Abstract
Cancer stem cells (CSCs) represent a small subpopulation of tumor cells that play a critical role in initiating and sustaining tumor growth. However, we currently have an incomplete understanding of the expression patterns of CSC antigens in tumors of dogs, nor do we understand how expression of these antigens vary between tumor cell lines and tumor biopsy specimens. Therefore, we used flow cytometry and commonly reported CSC surface and intracellular markers to evaluate the phenotype and overall frequency of CSC subpopulations in tumor cell lines and primary tumor biopsy samples from dogs with melanoma and osteosarcoma. We found that cells expressing common CSC antigens were rare in tumor cell lines, with the exception of tumor cells expressing CD44 and CD90. In contrast, tumor cells expressing conventional CSC antigens such as CD133, CD34, CD44, CD24 and Oct3/4 were much more common in tumor biopsy samples. Notably, the frequency and types of putative CSC subpopulations were very similar in biopsy samples from dogs with either melanoma or osteosarcoma. Our results suggest that the tumor microenvironment significantly influences CSC subpopulations within tumors and that tumor cell lines may not accurately reflect the actual frequency or types of CSC subpopulations present in tumor tissues in vivo.
Keywords: Flow cytometry, Cancer, Stem cells, Melanoma, Osteosarcoma
1. Introduction
Tumors are known to consist of a heterogeneous population of cells with a number of distinct properties. Recent studies have demonstrated that within all tumors there exists a small population of self-renewing cells known as cancer stem cells (CSCs) (Lapidot et al, 1994; Kruger et al., 2006; Dalerba et al., 2007; Croker and Allan, 2008). CSCs were initially defined by their increased ability to initiate tumor growth when transplanted in very small numbers into mice (Lapidot et al., 1994). The CSCs have also been referred to as tumor-propagating cells (TPCs) and cancer-initiating cells (CICs) (Lapidot et al., 1994; Kruger et al., 2006; Al-Hajj et al., 2003; Quintana et al., 2008; Schatton et al., 2008). CSCs are now believed to play critical roles in initiating and sustaining tumor formation and metastasis (Croker and Allan, 2008; Aguirre-Ghiso, 2007; Malanchi et al., 2012). Because CSCs typically have very slow rates of cell division, they are relatively resistant to radiation therapy (Bao et al., 2006). They also exhibit the ability to rapidly efflux or metabolize many chemotherapeutic drugs (Bao et al., 2006; Li et al., 2008; Diehn et al., 2009; O’Hare et al., 2006; Masters and Koberle, 2003). Finally, it has also been hypothesized that CSCs play a role in the recurrence of primary tumors or metastases at long intervals following apparent tumor cures (Aguirre-Ghiso, 2007; Malanchi et al., 2012).
CSCs were initially identified in hematopoietic tumors using cell surface and intracellular molecules (Lapidot et al., 1994). Since this discovery, numerous other tumors in various species have been examined for populations of CSCs, using both phenotypic and functional assays (Kruger et al., 2006; Al-Hajj et al., 2003; Schatton et al., 2008; Bao et al, 2006; Christgen et al., 2007; Wilson et al., 2008; Cocolaet al, 2009; Awadet al., 2010; Cameron et al., 2010; Michishita et al., 2010; Wang and Shen, 2010; Smith et al., 2011). Many studies have used surface marker expression to characterize the subpopulations of CSCs. Most of the markers used to define CSCs are also expressed on normal stem cells, thus suggesting that CSCs may arise directly from normal stem cells. Among the surface markers typically examined are CD19, CD20, CD24, CD34, CD38, CD44, CD90, and CD133 (Clevers, 2011; Overdevest et al., 2011; Tang et al., 2013) Oct3/4, a transcription factor, is also a commonly used intracellular CSC marker (Blacking et al., 2012).
CSCs have also been characterized using functional assays. The two major assays used to assess CSC populations include determination of cells with aldehydedehydrogenase (ALDH) activity and cells that efflux DNA dyes efficiently (also known as side population cells) (Awad et al., 2010; Golebiewska et al, 2011; Ginestier et al., 2007). ALDH is an enzyme that is known to breakdown chemotherapy drugs inside a cell and can be measured using a commercially available kit. Side population is defined as those cells that possess the ability to exclude Hoechst dye staining via various efflux pumps. ALDH activity and the ability to efflux chemicals, such as Hoeschst dye out of the cells, impart significant chemoresistance to these cells.
Despite the identification and classification of various CSCs in many tumor types, the CSC concept remains controversial (Magee et al., 2012). This controversy revolves around the fact that vast differences in CSC numbers and populations have been observed by investigators studying the same tumor type. For example, initial studies of human melanoma tumors suggested that melanoma CSCs were rare, with a frequency of approximately one in a million (Schatton et al., 2008). However, a recent study using a more rigorous xenotransplantation model demonstrated that the frequency of CSCs in human melanoma was actually rather high, with about 30% of single cell transplantations resulting in tumor growth (Quintana et al., 2008). Thus, differences in techniques used to identify CSCs can lead to disparate results.
CSCs subpopulations have also been assessed in tumors of dogs (Wilson et al., 2008; Cocola et al., 2009; Michishita et al., 2010; Blacking et al., 2012; Blacking et al., 2011; Pang and Argyle, 2009; Stoica et al., 2009; Cogliati et al., 2010). In one study examining single tumor cell line samples and biopsies from various cancers, it was observed that cultured canine tumor cell lines lacked discrete CSC subpopulations while cells derived from spontaneous tumors had more obvious CSC subpopulations (Blacking et al., 2012). In a second study examining CSC subpopulations within a small cell carcinoma cell line of dogs, it was noted that there was a high variation in CSC populations in clones derived from the cell line (Cameron et al., 2010).
To better understand the frequency of potential CSC subpopulations in canine tumors, we conducted a study of CSC subpopulations in 7 melanoma and 7 osteosarcoma cell lines and 11 melanoma and 17 OSA tumor biopsies from dogs using antibodies to detect cell surface antigens known to be associated with CSC populations in other species. This study was designed to address questions with respect to the relative prevalence of CSC subpopulations in tumor cell lines versus tumor biopsies, and to compare CSC subpopulations between different tumor types-in our case between canine melanoma and osteosarcoma tumor cell lines. Flow cytometry and a panel of established CSC markers were used to characterize the phenotype and frequencies of CSC in the tumor samples. Our study uncovered significant differences in CSC subpopulations between tumor cell lines and tumor biopsy specimens, as well as between different tumor types and the implications of these findings are discussed.
2. Materials and methods
2.1. Cell lines
Cell lines used in this study were derived from several sources, including ATCC (Manassas, VA) (osteosarcoma D17 cells), from collaborators (Dr. Doug Thamm, CSU; osteosarcoma cell lines Abrams, Gracie, Moresco, McKinley, and Vogel and melanoma cell line Jones) and Dr. Lauren Wolfe, Auburn University (melanoma cell lines CML10C2 and CML6M) and from osteosarcoma (COSDAm) and melanoma (CMSDJe, CMSDSc, CMSDTa, and CMSDSh) cell lines established in our laboratory. The osteosarcoma lines D17 (Riggs et al., 1974) and Abrams were derived from pulmonary metastases while the OSA line COSDAm, established in our lab, was generated from a primary bone tumor specimen. We were unable to determine the historical origin of the canine OSA lines Gracie, Moresco, McKinley, and Vogel. All of the melanoma lines established in our lab were derived from oral melanoma tumor biopsies and were either Stage II (CMSDSc and CMSDTa) or Stage III (CMSDJe and CMSDSh) melanomas. The melanoma cell lines CML-10C2 and CML-6M were derived from a cutaneous primary tumor and a lymph node metastatic lesion, respectively (Chon et al., 2013). The Jones melanoma cell line was derived from a primary tumor. All cell lines were verified as canine based on canine-specific CD90 staining. All studies described as “cell lines” were performed using the above-mentioned cell lines, which have been passaged greater than 20 times in culture. For the “early passage” experiments, passage numbers are indicated in the text/figure legends. Cells were cultured in MEM medium (Invitrogen, Carlsbad, CA) with essential and non-essential amino acids, l-glutamine, penicillin/streptomycin and 10% FBS (Gemini, West Sacramento, CA). In total, we examined 7 OSA and 7 Mel cell lines. Cell lines used in this study were routinely treated with Mycoplasma removal agent (MP Biomedicals, Solon, OH) and/or enrofloxacin to prevent Mycoplasma contamination and routinely tested for contamination.
2.2. Ex vivo tumor samples
Tumor biopsy samples were obtained from canine patients with histologically confirmed melanoma via fine-needle aspiration (FNA) of the primary lesion. These patients were not treated with any chemotherapy, however, one patient had received carprofen for one week prior to obtaining a FNA. In total, FNA samples were obtained from 11 different melanoma tumors. Tumor punch biopsies of primary OSA tumors were obtained from 17 dogs that had undergone limb amputation for confirmed OSA. These dogs were naïve to any tumor therapy. Not all melanoma FNA samples or OSA tumor biopsy samples were evaluated with every antibody due to the small amount of cells obtained with some biopsy samples. All animal studies were approved by the Animal Care and Use Committee at Colorado State University.
2.3. Flow cytometry
Single cell suspensions of cell lines for flow cytometry were prepared by cell detachment via light trypsinization. Melanoma FNA samples were washed in PBS and prepared as single cell suspensions. OSA tumor biopsies were subjected to collagenase digestion as described previously (Dow et al, 1999) to obtain a single cell suspension. Cells were immunostained with primary antibodies for 20 min at RT in FACs buffer (PBS with 2% fetal bovine serum and 0.05% sodium azide) following a 5 min incubation with normal dog serum (Jackson ImmunoResearch) to block non-specific binding. Antibodies used were as follows: anti-canine CD45 (YKIX716.13, Serotec, Raleigh, NC), anti-canine CD#4 (1H6, BD Pharmingen, San Jose, CA), anti-mouse CD24 (M1/69, eBioscience, San Diego, CA), anti-mouse CD133 (AC133, Miltenyi Biotec, San Diego, CA), anti-mouse/human CD44 (1M7, eBioscience), and anti-canine CD90 (YK1X337.217, eBioscience). The cells were then washed and stained with a secondary conjugate if necessary. 7-AAD (eBioscience, San Diego, CA) was added to each sample to exclude dead cells from the analysis. Isotype control antibodies were used to determine gating parameters (staining of isotype was set between 0.05% and 0.1%) and anti-CD45 antibody was added to tumor biopsy samples to remove contamination by differentiated hematopoietic cells. For intracellular immunostaining to detect Oct3/4, cells were initially labeled for surface determinants, then placed in fixation/permeabilization buffer, following manufacturer’s protocol (eBioscience), and incubated overnight at 4°C. The next day, cells were washed with 1 × permeabilization buffer and stained with the anti-Oct3/4 antibody (anti-mouse/human Oct3/4, clone EM92, eBioscience). Each cell line was analyzed by flow cytometry at least 2–3 times to ensure assay reproducibility. All samples were analyzed using a Cyan ADP instrument (Beckman Coulter, Miami, FL) and data were analyzed using FlowJo software (Tree Star, Ashland, OR).
2.4. Aldehyde dehydrogenase assay
ALDH content of tumor cells was measured using a commercially available kit (ALDEFLUOR, Stem Cell Technologies, Vancouver, Canada) according to manufacturer guidelines. Cells were washed, counted and resuspended in buffer plus 50 µg/ml verapamil at a concentration of 2 × 105 cells/ml. The ALDEFLUOR substrate was added to the cells, then half of the sample was added to another tube containing 5 µl of diethylaminobenzaldehyde (DEAB) as a positive control. Samples were incubated covered at 37°C for exactly 45 min. They were then transferred to ice and washed with chilled kit buffer plus verapamil prior to additional staining, which occurred while keeping the samples in the dark and on ice. After staining, 7-AAD was added to all the samples, incubated 5 min on ice and the samples were then immediately analyzed on the flow cytometer. The DEAB control was used to set sample gates at <0.05% FITC+.
2.5. Statistics
Differences between two groups were compared using a non-parametric t-test (Mann–Whitney). Statistical analysis was performed using GraphPad Prism software (La Jolla, CA) and a p-value of <0.05 was considered statistically significant.
3. Results
3.1. CSCs in canine cell lines
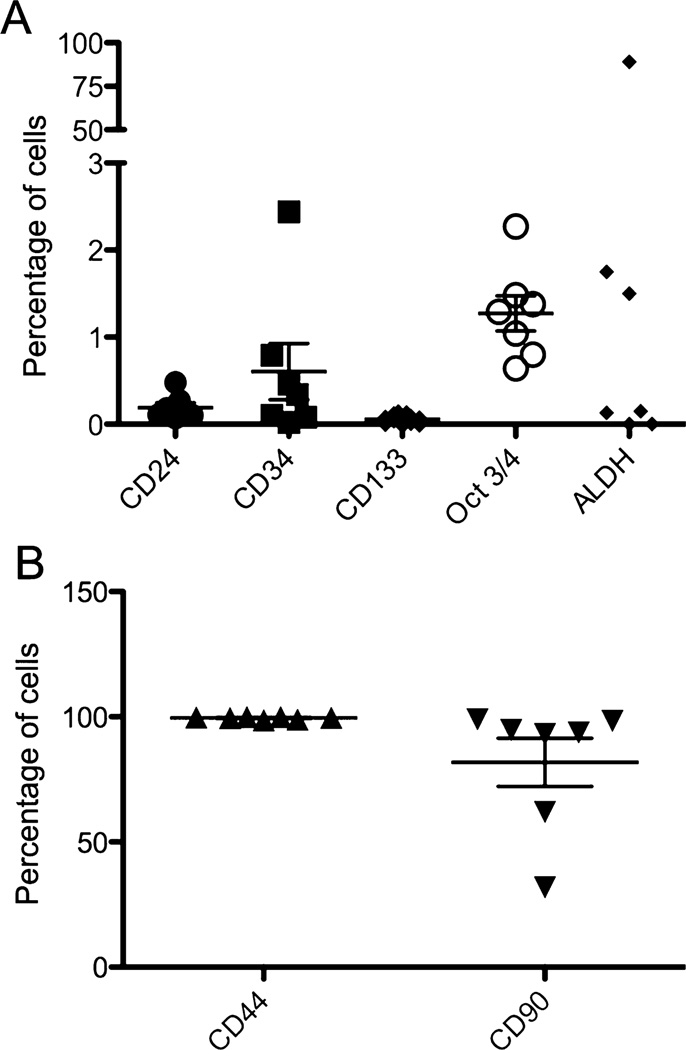
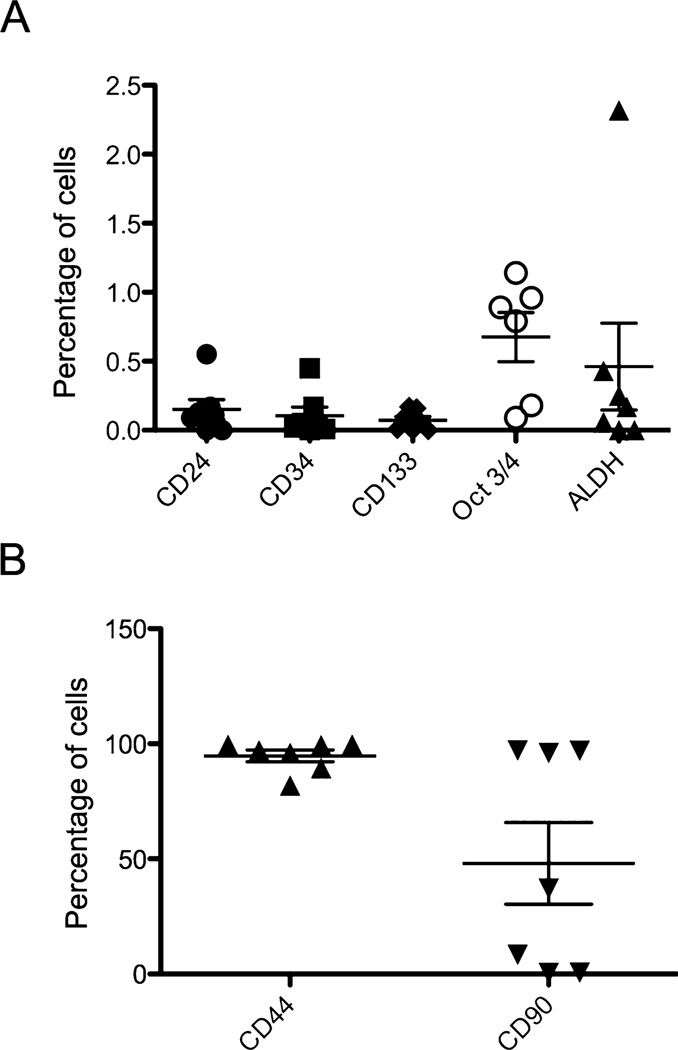
Canine melanoma and OSA cell lines were assessed for expression of the CSC surface antigens CD44, CD34, CD24, CD133 and CD90. As shown in Figs. 1 and 2 and Supplementary Fig. 1 (S1) and 2 (S2), the melanoma and OSA cell lines showed very low levels surface expression (defined as a discrete population with more than a log shift in fluorescence intensity verses the isotype control) for the CSC antigens examined, except for CD44, CD90, and CD24. Melanoma and OSA cell lines were strongly positive for CD44: (melanoma-average 99.5% CD44+ CSC and OSA-average 94.7% CD44+ CSC). The tumor cell lines also strongly expressed CD90: (average 81.7% CD90+ CSC for melanoma and average 47.9% CD90+ CSC for OSA), while expression of CD24 by CSC was low (average 0.2% CD24+ CSC for both melanoma and OSA cell lines). CD24 expression by individual cell lines was variable, with some cell lines negative while others demonstrated small but discrete CD24+ populations of CSC (Figs. 1 and 2). Very low levels of expression of CD34 and CD133 were also observed.
Fig. 1.
Expression of CSC markers by melanoma cells from cell lines. Melanoma cells were immunostained with the indicated antibodies as described in Methods. In (A), the expression of putative CSC markers in a small percentage of cells was plotted. In (B), the percentage of cells positive for CD44 and CD90 expression was plotted.
Fig. 2.
Expression of CSC markers by OSA cells from cell lines. OSA cells were immunostained with the indicated antibodies as described in Methods In (A), the expression of putative CSC markers in a small percentage of cells was plotted. In (B), the percentage of cells positive for CD44 and CD90 expression was plotted.
In addition, OSA and melanoma cell lines were immunostained for expression of the intracellular transcription factor Oct3/4, an embryonically expressed transcription factor that has previously been demonstrated to identify CSC in canine mammary carcinoma biopsies (Webster et al., 2007). Interestingly, overall Oct3/4 expression was upregulated by all CSC in a given cell line, rather than by discrete CSC subpopulations as observed for CD90 and CD44, similar to what was observed for CD34 expression (Fig. S1B and S2B). All surface molecule staining and intracellular Oct3/4 identification using flow cytometry was repeated at least twice at different time points for each cell line to assess the stability of the CSC populations during periods of culture. Significant variation of expression of these 4 CSC antigens (ie, CD44, CD90, CD24, and Oct3/4) over time was not observed (data not shown), thus suggesting that CSC subpopulations were stable within the tumor cell lines used in this study.
OSA and melanoma cell lines were also evaluated for ALDH activity. As shown in Fig. S1C, 1, S2C and 2, most cell lines did not appear to have large subpopulations of ALDH+ CSC and in fact many cells lines exhibited a slight shift of all the cells in the ALDH treated verses DEAB control suggesting that perhaps all of the tumor cells express some degree of ALDH activity, except for 2 of the 7 OSA and 3 of the 7 melanoma cell lines where no shift was observed (data not shown). Interestingly, one cell line (CMSDJe) contained close to 100% ALDH+ high expressing CSC (Fig. S1D). Therefore, we concluded overall that ALDH expression by CSC was distinctly uncommon in canine OSA and melanoma cell lines.
3.2. CSC subpopulations in melanoma FNAs
We next examined CSC subpopulations in fresh biopsies of melanoma and OSA tumor tissues. Samples for analysis of CSC subpopulations in melanoma were obtained by fine-needle aspiration (FNA), whereas samples from OSA were obtained by enzymatic digestion of small tumor tissue fragments obtained by punch biopsy from freshly amputated limbs of treatment-naïve dogs.
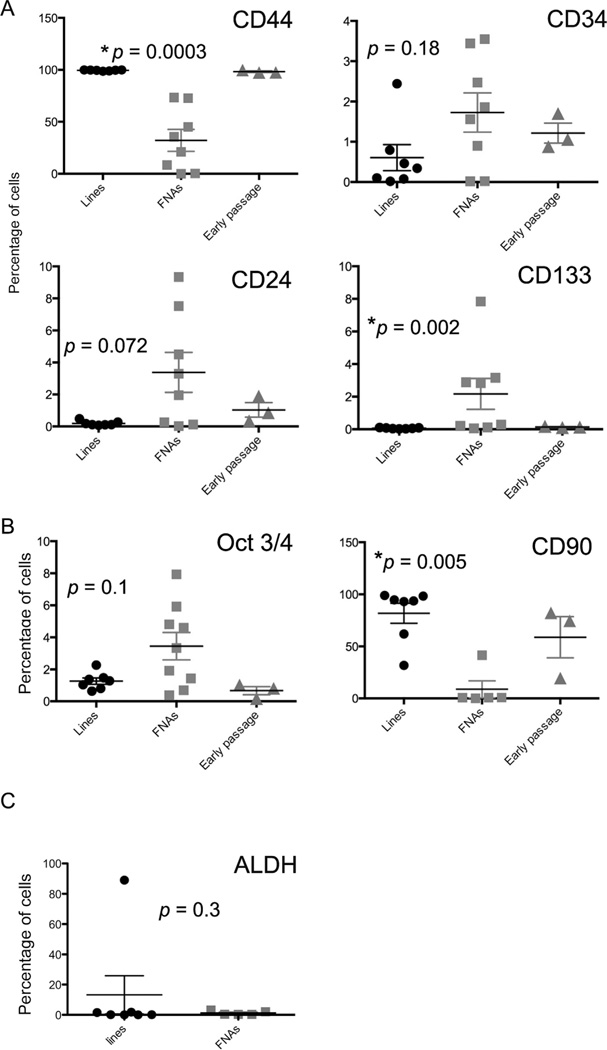
Eleven melanoma biopsies were examined for expression of CSC surface and/or intracellular antigens (Fig. 3 and Supplementary Fig. 3 (S3)). Analysis of these samples revealed striking differences in CSC subpopulations between fresh biopsies and cell lines (Fig. 3). For example, only approximately 32% of tumor cells (defined as CD45 negative cells) expressed CD44 in FNA samples, compared to virtually 100% of tumor cell line cells expression CD44 (Fig. 3). Interestingly, the dot plot and histogram analysis of CD44 expression in tumor biopsy samples reveals CD44-high and CD44-low sub-populations, whereas in typical cell lines there was a single peak of high positive staining. One caveat of the CD44 expression analysis of tumor biopsy samples is that our analysis criteria (i.e., only CD45− cells analyzed) did not necessarily exclude host stromal cells (e.g., fibroblasts and endothelial cells) from analysis.
Fig. 3.
Analysis of CSC marker expression by melanoma cells from cell lines versus FNA biopsy samples. (A) Comparison of percentage of surface marker staining of melanoma cell lines (“lines”, passaged >20 times), fresh melanoma tumor biopsy samples (“FNAs”), and “Early passage”, consisting of cell lines in passage numbers 1,6 and 7.p-Values are shown above each graph and reflect the comparison of the cell lines and the FNAs. (B) Comparison of percentage of Oct3/4 staining (left) and CD90 staining (right). n = 7 cell line samples, 8 FNA samples, and 3 Early passage samples for Oct3/4 and n = 7 cell line samples, n = 5 FNA samples, and n=3 Early passage samples for CD90. (C) ALDH staining of melanoma cell lines (n = 7) and FNA samples (n = 5). *p-Values less than 0.05 were considered significant.
Differences in CD90 expression by tumor biopsies versus cell line samples were also noted, with FNA samples containing a much smaller percentage of CD90+ CSC than cell line CSC (Fig. 3). Overall, in melanoma samples, the major significant differences in CSC subpopulations were noted between tumor biopsy samples and cell lines with respect to all of the CSC antigens evaluated (Fig. 3). For example, melanoma tumor biopsy specimens had significantly more CD133+, CD24+, and Oct3/4+ CSC than cell lines, whereas cell lines contained significantly greater numbers of CD44+ and CD90+ CSC.
3.3. CSC subpopulations in OSA biopsy samples
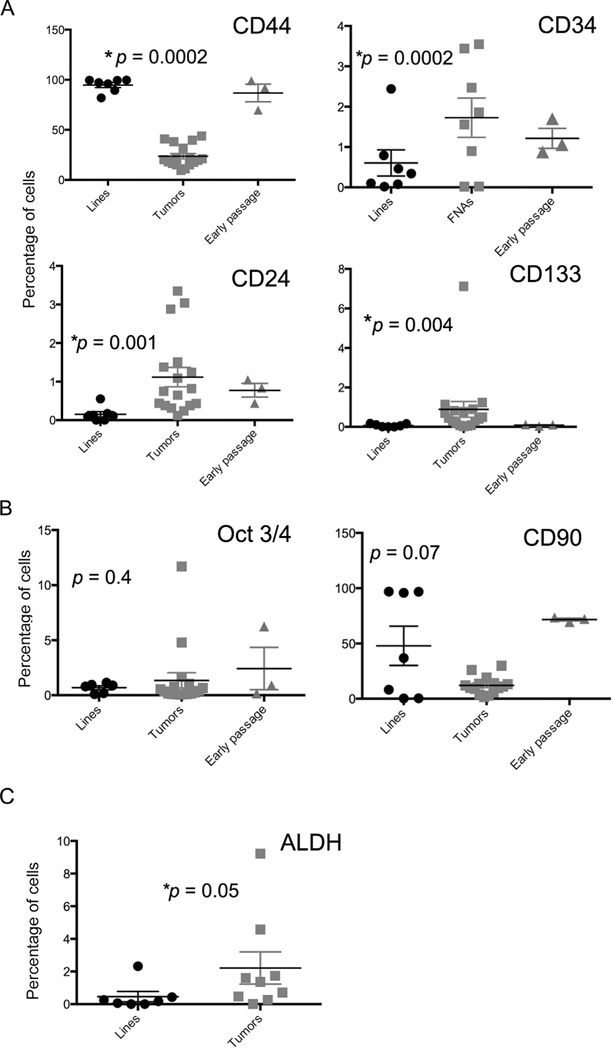
A similar analysis was done with 17 fresh, biopsy-isolated canine OSA samples compared to OSA cell lines (Fig. 4 and Supplementary Fig. 4 (S4)). Similar to melanoma, significant differences between the cell lines and the tumor biopsy samples were observed. The major differences were similar to those noted in melanoma samples, with tumor biopsy samples containing significantly more CD24+, CD133+, ALDH+, and CD34+ CSC than cell line samples, whereas cells lines contained significantly more CD144+ and CD90+ CSC than tumor biopsy specimen, while there was no difference in Oct3/4 expression between tumor cell lines and tumor biopsy samples for OSA. Of note, two of the cell lines we examined, D17 and Abrams, were derived from pulmonary tumor metastases. However, these cell lines were not different in their expression of putative CSC markers in our analysis compared to tumor cell lines derived from primary tumors, suggesting that either CSC subpopulations are similar between primary and metastatic lesions, or that culturing the cells over numerous passages leads eventually to similar levels of expression of putative CSC markers.
Fig. 4. gfmlmgnlmgln.
Analysis of CSC marker expression by OSA cells from cell lines versus FNA biopsy samples. (A) Comparison of percentage of surface marker staining of OSA cell lines (“lines”, passaged >20 times) and fresh OSA tumor biopsy samples (“FNAs”), and “Early passage”, consisting of cell lines in passage numbers 8, 7, and 10. p-Values are shown above each graph and reflect the comparison of cell lines and the FNAs. (B) Comparison of percentage of Oct3/4 staining (left) and CD90 staining (right). n = 7 cell line samples, 8 FNA samples, and 3 Early passage samples forOct3/4 and n = 7 cell line samples, 3 FNA samples, and 3 Early passage samples for CD90. (C) ALDH staining of OSA cell lines (n = 7) and FNA samples (n = 5). *p-Value less than 0.05 was considered significant.
3.4. Analysis of early passage cell lines as compared to ex vivo samples and established cell lines
We examined early passages of cell lines for OSA (n = 3) and melanoma (n = 3). OSA and melanoma cell lines passaged between 1 and 10 times were assessed for surface markers and intracellular staining (Figs. 3 and 4). Our analysis suggested that for melanomas, the percentages of the CSC subpopulations expressing CD44, CD133, and CD90 were very similar to those observed in high passage number cell lines, while the expression of CD34, CD24, and Oct3/4 was similar to that observed in FNA specimens (Fig. 3). For OSA, a similar pattern was observed whereby the subpopulations expressing CD44, CD133, and CD90 resembled those of high-passage OSA lines, while expression of CD34, CD24, and Oct3/4 was similar to expression by tumor biopsy samples (Fig. 4). Thus, we concluded from these preliminary studies that time in culture and passage number preferentially affected CD44, CD133, and CD90 subset expression, whereas expression of CD34, CD24, and Oct3/4 was relatively resistant to the effects of cell passage.
3.5. Tumor-specific differences in CSC subpopulations in melanoma versus OSA tumors
Tumor-specific differences in melanoma versus OSA CSC subpopulations were also assessed (Table 1). In comparing cell lines, only expression of CD44 and Oct3/4 were significantly different between the melanoma and OSA cell lines, melanoma cell lines containing significantly more of both CSC subpopulations. For tumor biopsies, significant differences in percentages of CSC subpopulations were noted for Oct3/4+ and CD90+ subpopulations, though these differences were not numerically large. Thus, these results indicated that the heterogeneity of CSC subpopulations in vivo in melanoma versus OSA tumors was relatively small.
Table 1.
Comparison of percentages of putative CSC populations between the two tumor types. Top: comparison of the OSA and melanoma tumor cell lines, bottom: comparison of ex vivo OSA tumor biopsies and melanoma FNAs.
| Marker | OSA-average | OSA-min/max | Mel-average | Mel-min/max | p-Value |
|---|---|---|---|---|---|
| Cell line analysis | |||||
| CD44 | 94.7% | 81.9–99.6% | 99.5% | 98.7–99.9% | 0.04* |
| CD34 | 0.1% | 0–0.5% | 0.6% | 0.02–2.4% | 0.06 |
| CD24 | 0.2% | 0–0.6% | 0.2% | 0.1–0.5% | 0.52 |
| CD133 | 0.07% | 0–0.2% | 0.06% | 0.02–0.1% | 0.94 |
| CD90 | 47.9% | 0.4–96.9% | 81.8% | 31.8–98.9% | 0.31 |
| Oct 3/4 | 0.6% | 0–1.1% | 1.3% | 0.8–2.3% | 0.03** |
| ALDH | 0.5% | 0–2.3% | 13.2% | 0–89% | 0.80 |
| Tumor biopsy analysis | |||||
| CD44 | 23.5% | 9.3–43.8% | 32% | 0.1–73.4% | 0.77 |
| CD34 | 4.4% | 0.6–14.6% | 1.7% | 0.02–3.6% | 0.12 |
| CD24 | 1.1% | 0.1–3.4% | 3.4% | 0.02–9.3% | 0.38 |
| CD133 | 0.9% | 0.02–1.2% | 2.2% | 0.1–7.8 | 0.44 |
| CD90 | 12% | 1.5–30% | 8.8% | 0.06–41.6 | 0.05* |
| Oct 3/4 | 1.3% | 0.1–11.7% | 3.5% | 0.4–7.9% | 0.006* |
| ALDH | 2.2% | 0.01–9.2% | 1.3% | 0.3–3.2% | 0.79 |
Statistically significant (p ≥ 0.05).
4. Discussion
The primary goal of this study was to assess the relative frequency and diversity of putative CSC subpopulations in canine melanoma and OSA tumor cell lines and tissue biopsies. The study major findings were that substantial heterogeneity in tumor CSC subpopulations existed between canine tumor cell lines and tumor biopsies, while between-tumor type differences in CSC subpopulations were relatively minor. This study was novel in that relatively large sample sizes were utilized, and because two different common tumor types of dogs were compared, using a relatively large panel of potential CSC antibodies. Thus, our results may more closely reflect results that can be expected for analysis of different canine CSC subpopulations using flow cytometery.
One limitation to our study is that we only examined singly stained cells, as opposed to CSC immunostained concurrently with multiple markers for examination of specific subpopulations of cells. For example, in humans, breast cancer stem cells are defined as CD44+/CD24−. Hence, while our study provides a template for determining what CSC markers identify subpopulations of tumor cells in dogs, it would be of interest to further characterize these subpopulations looking at the expression of multiple markers on these cells.
Blacking et al. also recently compared cell line and tumor biopsy CSC populations in smaller numbers of tumors of dogs with melanoma (Blacking et al., 2012). In that study, differences in CSC populations in tumor samples and tumor cell lines were noted. Moreover, this study also demonstrated that when fresh tumor samples were cultured in vitro, the CSC subpopulations changed over time and came to resemble those of more established canine tumor cell lines, though this analysis was only done using 2 low-passage tumor cell lines. However, the study by Black-ling et al. was limited by the small numbers of each tumor type examined (only 1 OSA sample, 1 melanoma cell line, and only 1 ex vivo OSA sample was examined). In addition, that study also evaluated a more limited set of CSC markers (CD44, CD34, CD117, CD133 and ALDH). Nonetheless, our data are consistent with the preliminary results presented in that earlier paper.
Other studies have examined CSC subpopulations in various canine tumors (Wilson et al, 2008; Cocola et al, 2009; Michishita et al., 2010; Pang and Argyle, 2009; Stoica et al, 2009; Cogliati et al, 2010; Blacking et al., 2011; Webster et al., 2007). One of these studies (Wilson et al, 2008) examined canine OSA cell lines for embryonic gene expression, including analysis of Oct4 expression. Interestingly, little expression of Oct 4 was noted in adherent OSA lines, but Oct4 expression increased when the cells were cultured in non-adherent media forming sarcospheres. We also identified a small subpopulation (average = 0.6%) of Oct3/4+ OSA cells in our OSA cell lines. The percentage of Oct3/4+ cells we noted in our OSA tumor biopsy samples were similar to those noted in the sarcosphere cultures, suggesting that perhaps in vitro sarcosphere culture may select for cells with increased expression of Oct4.
Our results and those of others demonstrate that certain canine CSC subpopulations, especially CD44+ and CD90+ CSC, are increased when tumor cell lines are compared to tumor biopsy samples (Blacking et al., 2012). On the other hand, other CSC populations are significantly reduced or lost in canine tumor cell lines, including CD34+, CD133+, Oct3/4+, CD24+, and ALDH+ CSC. These differences in CSC subpopulations in cultured cell lines versus primary tumors may occur because of the effects of the tumor microenvironment in shaping CSC populations (Smith et al., 2011; Jinushi et al., 2011) and because it is likely that cells cultured over time select for dominant subpopulations, thus diminishing overall CSC heterogeneity (Blacking et al., 2012). In regards to the tumor microenvironment, evidence exists that macrophages can help protect CSCs from the effects of chemotherapy via expression of the factor milk-fat globule-epidermal growth factor-VIII (MFG-E8) (Jinushi et al., 2011) and that immune stimulation via vaccination selects for a population of immune-resistant CSCs that display increased expression of Nanog, a transcription factor involved in conferring some stem cell properties to tumor cells (Noh et al., 2012). Culturing of tumor cells has been demonstrated to result in decreased CSC heterogeneity (Blacking et al., 2012) and may likely explain why almost all of the cultured canine tumor cell lines we examined had high expression of CD44 and CD90, as these markers may have been selected for over time.
Surprisingly, we observed limited CSC heterogeneity between OSA and melanoma tumor biopsies. These results suggest that the different CSC subpopulations may be fairly conserved between various tumor types in dogs, even in tumors of different tissue origin, though this supposition remains to be confirmed by additional studies. Thus, additional evaluation of CSC subpopulations, especially those that express CD90, CD24, CD34 and/or CD133 is warranted to determine the degree to which these CSC antigens are conserved between different tumor types and whether these CSC subpopulations exhibit CSC function properties such as spheroid growth in vitro. At present, it may be preferable whenever possible to study CSC populations in fresh tumor biopsies rather than in tumor cell lines.
Supplementary Material
Acknowledgements
We would like to thank Dr. Robyn Elmslie and Dr. Douglas Thamm for providing some of the cell lines used in this study as well as the clinical trials team at Colorado State University for collection of tumor biopsy samples. This work was supported by a grant by the National Center for Research Resources (5K01OD010911-04) to (A.M.G.) that is currently supported by the Office of Research Infrastructure Progarms/OD, by the Merck-Merial Summer Research Program (to M.D.) by a grant provided by the Colorado State University Supercluster fund (A.M.G. and S.W.D.) and by a grant from the Shipley Foundation (S.W.D.).
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vetimm.2014.07.006.
References
- Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat. Rev. Cancer. 2007;7(11):834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad O, Yustein JT, Shah P, Gul N, Katuri V, O’Neill A, Kong Y, Brown ML, Toretsky JA, Loeb DM, High ALDH activity identifies chemotherapy-resistant Ewing’s sarcoma stem cells that retain sensitivity to EWS-FLI1 inhibition. PLoS ONE. 2010;5(11):e13943. doi: 10.1371/journal.pone.0013943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radiore-sistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Blacking TM, Waterfall M, Samuel K, Argyle DJ. Flow cytometric techniques for detection of candidate cancer stem cell subpopulations in canine tumour models. Vet. Comp. Oncol. 2012;10(4):252–273. doi: 10.1111/j.1476-5829.2011.00293.x. [DOI] [PubMed] [Google Scholar]
- Blacking TM, Waterfall M, Argyle DJ. CD44 is associated with proliferation, rather than a specific cancer stem cell population, in cultured canine cancer cells. Vet. Immunol. Immunopathol. 2011;141(1–2):46–57. doi: 10.1016/j.vetimm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Cameron SR, Dahler AL, Endo-Munoz LB, Jabbar I, Thomas GP, Leo PJ, Poth K, Rickwood D, Guminski A, Saunders NA. Tumor-initiating activity and tumor morphology of HNSCC is modulated by interactions between clonal variants within the tumor. Lab. Investig. J. Tech. Methods Pathol. 2010;90(11):1594–1603. doi: 10.1038/labinvest.2010.131. [DOI] [PubMed] [Google Scholar]
- Chon E, Thompson V, Schmid S, Stein TJ. Activation of the canonical Wnt/beta-catenin signalling pathway is rare in canine malignant melanoma tissue and cell lines. J. Comp. Pathol. 2013;148(2–3):178–187. doi: 10.1016/j.jcpa.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christgen M, Ballmaier M, Bruchhardt H, von Wasielewski R, Kreipe H, Lehmann U. Identification of a distinct side population of cancer cells in the Cal-51 human breast carcinoma cell line. Mol. Cell Biochem. 2007;306(1–2):201–212. doi: 10.1007/s11010-007-9570-y. [DOI] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat. Med. 2011;17(3):313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Cocola C, Anastasi P, Astigiano S, Piscitelli E, Pelucchi P, Vilardo L, Bertoli G, Beccaglia M, Veronesi MC, Sanzone S, Barbieri O, Reinbold RA, Luvoni GC, Zucchi I. Isolation of canine mammary cells with stem cell properties and tumour-initiating potential. Reprod. Domest. Anim. 2009;44(Suppl. 2):214–217. doi: 10.1111/j.1439-0531.2009.01413.x. [DOI] [PubMed] [Google Scholar]
- Cogliati B, Aloia TP, Bosch RV, Alves VA, Hernandez-Blazquez FJ, Dagli ML. Identification of hepatic stem/progenitor cells in canine hepatocellular and cholangiocellular carcinoma. Vet. Comp. Oncol. 2010;8(2):112–121. doi: 10.1111/j.1476-5829.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- Croker AK, Allan AL. Cancer stem cells: implications for the progression and treatment of metastatic disease. J. Cell. Mol. Med. 2008;12(2):374–390. doi: 10.1111/j.1582-4934.2007.00211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu. Rev. Med. 2007;58:267–284. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, Joshua B, Kaplan MJ, Wapnir I, Dirbas FM, Somlo G, Garberoglio C, Paz B, Shen J, Lau SK, Quake SR, Brown JM, Weissman IL, Clarke MF. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow SW, Fradkin LG, Liggitt DH, Willson AP, Heath TD, Potter TA. Lipid-DNA complexes induce potent activation of innate immune responses and antitumor activity when administered intravenously. J. Immunol. 1999;163(3):1552–1561. [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, Schott A, Hayes D, Birnbaum D, Wicha MS, Dontu G. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell. 2011;8(2):136–147. doi: 10.1016/j.stem.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Jinushi M, Chiba S, Yoshiyama H, Masutomi K, Kinoshita I, Dosaka-Akita H, Yagita H, Takaoka A, Tahara H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108(30):12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger JA, Kaplan CD, Luo Y, Zhou H, Markowitz D, Xiang R, Reisfeld RA. Characterization of stem cell-like cancer cells in immune-competent mice. Blood. 2006;108(12):3906–3912. doi: 10.1182/blood-2006-05-024687. [DOI] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC, Wong H, Rosen J, Chang JC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21(3):283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481(7379):85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- Masters JR, Koberle B. Curing metastatic cancer: lessons from testicular germ-cell tumours. Nat. Rev. Cancer. 2003;3(7):517–525. doi: 10.1038/nrc1120. [DOI] [PubMed] [Google Scholar]
- Michishita M, Akiyoshi R, Yoshimura H, Katsumoto T, Ichikawa H, Ohkusu-Tsukada K, Nakagawa T, Sasaki N, Takahashi K. Characterization of spheres derived from canine mammary gland adenocarcinoma cell lines. Res. Vet. Sci. 2010 doi: 10.1016/j.rvsc.2010.11.016. [DOI] [PubMed] [Google Scholar]
- Noh KH, Lee YH, Jeon JH, Kang TH, Mao CP, Wu TC, Kim TW. Cancer vaccination drives Nanog-dependent evolution of tumor cells toward an immune-resistant and stem-like phenotype. Cancer Res. 2012;72(7):1717–1727. doi: 10.1158/0008-5472.CAN-11-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare T, Corbin AS, Druker BJ. Targeted CML therapy: controlling drug resistance, seeking cure. Curr. Opin. Genet. Dev. 2006;16(1):92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Res. 2011;71(11):3802–3811. doi: 10.1158/0008-5472.CAN-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang LY, Argyle DJ. Using naturally occurring tumours in dogs and cats to study telomerase and cancer stem cell biology. Biochim. Biophys. Acta. 2009;1792(4):380–391. doi: 10.1016/j.bbadis.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs JL, McAllister RM, Lennette EH. Immunofluorescent studies of RD-114 virus replication in cell culture. J. Gen. Virol. 1974;25(1):21–29. doi: 10.1099/0022-1317-25-1-21. [DOI] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH. Identification of cells initiating human melanomas. Nature. 2008;451(7176):345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BH, Gazda LS, Conn BL, Jain K, Asina S, Levine DM, Parker TS, Laramore MA, Martis PC, Vinerean HV, David EM, Qiu S, Cordon-Cardo C, Hall RD, Gordon BR, Diehl CH, Stenzel KH, Rubin AL. Three-dimensional culture of mouse renal carcinoma cells in agarose macrobeads selects for a subpopulation of cells with cancer stem cell or cancer progenitor properties. Cancer Res. 2011;71(3):716–724. doi: 10.1158/0008-5472.CAN-10-2254. [DOI] [PubMed] [Google Scholar]
- Stoica G, Lungu G, Martini-Stoica H, Waghela S, Levine J, Smith R., 3rd Identification of cancer stem cells in dog glioblastoma. Vet. Pathol. 2009;46(3):391–406. doi: 10.1354/vp.08-VP-0218-S-FL. [DOI] [PubMed] [Google Scholar]
- Tang KH, Dai YD, Tong M, Chan YP, Kwan PS, Fu L, Qin YR, Tsao SW, Lung HL, Lung ML, Tong DK, Law S, Chan KW, Ma S, Guan XY. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73(7):2322–2332. doi: 10.1158/0008-5472.CAN-12-2991. [DOI] [PubMed] [Google Scholar]
- Wang ZA, Shen MM. Revisiting the concept of cancer stem cells in prostate cancer. Oncogene. 2010 doi: 10.1038/onc.2010.530. [DOI] [PubMed] [Google Scholar]
- Webster JD, Yuzbasiyan-Gurkan V, Trosko JE, Chang CC, Kiupel M. Expression of the embryonic transcription factor Oct4 in canine neoplasms: a potential marker for stem cell subpopulations in neoplasia. Vet. Pathol. 2007;44(6):893–900. doi: 10.1354/vp.44-6-893. [DOI] [PubMed] [Google Scholar]
- Wilson H, Huelsmeyer M, Chun R, Young KM, Friedrichs K, Argyle DJ. Isolation and characterisation of cancer stem cells from canine osteosarcoma. Vet. J. 2008;175(1):69–75. doi: 10.1016/j.tvjl.2007.07.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.