NRIF3 acts as a “stress” response gene that may determine cell fate in a cell type– specific manner. Interestingly, α-amanitin, a polymerase II inhibitor, blocks DD1-mediated apoptosis in breast cancer cells (Fig. 4A). Taken together, our findings suggest that DIF-1 acts to repress one or more proapoptotic genes in breast cancer cells and the binding of NRIF3/DD1 to DIF-1 in breast cancer cells reverses repression and leads to the expression of these proapoptotic genes (Fig. 6).
Figure 6.
Mechanism by which NRIF3/DD1 and DIF-1 regulate apoptosis of breast cancer cells. The figure depicts a model that is consistent with our findings that DIF-1 is a transcriptional repressor that can bind DNA whose activity is reversed by NRIF3/DD1 and that preincubation with α-amanitin blocks DD1-mediated apoptosis of breast cancer cells. Our findings suggest that one or more proapoptotic genes bind to and are repressed by DIF-1 in breast cancer cells. Expression of NRIF3/DD1 binds DIF-1 and mediates a conformational change that reverses its ability to act as a repressor. We do not know whether the NRIF3/DIF-1 complex remains bound to the proapoptotic gene(s) or whether the binding of NRIF3/DD1 to DIF-1 results in a dissociation of DIF-1 from the proapoptotic gene. By either mechanism, reversal of repression of one or more proapoptotic genes leads to gene expression and the initiation of apoptosis.
Because expression of NRIF3/DD1 seems to selectively lead to apoptosis in breast cancer cells, we examined the relative mRNA levels of NRIF3 and DIF-1 by quantitative RT-PCR in the cell lines used in this study. All breast cancer cell lines expressed ~5-fold to 10-fold less NRIF3 mRNA than HeLa and 293 cells. U2OS and MCF10A cells expressed ~3-fold less NRIF3 mRNA than HeLa cells (Supplementary Fig. S6). Supplementary Fig. S7 indicates that (a) HeLa, 293, SKBR3, and MCF-7 cells express similar amounts of DIF-1 mRNA whereas U2OS cells express 7-fold less, (b) T-47D cells express 2-fold less, and (c) MCF10A cells express 2-fold higher DIF-1 mRNA levels. The increase in DIF-1 in MCF10A cells does not likely account for their resistance to DD1-mediated apoptosis because the 67NR and 4T1 cells express 5-fold to 10-fold higher levels and exhibit DD1-mediated apoptosis. Furthermore, DIF-1 mRNA levels in the DD1-resistant C57MG cells is similar to that found in 67NR cells, suggesting that differences in DIF-1 expression does not account for the difference in response of the 67NR tumor cells and the normal C57MG breast epithelial cells.
The specificity of the DIF-1 pathway in mediating apoptosis in breast cancer cells suggests that genes, which are silenced in breast cancer cells by DIF-1, are silenced by other mechanisms in other cell types. Very few studies have examined the role of SirT1 on mammary epithelium or breast cancer. Knockout of the SirT1 gene identified an abnormality in mammary gland development (23), whereas another study noted that the SirT1 inhibitor Sirtinol leads to growth arrest of MCF-7 cells, although apoptosis was not examined (24). In addition, siRNA knockdown of SirT1 leads to apoptosis or senescence of epithelial-derived tumor cells, including MCF-7 (25).
Although DIF-1 seems to repress through SirT1, its antiapoptotic activity is not fully reproduced when we knocked down SirT1 in breast cancer cells. Thus, knockdown of SirT1 by siRNA in T-47D cells leads to 30% of the cells exhibiting apoptosis1 compared with knockdown of DIF-1 or expression of DD1/NRIF3 (>90%). Although this might suggest that DIF-1 acts as an antiapoptotic factor via a mechanism independent of SirT1, the extent of knockdown of SirT1 (~60–70%) is less than that found for DIF-1 (>90%). In addition, if DIF-1 selectively repressed one or more proapoptotic genes, knockdown of DIF-1 would be expected to have a greater effect than knockdown of SirT1, unless knockdown of SirT1 was complete. Preliminary studies indicate that DIF-1 is part of a large nuclear protein complex.2 Identification of the proteins that interact with DIF-1 may shed light on why the antiapoptotic function of DIF-1 seems selective for breast cancer cells. Irrespective of the mechanism by which DIF-1 functions, our studies indicate that DIF-1 is a novel factor that can act as a death switch that plays a central and specific role in determining whether breast cancer cells survive or undergo programmed cell death.
Supplementary Material
Acknowledgments
Grant support: NIH grant DK16636 and Entertainment Industry Foundation grant (H.H. Samuels).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
We especially thank Kay Childs and Stephen Goodbourn, Department of Biochemistry and Immunology, St George's Hospital Medical School, University of London, for providing us with an IRF-2BP2A cDNA clone.
Footnotes
A.A. Tinnikov and K.T. Yeung are co–first authors.
S. Das and H.H. Samuels, unpublished data.
K.T. Yeung and H.H. Samuels, unpublished data.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–6. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 2.Lahm A, Paradisi A, Green DR, Melino G. Death fold domain interaction in apoptosis. Cell Death Differ. 2003;10:10–2. doi: 10.1038/sj.cdd.4401203. [DOI] [PubMed] [Google Scholar]
- 3.Afonja A, Raaka BM, Huang A, et al. RAR agonists stimulate SOX9 gene expression in breast cancer cell lines: evidence for a role in retinoid-mediated growth inhibition. Oncogene. 2002;21:7850–60. doi: 10.1038/sj.onc.1205985. [DOI] [PubMed] [Google Scholar]
- 4.Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene. 2004;23:8135–45. doi: 10.1038/sj.onc.1207983. [DOI] [PubMed] [Google Scholar]
- 5.Li D, Das S, Yamada T, Samuels HH. The NRIF3 family of transcriptional coregulators induces rapid and profound apoptosis in breast cancer cells. Mol Cell Biol. 2004;24:3838–48. doi: 10.1128/MCB.24.9.3838-3848.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, Nwachukwu JC, Li D, et al. The nuclear receptor interacting factor-3 transcriptional coregulator mediates rapid apoptosis in breast cancer cells through direct and bystander-mediated events. Cancer Res. 2007;67:1775–82. doi: 10.1158/0008-5472.CAN-06-4034. [DOI] [PubMed] [Google Scholar]
- 7.Childs KS, Goodbourn S. Identification of novel corepressor molecules for interferon regulatory factor-2. Nucleic Acids Res. 2003;31:3016–26. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Wang F, Samuels HH. Domain structure of the NRIF3 family of coregulators suggests potential dual roles in transcriptional regulation. Mol Cell Biol. 2001;21:8371–84. doi: 10.1128/MCB.21.24.8371-8384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahajan MA, Samuels HH. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Mol Cell Biol. 2000;20:5048–63. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samuels HH, Stanley F, Casanova J. Depletion of L-3,5,3′-triiodothyronine and L-thyroxine in euthyroid calf serum for use in cell culture studies of the action of thyroid hormone. Endocrinology. 1979;105:80–5. doi: 10.1210/endo-105-1-80. [DOI] [PubMed] [Google Scholar]
- 11.Witzgall R, O'Leary E, Gessner R, Ouellette AJ, Bonventre JV. Kid-1, a putative renal transcription factor: regulation during ontogeny and in response to ischemia and toxic injury. Mol Cell Biol. 1993;13:1933–42. doi: 10.1128/mcb.13.3.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S, Schapira M, Tomic-Canic M, Goyanka R, Cardozo T, Samuels HH. Farnesyl pyrophosphate is a novel transcriptional activator for a subset of nuclear hormone receptors. Mol Endocrinol. 2007;21:2672–86. doi: 10.1210/me.2007-0080. [DOI] [PubMed] [Google Scholar]
- 13.Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–65. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 15.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–20. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 16.Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–6. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc Natl Acad Sci U S A. 2007;104:7217–22. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–86. [PubMed] [Google Scholar]
- 19.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–405. [PubMed] [Google Scholar]
- 20.Vaidya AB, Lasfargues EY, Sheffield JB, Coutinho WG. Murine mammary tumor virus (MuMTV) infection of an epithelial cell line established from C57BL/6 mouse mammary glands. Virology. 1978;90:12–22. doi: 10.1016/0042-6822(78)90328-8. [DOI] [PubMed] [Google Scholar]
- 21.Manna PP, Frazier WA. CD47 mediates killing of breast tumor cells via Gi-dependent inhibition of protein kinase A. Cancer Res. 2004;64:1026–36. doi: 10.1158/0008-5472.can-03-1708. [DOI] [PubMed] [Google Scholar]
- 22.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-g. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Rajendran GK, Liu N, Ware C, Rubin BP, Gu Y. SirT1 modulates the estrogen-insulin-like growth factor-1 signaling for postnatal development of mammary gland in mice. Breast Cancer Res. 2007;9:R1. doi: 10.1186/bcr1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ota H, Tokunaga E, Chang K, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–85. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
- 25.Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–63. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.