Abstract
Natural killer (NK) cells contribute to clinical responses in patients treated with rituximab, but the rules determining NK cell responsiveness to mAb therapies are poorly defined. A deeper understanding of the mechanisms responsible for antibody-dependent cellular cytotoxicity (ADCC) could yield useful biomarkers for predicting clinical responses in patients. Unlicensed NK cells, defined as NK cells lacking expression of an inhibitory KIR for self-HLA class I ligands, are hypo-responsive in steady-state, but are potent effectors in inflammatory conditions. We hypothesized that antitumor antibodies such as rituximab can overcome NK cell dependence on licensing, making unlicensed NK cells important for clinical responses. Here we examined the influences of variations in KIR and HLA class I alleles on in vitro responses to rituximab. We tested the clinical significance in a cohort of follicular lymphoma patients treated with rituximab-containing mAb combinations and show that rituximab triggers responses from all NK cell populations regardless of licensing. Neither IL-2 nor accessory cells are required for activating unlicensed NK cells, but both can augment rituximab-mediated ADCC. Moreover, in 101 follicular lymphoma patients treated with rituximab-containing mAb combinations, a “missing ligand” genotype (predictive of unlicensed NK cells) is associated with higher progression-free survival. Our data suggest that the clinical efficacy of rituximab may be driven, in part, by its ability to broaden the NK cell repertoire to include previously hypo-responsive, unlicensed NK cells. A “missing ligand” KIR and HLA class I genotype may be predictive of this benefit, and useful for personalizing treatment decisions in lymphomas and other tumors.
Keywords: natural killer, KIR, ADCC, lymphoma, rituximab
Introduction
Curing patients while sparing unnecessary toxicity remains the ultimate goal in cancer treatment. Monoclonal antibodies (mAb) hold this potential by recruiting a patient’s immune system to destroy cancer cells with minimal toxicity, but responses are unpredictable and rarely curative (1–5). Identifying the genetic and cellular factors that drive immune responses in mAb therapy may make clinical responses more predictable, and allow the identification of strategies for augmenting mAb-induced responses (6). Natural killer (NK) cells contribute to clinical responses in patients treated with rituximab, but the rules governing how NK cells respond to rituximab are unclear, thus limiting our ability to clinically manipulate and predict NK cell behavior in rituximab-treated patients (3, 7–9).
The number and ligand-specificity of inhibitory receptors expressed by an NK cell clone have been shown to modify NK cell-mediated cytotoxic responses (10–13). NK cells lacking inhibitory receptors specific for self-MHC class I are weakly responsive compared to fully competent (i.e., licensed) NK cells expressing one or more inhibitory receptors for self-MHC class I (10–14). However, these same inhibitory receptors can suppress the licensed NK cells through interactions with MHC class I ligands expressed by tumors. In human hematopoietic stem-cell transplantation and mouse CMV infection, limitations of NK cell licensing can be overcome by harnessing hypo-responsive NK cells lacking inhibitory receptors for self-MHC class I that may contribute to clinical responses, although data are conflicting (15–20). It is unknown whether rules of licensing similarly govern NK cell responses to antibody-coated hematologic malignancies and whether mAb alone can trigger these responses (12, 21). Because rituximab activates NK cells through CD16, and CD16 does not require co-activating signals to trigger NK cell responses (22), we hypothesize that rituximab can convert tolerant human NK cells lacking inhibitory Killer cell Ig-like Receptors (KIR) for self-MHC class I into potent killers. This may be a fundamental and potentially exploitable mechanism contributing to antitumor antibody clinical responses.
In the present study, we examined the influences of variations in human KIR and HLA class I alleles on in vitro responses to rituximab. We further tested the clinical significance of our in vitro findings in a cohort of follicular lymphoma patients treated with rituximab-containing antibody combinations (23, 24).
Materials and Methods
Follicular lymphoma patients
One hundred and two patients with previously untreated follicular lymphoma (FL) provided IRB-approved informed consent for collection of blood and biospecimens to be used for research related to his or her cancer, such as the correlative science aims of CALGB protocol #150905 (NCT01057459; NCT01749969). Eligible FL patients had previously untreated, stage III, IV, or bulky stage II disease and WHO tumor grade 1, 2, or 3a. Patients were treated with a non-cytotoxic strategy of rituximab-containing antibody combinations on CALGB protocols 50402 (rituximab with galiximab) and 50701(rituximab with epratuzumab) from 2005 through 2009. (23, 24) Forty-six of 62 patients (74%) from CALGB 50402 consented and had samples available, although one patient never began treatment and was excluded from the analysis. Fifty-six of 60 patients (93%) from CALGB 50701 consented and had samples available. One patient from 50701 was determined to have stage I disease at baseline during final chart review and was excluded from analysis. In all, 101 patients are included in this analysis (Table 2, Supplemental Figure 6).
Table 2.
Characteristics of patients from CALGB 50402 and CALGB 50701
| Treatment Protocol | p-value* | ||
|---|---|---|---|
| Rituximab + Epratuzimab | Rituximab + Galiximab | ||
| Number of patients | 55 | 46 | |
| Ethnicity | 1.00 | ||
| Caucasian | 50/53 (94.3%) | 45 (97.8%) | |
| Non-Caucasian | 3/53 (5.7%) | 1 (2.2%) | |
| Median age, y (range) | 54 (32–90) | 58 (22–84) | 0.75 |
| Female Gender | 32 (58.1%) | 16 (34.8%) | 0.03 |
| LDH > ULN | 5 (9.0%) | 7 (15.2%) | 0.37 |
| “B” symptoms | 4/53 (7.5%) | 4/43 (9.3%) | 1.00 |
| Follicular Lymphoma Stage | 0.74 | ||
| I | - | - | |
| II | 2 (3.6%) | 3/45 (6.7%) | |
| III | 18 (32.7%) | 16/45 (35.6%) | |
| IV | 35 (63.3%) | 26/45 (57.8%) | |
| FLIPI | 0.97 | ||
| 0–1 | 12/54 (22.2%) | 9/45 (20.0%) | |
| 2 | 24/54 (44.4%) | 20/45 (44.4%) | |
| ≥3 | 18/54 (33.3%) | 16/45 (45.6%) | |
| Histologic grade | 0.82 | ||
| Grade 1 | 30/53 (56.6%) | 27/45 (60.0%) | |
| Grade 2 | 20/53 (37.7%) | 17/45 (37.8%) | |
| Grade 3a | 3/53 (5.7%) | 1/45 (2.2%) | |
| Bone marrow | 0.69 | ||
| Positive | 30 (54.6%) | 23 (50.0%) | |
| Negative | 25 (45.4%) | 23 (50.0%) | |
| Bulky disease | 11/55 (20.0%) | 8/40 (20.0%) | 1.00 |
| ECOG PS | 0.05 | ||
| 0 | 33 (60.0%) | 33/45 (73.3%) | |
| 1 | 22 (40.0%) | 10/45 (22.2%) | |
| 2 | 0 (0.0%) | 2/45 (4.4%) | |
P-values for the categorical variables were computed using Fisher’s exact test; the Wilcoxon Rank Sums test was used to compute age differences between groups
KIR genotyping, HLA genotyping, and KIR ligand assignment
Genomic DNA was extracted from peripheral blood mononuclear cells (PBMC) by using a QIAmp DNA Blood Mini Kit (Qiagen). KIR genotyping was performed as previously described (25, 26). HLA class I genotyping was performed by PCR using a combination of sequence-specific primers and PCR-specific oligonucleotide probes. The CLIA-approved Immunogenetics Laboratory at the University of California, San Francisco, performed the genotyping. HLA-C and -B alleles were segregated into KIR ligand groups: C1 (HLA-CAsn80), C2 (HLA-CLys80), and HLA-Bw4 or -Bw6, and KIR haplotypes were assigned as previously described (25).
In vitro culture conditions
For functional experiments, blood was obtained from healthy volunteer de-identified leukocyte reduction filters (Blood Centers of the Pacific, San Francisco, CA). PBMCs were separated by density gradient centrifugation (Histopaque-1077 Sigma, St. Louis, MO) and were suspended in 10% dimethyl sulfoxide (DMSO, Fisher Scientific, Pittsburgh, PA) and 90% fetal bovine serum (FBS; Omega, Tarzana, CA), and then stored in liquid nitrogen. For NK cell recovery, cryovials of PBMCs were transferred to a 37°C water bath, thawed quickly in RPMI-1640 media (with 20% FBS, warmed to 37°C), and then washed in complete cell culture media (RPMI-1640 with 10% FBS, 2 mM glutamine, 100 U/ml penicillin and 100 ug/ml streptomycin; Cell Culture Facility, University of California, San Francisco). Cells were counted and viability was confirmed using a Vi-Cell XR (Beckman Coulter Inc, Brea, CA). Cells were cultured overnight at 37°C with 5% CO2 in a 24-well plate at a concentration of 3x106/ml of RPMI-1640 media. Exogenous IL-2 was not routinely added to the culture conditions, but only added (1,000 U/ml of IL-2, Biovision, Milpitas, CA) in experiments testing the specific contribution of IL-2. NK cells were isolated from PBMCs using a MACS NK Isolation kit (Miltenyi Biotec Inc., Auburn, CA).
Tumor cells
Three cell lines were used: human CD20+ HLA class I-deficient EBV-transformed B lymphoblastoid cell line 721.221, Raji Burkitt’s lymphoma cell line, and the human erythroleukemia cell line K562. All cell lines were cultured in complete RPMI-1640 media. CD20 expression was confirmed by staining with Brilliant Violet 421-conjugated anti-human CD20 (clone 2H7, BioLegend, San Diego, CA). HLA-A, -B, -C expression was monitored weekly, and before each experiment, by using FITC-conjugated anti-human HLA-A, -B, -C antibody (clone G46-2.6, BD Bioscience, Chicago, IL). The FITC-conjugated anti-human HLA-Bw4 mAb (One Lambda, Canoga Park, CA) was used to evaluate cell surface expression of HLA-Bw4. All three cell lines were validated to be mycoplasma-free; except for the expression of CD20 and other cell-surface markers, no other authentication assays were performed.
Antibody-induced NK cell activation assays
To model the physiologic interaction of NK cells with antibody-coated tumors, we developed a co-culture system using resting human PBMCs from KIR- and HLA-genotyped subjects added to antibody-coated tumor cell lines. To assess individual NK cell responses, we measured both degranulation of NK cells by staining for LAMP-1 (Pacific Blue-conjugated anti-CD107a; clone H4A3, BioLegend, San Diego, CA) (15, 27), the lysosome-associated membrane protein up-regulated on the NK cell surface after NK cell stimulation, and intracellular IFNγ (V450-conjugated anti-IFNγ, clone B27; BD) expression as a marker of cytokine secretion, which both correlate with target cell death (12, 27). We initially determined the percentage of CD3−CD56+ NK cells in the PBMCs using antibodies for CD3 (APC-eFluor 780-conjugated anti-CD3, clone SK7, eBioscience, San Diego, CA), and CD56 (Brilliant Violet 605-conjugated anti-CD56, clone HCD56, BioLegend, San Diego, CA). Based on the percentage of CD3−CD56+ NK cells, PBMCs containing 1.2–3.0 x 105 NK cells were mixed with tumor cells coated with 0.1–1000 ug/ml of rituximab at an NK cell-to-target ratio (1:2) in sterile 24-well plates with RPMI-1640 +10% FBS. Cells were incubated for 4 hours at 37° C, 5% CO2. For CD107a detection, 5 ul/ml of Pacific Blue-conjugated anti-CD107a was added to the mixture of PBMCs and antibody-coated tumor cells in each well at the beginning of the incubation. For intracellular IFNγ staining, brefeldin A (BioLegend, San Diego, CA) was added to the mixture of PBMCs and antibody-coated tumor cells in each well after 1 hour of incubation. Fixation was performed after staining cell surface markers, and permeabilization was performed at the time of IFNγ staining, according to the manufacturer’s instructions (Fix & Perm Cell Permeabilization Kit, Invitrogen Life Technologies, Grand Island, NY).
NK cell subset analysis
To interrogate the relative responsiveness of individual NK cell subsets expressing a single KIR (spKIR), we designed a panel of fluorochrome-conjugated antibodies specific for individual NK cell receptors. To evaluate and control for the influence of cognate KIR and HLA interactions endowing effector function to individual NK cell subsets, we used HLA genotyping and HLA class I KIR ligand assignment to identify the presence or absence of the ligand for the inhibitory KIR. NK cells exclusively expressing an inhibitory KIR from a subject lacking the cognate HLA class I ligand were considered “unlicensed” spKIR-expressing NK cells. For NK cell phenotyping, a combination of the following antibodies was used: APC-eFluor 780-conjugated anti-CD3 (clone SK7, eBioscience, San Diego, CA), PerCp-conjugated anti-CD3 (clone SK7, BioLegend, San Diego, CA), Brilliant Violet 605-conjugated anti-CD56 (clone HCD56, BioLegend, San Diego, CA), APC-Cy7-conjugated anti-CD16 (clone 3G8, BioLegend, San Diego, CA), FITC-conjugated anti-KIR2DL1 (clone 143211, R&D Minneapolis, MN), Alexa fluor 700-conjugated anti-KIR3DL1 (clone DX9, BioLegend, San Diego, CA), PE-Cy7-conjugated anti-KIR2DL2/L3/S2 (clone GL183, Beckman Coulter, Brea, CA), APC-conjugated anti-NKG2A (clone Z199, Beckman Coulter, Brea, CA), and PE-conjugated anti-KIR3DL2 (clone DX31, UCSF Hybridoma Core). Unstained controls, isotype-matched Ig controls, single-color controls (BD Compbeads), and FMO controls were used for multicolor compensation and gating. The isotype-matched Ig control for CD107a is Pacific Blue-conjugated mouse IgG1 (MOPC-21, BioLegend, San Diego, CA); IFNγ staining was performed by using an isotype-matched Ig control or anti-IFNγ antibody combined with anti-CD3, anti-CD56, and anti-KIR antibodies, allowing precise gating of IFNγ+ cells.
Gating strategy
Cells were analyzed on an LSR Fortessa flow cytometer (BD Biosciences, San Jose, CA) using FACSDiva software. Data were further processed with FlowJo software (v9.5.2, Tree Star, Inc). Lymphocytes were gated on forward and side light scatter parameters after excluding doublets. NK cell subsets exclusively expressing a single inhibitory KIR were detected after gating on the CD3−CD56+ NK cell population (Supplemental Figure 1).
Cytotoxicity assay
We performed cytotoxicity assays using sorted NK cell populations and PKH-26-labeled 721.221 lymphoblasts to validate the degranulation and cytokine measurements, as previously described (12, 28). Both direct killing and ADCC were tested, in triplicates, using labeled target cells coated with or without 10 ug/ml of rituximab and mixed with unlicensed NK cells at an effector:target cell ratio of 1:1, 2:1, and 5:1. Dead 721.221 lymphoblast target cells were defined as TO-PRO3+PKH−26+, after subtracting spontaneous death of 721.221 cells cultured without NK cells.
Statistics
Patients were classified based on the presence or absence of cognate HLA class I ligand (determined by HLA genotype) for inhibitory KIR (determined by KIR genotype). HLA-A and -B alleles were grouped based on the presence or absence of the Bw4 epitope; HLA-C status was characterized by the presence of HLA-C1 alleles only, -C2 alleles only, or both. “Missing ligand” was defined as the absence of cognate HLA class I ligand in the presence of the gene encoding its corresponding inhibitory KIR. The primary outcomes for this study were complete response and overall best response (29). The secondary endpoint was progression-free survival (PFS), defined as time from study entry to progression, relapse, or death, whichever occurred first. Probabilities of PFS were estimated using the Kaplan-Meier method, and the log-rank test was used to evaluate differences in survival distributions based on ligand status. To investigate statistically significant differences in NK cell function, NK cell subsets were compared using the Mann-Whitney, Wilcoxon, or Kruskal-Wallis tests. All p-values reported in this paper are two-sided. Statistical analyses were conducted by the Alliance Statistics and Data Center, using data collected through November 2012.
Results
Estimation of the size of NK cell subpopulations
We used an exclusion gating technique to enumerate specific NK cell populations expressing a single KIR (Supplemental Figure 1). We focused on five NK cell subsets: spKIR2DL2/3, spKIR2DL1, spKIR3DL1, spKIR3DL2, and KIR−. We operationally defined “KIR−” NK cells as lacking KIR2DL1, 2DL2/S2, 2DL3, 3DL1, KIR3DL2, and NKG2A. We used HLA class I genotyping and KIR ligand assignment to further characterize spKIR+ NK cells (NKG2A−) as either “licensed” (cognate HLA class I ligand present in the subject) or “unlicensed” (subjects lacking cognate HLA class I ligand). The frequency of spKIR+ and KIR− NK cells in the 19 study subjects was variable (Table 1), consistent with prior reports (21, 30, 31). The proportion of immature CD56brightCD16dim and mature CD56dimCD16bright NK cells within KIR−, unlicensed, and licensed NK cell subsets was comparable (Supplemental Figure 4). To assess and compare NK cell subset responses within each subject, we measured tumor-triggered degranulation (CD107a) and cytokine production (IFNγ) of spKIR+ NK cell populations after excluding NKG2A and other KIR (Supplemental Figure 1).
Table 1.
Frequency of NK cell populations and HLA KIR ligand status in healthy study subjects
| Subject no. | Percentages of Single-Positive NK cell subsets (%)
|
HLA KIR Ligands
|
||||||
|---|---|---|---|---|---|---|---|---|
| KIR2DL2/L3 | KIR2DL1 | KIR3DL1 | KIR3DL2 | KIR− | HLA-B | HLA-C | HLA-A | |
| 1 | 3.23 | 2.82 | 1.97 | 3.06 | 8.22 | Bw4/Bw6 | C2C2 | 24,32 |
| 2 | 6.56 | 5.30 | 5.80 | 2.98 | 18.20 | Bw4/Bw6 | C1C1 | 03,26 |
| 3 | 5.52 | 0.41 | 0.78 | 1.52 | 2.78 | Bw6/Bw6 | C1C1 | 02,02 |
| 4 | 7.56 | 5.06 | 1.06 | 3.69 | 11.40 | Bw4/Bw6 | C2C2 | 03,26 |
| 5 | 22.4 | 4.78 | 0.00 | 2.75 | 17.30 | Bw6/Bw6 | C1C1 | 02,24 |
| 6 | 6.85 | 1.48 | 0.71 | 1.89 | 5.49 | Bw4/Bw6 | C1C2 | 02,11 |
| 7 | 9.45 | 1.43 | 0.97 | 3.27 | 6.13 | Bw4/Bw6 | C1C1 | 03,29 |
| 8 | 4.97 | 2.91 | 1.96 | 7.10 | 11.20 | Bw6/Bw6 | C1C1 | 02,24 |
| 9 | 4.41 | 1.81 | 3.85 | 1.37 | 12.70 | Bw4/Bw6 | C2C2 | 02,30 |
| 10 | 9.17 | 6.70 | 2.58 | 1.61 | 14.40 | Bw6/Bw6 | C1C1 | 02,11 |
| 11 | 2.32 | 1.24 | 1.54 | 0.44 | 2.30 | Bw6/Bw6 | C1C2 | 11,31 |
| 12 | 3.00 | 1.21 | 2.58 | 2.74 | 7.54 | Bw4/Bw6 | C1C2 | 02,31 |
| 13 | 5.78 | 9.56 | 0.00 | 4.11 | 16.70 | Bw4/Bw6 | C1C2 | 03,30 |
| 14 | 3.76 | 4.81 | 0.00 | 1.88 | 4.94 | Bw4/Bw6 | C1C2 | 02,68 |
| 15 | 8.01 | 1.23 | 1.02 | 1.09 | 5.17 | Bw4/Bw6 | C1C2 | 02,11 |
| 16 | 1.77 | 1.45 | 0.62 | 1.22 | 3.57 | Bw4/Bw4 | C1C2 | 01,02 |
| 17 | 9.51 | 5.20 | 5.61 | 1.53 | 16.80 | Bw4/Bw6 | C1C1 | 03,26 |
| 18 | 8.24 | 1.01 | 4.09 | 3.05 | 10.10 | Bw4/Bw6 | C1C1 | 03,24 |
| 19 | 5.12 | 2.01 | 1.44 | 3.61 | 9.35 | Bw4/Bw6 | C1C1 | 02,24 |
KIR ligands are assigned based on HLA class I genotyping.
NK cell subsets are NKG2A−.
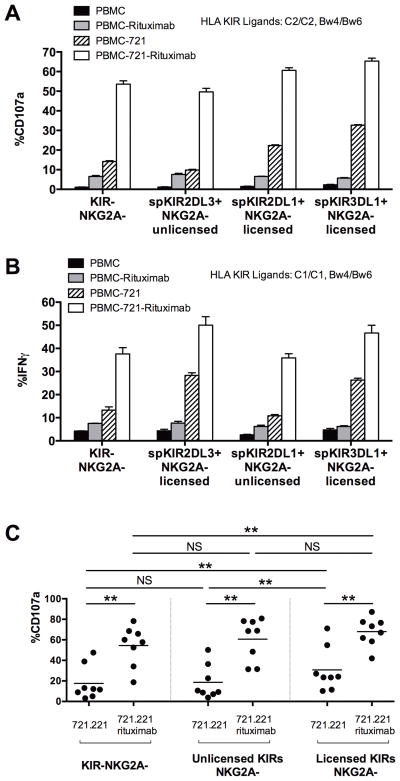
Rituximab stimulates robust degranulation and IFNγ secretion from KIR−NKG2A− and unlicensed spKIR+NKG2A− NK cells
To control for inter-individual variability in NK cell responses, we compared NK cell responses within subjects possessing both licensed and unlicensed spKIR+ populations. For the functional experiments, we used KIR and HLA class I genotyping to deliberately select subjects lacking a self-HLA class I ligand for an inhibitory KIR. As expected, the NK cell repertoire had variable responses to the CD20-expressing, HLA class I-deficient B-cell lymphoblast 721.221 cell line (and K562 cell line), consistent with a role for cognate NK cell KIR - HLA interactions in endowing a “missing self” response to HLA class I-deficient lymphoid and myeloid tumors (Figure 1A, 1B, 1C and Supplemental Figure 2). In direct 721.221 cell killing assays, licensed spKIR+ NK cells showed stronger responses than unlicensed spKIR+NKG2A− NK cells and KIR-NKG2A- NK cells. Adding rituximab to CD20+ 721.221 cells, however, allowed all NK cells to become activated, triggering NK cell degranulation from the hypo-responsive subsets of unlicensed NK cells expressing a spKIR2DL3 receptor (NKG2A−) and KIR−NKG2A− NK cells (Figure 1A). Confirming the NK cell degranulation responses, rituximab-coated B-lymphoblasts similarly triggered IFNγ release from hypo-responsive KIR−NKG2A− and unlicensed spKIR+NKG2A− NK cells, and increased cytotoxicity mediated by unlicensed NK cells (Figure 1B and Supplemental Figure 3). The presence of NKG2A minimally enhanced the responsiveness of NK cells to rituximab-coated 721.221 (data not shown). Aggregating the analysis for 8 subjects, rituximab consistently augmented the activity of previously tolerant, hypo-responsive NK cells and abolished the functional hierarchy observed when stimulating NK cells with tumor alone (Figure 1C). These data suggest that coating hematologic tumors with antitumor antibody may overcome intrinsic mechanisms of NK cell tolerance, unleashing the full repertoire of NK cells for a therapeutic benefit.
Figure 1. Rituximab triggers degranulation and IFNγ release from hypo-responsive KIR-NKG2A− and unlicensed spKIR+NKG2A− NK cells.
(A) The vertical axis depicts the percentage of NK cells expressing CD107a (a marker of degranulation) after 4 hours of incubation with CD20+ B-cell lymphoblasts (721.221) +/− rituximab. Identification of degranulation by discrete NK cell subsets was performed as shown in Figure 1, using freshly isolated, resting PBMCs from subject # 1 (HLA-C2/C2, Bw4/Bw6). The KIR−NKG2A− and unlicensed spKIR2DL3+NKG2A−NK cell subsets responded poorly to CD20+ lymphoblasts alone, but coating the lymphoblasts with 10 ug/ml of rituximab for 30 minutes activated these hypo-responsive NK cell subsets. (B) Intracellular IFNγ responses between KIR−NKG2A− and unlicensed spKIR2DL1+NKG2A− NK cells from subject #2 (HLA-C1/C1, Bw4/Bw6) were consistent with the degranulation responses, with a 2–3-fold increase in the percentage of hypo-responsive, KIR−NKG2A− and spKIR2DL1+NKG2A− NK cell subsets releasing IFNγ after the addition of rituximab. (C) Aggregate degranulation responses of NK cells exclusively expressing an unlicensed KIR, licensed KIR, or KIR−NKG2A− from 8 subjects with distinct KIR - HLA genotypes and circulating NK cell repertoires. Data were combined into three functional groups: NK cells lacking KIR2DL1, 2DL2, 2DL3, 3DL1, and NKG2A (KIR−NKG2A−); NK cells expressing unlicensed KIR (spKIR3DL1+HLA-Bw6/6, KIR2DL1+HLA-C1/C1, or KIR2DL2/3+HLA-C2/C2); and NK cells expressing licensed KIR (spKIR3DL1+HLA-Bw4, KIR2DL1+HLA-C2, or KIR2DL2/3-HLA-C1). Statistical comparisons were made using the Wilcoxon test, with **P<0.01. In subjects with more than 1 unlicensed or licensed KIR+ NK cell subset, the dot represents the total number of CD107a+ spKIR NK cells divided by the number of spKIR NK cells. Experiments were performed in triplicates; horizontal bar represents mean, with standard errors of mean.
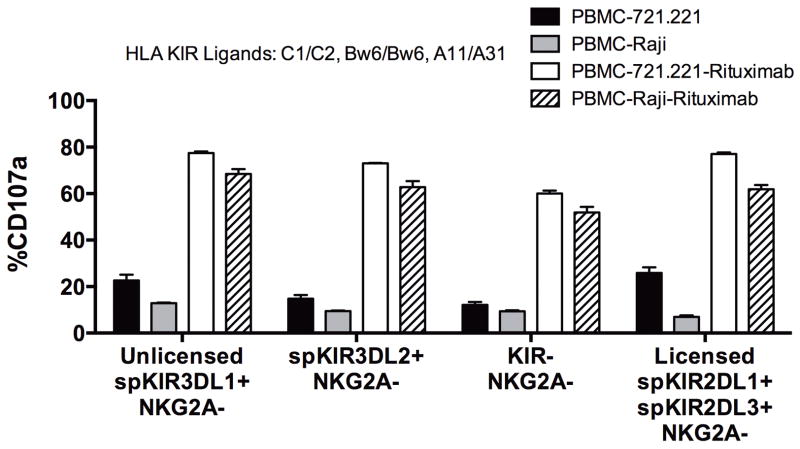
Rituximab augments the activity of hypo-responsive NK cells against the Raji Burkitt’s lymphoma cell line
To test whether the antibody-mediated triggering of hypo-responsive NK cells was restricted to rituximab-coated HLA class I-deficient 721.221 B-lymphoblasts, we evaluated NK cell activity in KIR-ligand compatible HLA-Bw6/6 subjects against the patient-derived, CD20-expressing, HLA-Bw6/6 Raji Burkitt’s lymphoma cell line, which we confirmed lacks expression of HLA-Bw4 by staining with anti-HLA-Bw4 specific mAb (data not shown). Consistent with results using the 721.221 cell line, unlicensed spKIR3DL1+NKG2A− NK cells from an HLA-Bw6/Bw6, C1/C2 subject were tolerant of Raji B-lymphoblast cells (Figure 2). By contrast, coating Raji tumors with rituximab stimulated robust degranulation by the unlicensed spKIR3DL1+NKG2A− NK cells (Figure 2). Unlicensed spKIR3DL2+NKG2A− NK cells had a similar response. This antibody-induced activation of hypo-responsive NK cells by rituximab likely reflects the uniquely potent calcium flux and signaling cascade triggered by CD16 ligation (22).
Figure 2. Coating Raji B-cell lymphoblast with rituximab overcomes intrinsic NK cell tolerance of the unlicensed spKIR3DL1+NKG2A−, KIR−NKG2A− and hypo-responsive spKIR3DL2+NKG2A− NK cell subsets.
Percentage of NK cells expressing cell surface CD107a exclusively expressing spKIR3DL1, spKIR3DL2, and NK cells lacking inhibitory KIR expression for self-HLA using subject 11 (HLA-C1/C2, Bw6/6, A11/A31) after 4 hours of incubation with a different CD20-expressing B-lymphoblast cell line (Raji) confirmed to match the HLA-B KIR ligand status of subject 11. Unlicensed spKIR3DL1+NKG2A−, spKIR3DL2+NKG2A−, and KIR−NKG2A− NK cells have minimal “missing self” activity against CD20+ HLA-class I-deficient 721.221 and HLA-Bw6/6 Raji B-lymphoblasts. Incubating both CD20+ B-lymphoblasts with 10ug/ml of rituximab stimulates robust degranulation from these hypo-responsive unlicensed NK cell subsets.
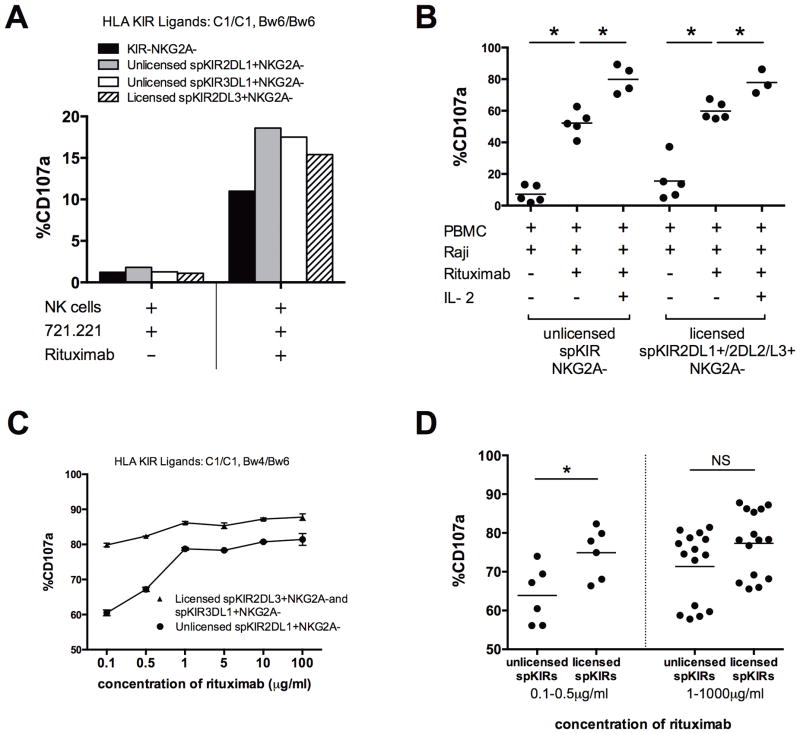
Unlicensed NK cell ADCC activity does not require accessory cells, and is modified by antibody dose and IL-2
Cytokines can overcome NK cell requirements for licensing and may independently augment the effector function of hypo-responsive NK cells (11, 13, 32). To examine the requirement of cytokine signaling, and to eliminate the confounding influence of cytokine production by accessory cells, we sorted NK cells prior to incubation with antibody-coated tumors and found similar rituximab-mediated triggering of unlicensed spKIR2DL1+NKG2A−, unlicensed spKIR3DL1+NKG2A−, and KIR−NKG2A− NK cells (Figure 3A). To address the specific contribution of IL-2, and to ask whether cytokines can enhance the antibody-dependent effector function of previously tolerant NK cells, PBMCs were primed overnight with 1,000 U/ml of IL-2 prior to the addition of tumor targets. Hypo-responsive NK cells expressing KIR without self-MHC class I ligands did not require IL-2 to degranulate in response to rituximab-coated Raji tumor cells, but IL-2 augmented the unlicensed NK cell ADCC activity compared with resting unlicensed NK cells (Figure 3B). IL-2 alone was sufficient to endow a degranulation response of hypo-responsive NK cells to direct stimulation by CD20+ Raji tumors, in the absence of rituximab (Supplemental Figure 5). Titration of rituximab revealed that the majority (≥ 55%) of both licensed and unlicensed NK cells can be activated with low and sub-therapeutic concentrations of rituximab (Figure 3C), but higher rituximab concentrations were required to generate comparable responses from unlicensed and licensed NK cells (Figure 3D) (33, 34). These data confirm that cytokines and accessory cells are not required for rituximab-dependent activation of unlicensed NK cells, and that activation of unlicensed NK cells may contribute to the clinical dose-response seen in patients treated with rituximab (35).
Figure 3. Rituximab alone is sufficient to activate unlicensed NK cells, which is augmented by IL-2 and higher doses of rituximab.
(A) To determine whether cytokines secreted from other mononuclear cells within the PBMC population are required for rituximab-dependent activation of unlicensed NK cells, NK cells from subject #8 (HLA-C1/C1, Bw6/Bw6) were purified by antibody-coated magnetic bead sorting and incubated for 4 hours with rituximab-coated 721.221 B-lymphoblasts. The CD107a response of NK cell subsets is shown; similar results were seen using subject #2 (data not shown). (B) To evaluate requirements for IL-2, PBMCs from subjects #2, #1, #8, and #11 were incubated overnight with or without 1,000 U/ml of IL-2 prior to co-culture with rituximab-coated Raji cell lines. (C) Escalation of rituximab concentration demonstrates dose-dependent activation of unlicensed NK cells (subject #17). (D) NK cells from 3 study subjects (#17, #18, and #19) were stimulated in vitro with increasing concentrations of rituximab and a fixed NK cell to 721.221 cell ratio of 1:2. CD107a was measured on individual NK cell subsets and licensing was determined based on KIR expression and HLA class I genotyping. Statistical significance was defined as p<0.05. Experiments were performed in duplicates for each study subject.
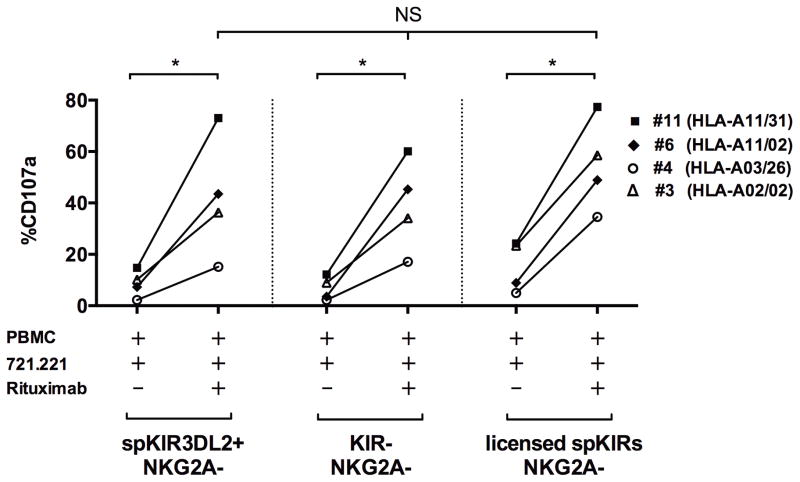
KIR3DL2+ NK cells are activated by rituximab
KIR3DL2 is encoded by a framework KIR gene present in all humans, expressed on 18–27% of NK cells, and reported to bind HLA-A3 and -A11 ligands in the presence of specific peptides (36). The function and contribution of spKIR3DL2+ NK cells, however, is unclear (36, 37). NK cells exclusively expressing KIR3DL2 from HLA-A3 or A11 healthy individuals are minimally responsive (37, 38). To determine whether KIR3DL2+ NK cells contribute to the ADCC response, we performed exclusion gating to isolate the function of spKIR3DL2+NKG2A− NK cells lacking KIR3DL1, KIR2DL1, KIR2DL2/3/S2, and NKG2A using NK cells from subjects with and without the cognate HLA-A3 or -A11 ligand. Similar to KIR−NKG2A− and unlicensed spKIR+NKG2A− NK cells, the 721.221 B-lymphoblasts alone triggered minimal degranulation, but the addition of rituximab activated the hypo-responsive spKIR3DL2+NKG2A− NK cell subset independent of HLA-A3 or –A11 (Figure 4). The expression of KIR3DL2 on unlicensed NK cells expressing another KIR without a self-MHC class I ligand did not significantly alter the responsiveness (data not shown). The abundant KIR3DL2+ NK cell subset, therefore, may require potent stimulation through CD16 for activation, and may contribute to the aggregate NK cell response in patients treated with rituximab.
Figure 4. Hypo-responsive NK cells exclusively expressing KIR3DL2 are activated by rituximab.
Shown is the degranulation (CD107a) response by NK cells exclusively expressing KIR3DL2 induced by 721.221 B-lymphoblasts with or without rituximab. CD107a results from 4 subjects were aggregated: subject #11 (HLA-A11/31), #6 (HLA-A11/02), #4 (HLA-A3/26), and #3 (HLA-A02/02). Similar to the KIR−NKG2A− (shown) and unlicensed spKIR+NKG2A− (not shown) NK cells, NK cells exclusively expressing KIR3DL2 were hypo-responsive to 721.221 B-lymphoblast tumor cells alone, but degranulated after the addition of rituximab, comparable to the licensed NK cell subset. Statistical comparisons were made using the Mann-Whitney and Kruskall-Wallis tests, with *P<0.05. The presence of the HLA-A11 (subject #11, #6) or -A3 (subject #4) ligand did not influence the antibody-dependent activation of the spKIR3DL2+ NK cell subset.
“Missing KIR ligand” is associated with PFS in follicular lymphoma patients treated with rituximab
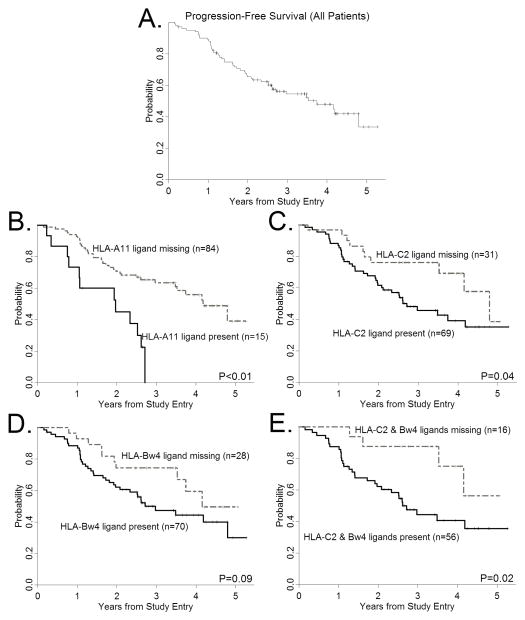
To test whether the ability of rituximab to activate unlicensed NK cells (without cytokines, transplantation, or chemotherapy) is clinically significant, we genotyped one hundred and one FL patients treated with mAb alone [CALGB protocols 50402 (rituximab with galiximab) and 50701 (rituximab with epratuzumab)] (23, 24) and compared PFS between patients with and without a “missing KIR ligand” genotype (Table 2, Supplemental Figure 6). The median PFS estimate for all patients was 3.74 years. We hypothesized that patients with a “missing KIR ligand” genotype will uniquely benefit from unlicensed NK cell ADCC. Consistent with this hypothesis, we found a statistically significant difference in PFS among the KIR ligand groups (Figure 5). FL patients lacking the HLA-A11 ligand for KIR3DL2 had a higher probability of remaining alive and progression-free compared with patients possessing HLA-A11 for the inhibitory KIR3DL2 [Figure 5B, hazard ratio (HR)=0.29, P<0.01]. We observed a similar association for patients lacking the HLA-C2 ligand for KIR2DL1 (Figure 5C, HR=0.48, P=0.04). A difference in PFS among patients lacking the HLA-Bw4 ligand for KIR3DL1 was marginally statistically significant (PFS 74% vs. 47%, HR 0.55, P=0.09; Figure 5D). Patients lacking both the HLA-C2 and HLA-Bw4 ligands had the highest probability of remaining alive and progression-free (PFS 88%, HR 0.75, P=0.02; Figure 5E). We observed an association of PFS with HLA-C2 gene dose, consistent with prior reports (39–41), such that HLA-C1/C1 homozygosity is associated with longer PFS, followed by HLA-C1/C2 heterozygosity, and HLA-C2 homozygosity was associated with the highest probability of disease progression (76% vs. 49% vs. 37%, HR 0.60, P=0.04). Differences in response rates between the different groups were not statistically significant (data not shown).
Figure 5. Influence of a “missing ligand” genotype on PFS in follicular lymphoma patients treated with rituximab antibody combinations.
(A) Progression-free survival among FL patients treated with rituximab-containing monoclonal antibody combinations, and stratified according to: (B) the presence or absence of HLA-A11, (C) missing the HLA-C2 ligand for KIR2DL1, (D) missing the HLA-Bw4 ligand for KIR3DL1, and (E) the presence or absence of both HLA-C2 and HLA-Bw4 ligands.
Discussion
ADCC is one of several mechanisms contributing to clinical responses in patients treated with rituximab, but the immunogenetic factors modulating cellular responses to rituximab remain unclear. A deeper understanding of the functional impact of KIR and HLA diversity on NK cell responses to rituximab-coated tumors may help predict clinical responses to rituximab and other mAbs. We specifically sought to determine how the rules of NK cell licensing modulate the responsiveness of NK cell subsets to rituximab. The licensing model predicts a hierarchical NK cellular response, dominated by the NK cell subset expressing an inhibitory KIR for self-HLA, which we confirmed in the absence of rituximab, when NK cells encounter HLA class I-deficient cells, consistent with prior studies (11–14, 42). Coating CD20-expressing transformed B-cells with rituximab, however, activates NK cells lacking an inhibitory KIR for self-HLA. Activation of these unlicensed NK cells may contribute to the “missing KIR ligand” benefit we observed in FL patients treated with rituximab-containing mAb combinations.
Rituximab and other antitumor antibodies provide a potent NK cell stimulus through CD16 (FcγRIII), the low-affinity IgG receptor, and unlike other activating NK cell receptors, CD16 ligation alone triggers fast calcium flux, cytokine secretion, and cytotoxicity of resting NK cells (22). We, therefore, hypothesized that rituximab exploits this unique signaling feature of CD16. We show that IL-2 is not required for mAb to activate unlicensed NK cells as unlicensed NK cells sorted from supportive accessory cells can mediate a similar effect, and that antitumor ADCC mediated by unlicensed KIR3DL2+ NK cells may contribute to antibody responses. These data shed light on the licensing model, and suggest that the rules governing NK cell responses to antibody-coated tumors may differ from the rules governing direct antitumor responses. Because the population of KIR− and unlicensed NK cells within the total NK cell repertoire is large, rituximab’s ability to activate these previously tolerant, hypo-responsive NK cells may be a fundamental mechanism contributing to the rituximab clinical responses. Our analysis of the NK cell repertoire is consistent with the literature, demonstrating significant inter-individual variability in spKIR+ and KIR−NKG2A− NK cell populations (11, 12, 21, 31, 38, 43). By combining single-cell exclusion gating with KIR phenotyping, HLA genotyping and HLA class I KIR ligand assignment, we also demonstrate that licensed NK cells may comprise a minority of the NK cell population, depending on an individual’s HLA genotype. In the absence of rituximab, therefore, NK cell responses to CD20+ tumors may be limited by the small subpopulation of licensed NK cells that degranulate after direct exposure to tumors. Rituximab’s ability to increase the number of activated NK cells by circumventing a licensing requirement may augment the magnitude of NK cell responses to CD20+ tumors, similar to the role of naive T-cell frequencies in modulating the immune response to viruses and vaccines (44–47). Moreover, the potential for KIR−NKG2A− and unlicensed NK cells to evade inhibition by HLA class I ligand expression on tumors may result in superior clinical responses in patients with greater numbers of these uninhibited NK cell subsets, as suggested in other model systems (16, 21, 48).
Little is known about the functional role of spKIR3DL2+ NK cells, and published data suggest that spKIR3DL2+ NK cells are hypo-responsive regardless of the presence of the HLA-A3 or -A11 ligand (36–38). The ability of rituximab to activate NK cells expressing KIR3DL2 and the clinical association of a “missing KIR3DL2 ligand” with improved PFS suggest that KIR3DL2+ NK cells are competent, do not require cognate HLA-A3 or -A11 ligand recognition for antibody-dependent activation, and may mediate clinically meaningful responses in patients treated with rituximab. Expression of HLA-A ligands on tumors may impair KIR3DL2 NK cell function (38), such that patients lacking HLA-A ligands for KIR3DL2 may benefit from a lack of KIR3DL2 inhibition. Since we used MHC class I-low or MHC class I-negative tumor targets in our in vitro experiments, we were unable to specifically address the role of HLA expression on tumors. Moreover, whether differences in HLA-A3 and -A11 expression or binding to KIR3DL2 explain different clinical responses among HLA-A3+ and HLA-A11+ FL patients merit further investigation.
The in vivo NK cell response to rituximab likely occurs in phases, similar to T-cell immune responses (49), with NK cell expansion, contraction and memory formation after initial activation (50). Our studies focus on the early activation phase, in which unlicensed and licensed NK cells appear to have comparable early degranulation and IFNγ responses to rituximab-coated transformed cells. Whether an advantage of unlicensed NK cells emerges in later phases of the in vivo response to rituximab is unclear, but suggested by prior studies (16, 21). Our experiments, performed in the absence of cytokines and validated with sorted NK cells, suggest that a highly inflammatory environment like mouse CMV infection or combination chemotherapy with mAb therapy may not be necessary for circumventing the requirement of MHC education (15, 16, 21). Antitumor antibodies, therefore, may be a uniquely targeted strategy for breaking tolerance and triggering activity from hypo-responsive NK cells, while avoiding toxicities of cytokines and inflammation.
Our finding that rituximab activates a large population of NK cells by overcoming a requirement for HLA-dependent NK cell licensing may inform strategies for augmenting clinical remissions. Cognate inhibitory KIR - HLA binding plays a dual, paradoxical role in controlling NK cell responses: endowing effector function and inhibiting NK cell responses (12, 14, 16, 51). Because the presence of an HLA class I ligand for inhibitory KIR is not required for rituximab-dependent NK cell responses, binding cognate ligand may actually lead to NK cell exhaustion and impede clinical responses mediated by NK cells (52). In neuroblastoma patients treated with combination anti-GD2 chemoimmunotherapy, licensed NK cell inhibition is the dominant result of cognate KIR - HLA binding (21). Blocking inhibitory KIR - HLA interactions to trigger licensed NK cell responses is feasible in the clinic (53–55), and a rationale augmentation strategy for patients treated with rituximab, although responses likely depend on the frequency of licensed NK cells in patients, which was highly variable in our study subjects. IL-2 might also augment rituximab NK cell responses, as demonstrated previously (56), although prevention of regulatory T-cell expansion may be required for optimal clinical benefit (57, 58). In aggregate, these data highlight the importance and feasibility of understanding the requirements for capturing the antitumor potential of the NK cell repertoire, which may facilitate safer, more effective, and personalized treatment decisions for cancer patients.
Supplementary Material
Acknowledgments
Financial support: The research for this study was supported, in part, by grants from the National Cancer Institute (CA31946) to the Alliance for Clinical Trials in Oncology (Monica M. Bertagnolli, M.D., Chair) and to the Alliance Statistics and Data Center (Daniel J. Sargent, Ph.D., CA33601). This study was also supported by a grant from the National Institutes of Health (NIH AI068129), a Mentored Research Scholar Grant from the American Cancer Society (MRSG-13-044-01-LIB), a Junior Investigator Research Award from the CALGB Foundation, a V Scholar Award from the V Foundation for Cancer Research, a Mt. Zion Health Fund Research Award, a grant from the UCSF Stephen and Nancy Grand Multiple Myeloma Translational Initiative, and generous support from friends and family of Peter Peracca. L.L.L. is an American Cancer Society Professor and supported by NIH grant AI068129.
The following institutions participated in this study by enrolling patients in Alliance studies CALGB 50402 and CALGB 50701: Cancer Centers of the Carolinas, Greenville, SC, Jeffrey K. Giguere, M.D., supported by CA29165; Dana-Farber Cancer Institute, Boston, MA, Harold J. Burstein, M.D., Ph.D., supported by CA32291; Dartmouth Medical School-Norris Cotton Cancer Center, Lebanon, NH, Konstantin Dragnev, M.D., supported by CA04326; Georgetown University Medical Center, Washington, DC, Bruce Cheson, M.D., supported by CA77597; Hematology-Oncology Associates of CNY CCOP, Syracuse, NY, Jeffrey Kirshner, M.D., supported by CA45389; Memorial Sloan-Kettering Cancer Center, New York, NY, Clifford A. Hudis, M.D., supported by CA77651; Missouri Baptist Medical Center, St. Louis, MO, Alan P. Lyss, M.D., supported by CA114558-02; Mount Sinai Medical Center, Miami, FL, Michael A. Schwartz, M.D., supported by CA45564; New Hampshire Oncology-Hematology PA, Concord, NH, Douglas J. Weckstein, M.D.; Roswell Park Cancer Institute, Buffalo, NY, Ellis Levine, M.D., supported by CA59518; Southeast Cancer Control Consortium Inc. CCOP, Goldsboro, NC, James N. Atkins, M.D., supported by CA45808; The Ohio State University Medical Center, Columbus, OH, Clara D. Bloomfield, M.D., supported by CA77658; University of California at San Francisco, San Francisco, CA, Charles J. Ryan, M.D., supported by CA60138; University of Chicago, Chicago, IL, Hedy L. Kindler, M.D., supported by CA41287; University of Illinois MBCCOP, Chicago, IL, David J. Peace, M.D., supported by CA74811; University of Iowa, Iowa City, IA, Daniel A. Vaena, M.D., supported by CA47642; University of Minnesota, Minneapolis, MN, Bruce A. Peterson, M.D., supported by CA16450; University of North Carolina at Chapel Hill, Chapel Hill, NC, Thomas C. Shea, M.D., supported by CA47559; University of Vermont, Burlington, VT, Steven M. Grunberg, M.D., supported by CA77406; Wake Forest University School of Medicine, Winston-Salem, NC, David D. Hurd, M.D., supported by CA03927; Walter Reed Army Medical Center, Washington, DC, David C. Van Echo, M.D., supported by CA26806; Washington University School of Medicine, St. Louis, MO, Nancy Bartlett, M.D., supported by CA77440; Weill Medical College of Cornell University, New York, NY, John Leonard, M.D., supported by CA07968.
The authors thank the patients for contributing their samples; the Alliance Lymphoma Committee, the Alliance Translational Research Program, Dr. Andrew Zelenetz and Dr. Paula Friedman for assistance with patient sample procurement; Dr. Elisabeth Lasater, Dr. Deborah Hendricks, Ms. Doris Kim, and Ms. Janice Arakawa-Hoyt for technical assistance; Dr. Neil Shah for the use of the human erythroleukemia cell line K562; Dr. James Rubenstein for the use of the Raji cell line; Dr. Julie Schwenka for the rituximab; and Dr. Andrew Leavitt for manuscript comments.
Footnotes
Disclosure of Potential Conflicts of Interest: J. Venstrom has served as a consultant for Bristol Myers Squibb and has received research funding from Sanofi Oncology.
Authors’ Contributions
Conception and design: J. Venstrom, L. Lanier
Development of methodology: J. Venstrom, L. Lanier, J. Du, L. Zhou, S. Lopez-Verges, B. Pitcher, K. Hsu, J. Johnson
Acquisition of data: J. Du, L. Zhou, L. Kaplan, B. Cheson, K. Hsu, B. Pitcher, J. Johnson
Analysis and interpretation of data: J. Venstrom, L. Lanier, J. Du, B. Pitcher, J. Johnson
Writing, review and/or revision of the manuscript: J. Venstrom, L. Lanier, J. Du, L. Zhou, S. Lopez-Verges, B. Pitcher, J. Johnson, L. Kaplan, B. Cheson, K. Hsu
Administrative, technical or material support: J. Venstrom
Study supervision: J. Venstrom
References
- 1.Rafiq K, Bergtold A, Clynes R. Immune complex–mediated antigen presentation induces tumor immunity. J Clin Invest. 2002;110:71–9. doi: 10.1172/JCI15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grillo-López AJ. Monoclonal antibody therapy for B-cell lymphoma. Int J Hematol. 2002;76:385–93. doi: 10.1007/BF02982803. [DOI] [PubMed] [Google Scholar]
- 3.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor Fcgamma RIIIa gene. Blood. 2002;99:754–8. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 4.McLaughlin P, Grillo-Lopez A, Link B, Levy R, Czuczman M, Williams M, et al. Rituximab chimeric anti-CD20 monoclonal antibody therapy for relapsed indolent lymphoma: half of patients respond to a four-dose treatment program. J Clin Oncol. 1998;16:2825–33. doi: 10.1200/JCO.1998.16.8.2825. [DOI] [PubMed] [Google Scholar]
- 5.Clynes RA, Towers TL, Presta LG, Ravetch JV. Inhibitory Fc receptors modulate in vivo cytoxicity against tumor targets. Nat Med. 2000;6:443–6. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 6.Cheson BD, Leonard JP. Monoclonal antibody therapy for B-cell non-Hodgkin’s lymphoma. N Engl J Med. 2008;359:613–26. doi: 10.1056/NEJMra0708875. [DOI] [PubMed] [Google Scholar]
- 7.Weng W, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independelty predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Weng W-K, Levy R. Genetic polymorphism of the inhibitory IgG Fc receptor FcgammaRIIb is not associated with clinical outcome in patients with follicular lymphoma treated with rituximab. Leuk Lymphoma. 2009;50:723–7. doi: 10.1080/10428190902829441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohen SP, Troyanskaya OG, Alter O, Warnke R, Botstein D, Brown PO, et al. Variation in gene expression patterns in follicular lymphoma and the response to rituximab. Proc Natl Acad Sci U S A. 2003;100:1926–30. doi: 10.1073/pnas.0437875100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Joncker NT, Fernandez NC, Treiner E, Vivier E, Raulet DH. NK Cell Responsiveness Is Tuned Commensurate with the Number of Inhibitory Receptors for Self-MHC Class I: The Rheostat Model. J Immunol. 2009;182:4572–80. doi: 10.4049/jimmunol.0803900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anfossi N, Andre P, Guia S, Falk C, Roetnynck S, Stewart C, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Heller G, Chewning J, Kim S, Yokoyama W, Hsu K. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol. 2007;179:5977–89. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez N, Treiner E, Vance R, Jamieson A, Lemieux S, Raulet D. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–23. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Poursine-Laurent J, Truscott S, Lybarger L, Song Y, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Venstrom J, Liu X, O’Reilly R, Pring J, Hasan R, et al. Breaking tolerance to self, circulating natural killer cells expressing inhibitory KIR for non-self HLA exhibit effector function following T-cell depleted allogeneic hematopoietic cell transplantation. Blood. 2009;113:3875–84. doi: 10.1182/blood-2008-09-177055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11:321–7. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu K, Keever-Taylor C, Wilton A, Pinto C, Heller G, Arkun K, et al. Improved outcome in allogeneic hematopoietic stem cell transplantation in acute myelogenous leukemia (AML) predicted by donor KIR genotype and recipient HLA genotype in T-cell depleted HLA-identical sibling transplants. Blood. 2005;105:4878–84. doi: 10.1182/blood-2004-12-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venstrom J, Zheng J, Noor N, Danis K, Yeh A, Cheung I, et al. KIR and HLA genotypes are associated with disease progression and survival following autologous hematopoietic stem cell transplantation for high-risk neuroblastoma. Clin Cancer Res. 2009;15:7330–4. doi: 10.1158/1078-0432.CCR-09-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sungur CM, Tang-Feldman YJ, Ames E, Alvarez M, Chen M, Longo DL, et al. Murine natural killer cell licensing and regulation by T regulatory cells in viral responses. Proc Natl Acad Sci U S A. 2013;110:7401–6. doi: 10.1073/pnas.1218767110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Björklund AT, Schaffer M, Fauriat C, Ringdén O, Remberger M, Hammarstedt C, et al. NK cells expressing inhibitory KIR for non–self-ligands remain tolerant in HLA-matched sibling stem cell transplantation. Blood. 2010;115:2686–94. doi: 10.1182/blood-2009-07-229740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarek N, Le Luduec J-B, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest. 2012;122:3260–70. doi: 10.1172/JCI62749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryceson Y, March M, Ljunggren H, Long E. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 2006;107:159–66. doi: 10.1182/blood-2005-04-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Czuczman MS, Leonard JP, Jung S, Johnson JL, Hsi ED, Byrd JC, et al. Phase II trial of galiximab (anti-CD80 monoclonal antibody) plus rituximab (CALGB 50402): Follicular Lymphoma International Prognostic Index (FLIPI) score is predictive of upfront immunotherapy responsiveness. Annals of Oncology. 2012;23:2356–62. doi: 10.1093/annonc/mdr620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant BW, Jung S-H, Johnson JL, Kostakoglu L, Hsi E, Byrd JC, et al. A phase 2 trial of extended induction epratuzumab and rituximab for previously untreated follicular lymphoma: CALGB 50701. Cancer. 2013;119:3797–804. doi: 10.1002/cncr.28299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu KC, Liu X, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–29. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 26.Vilches C, Castaño J, Gómez-Lozano N, Estefanía E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–22. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 27.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294(1–2):15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Lee-MacAry AE, Ross EL, Davies D, Laylor R, Honeychurch J, Glennie MJ, et al. Development of a novel flow cytometric cell-mediated cytotoxicity assay using the fluorophores PKH-26 and TO-PRO-3 iodide. J Immunol Methods. 2001;252(1–2):83–92. doi: 10.1016/s0022-1759(01)00336-2. [DOI] [PubMed] [Google Scholar]
- 29.Cheson B, Bennett J, Kopecky K, Buchner T, Willman C, Estey E, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Meyeloid Leukemia. J Clin Oncol. 2003;21:4642–9. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 30.Cooley S, McCullar V, Wangen R, Bergemann T, Spellman S, Weisdorf D, et al. KIR reconstitution is altered by T cells in the graft and correlates with clinical outcomes after unrelated donor transplantation. Blood. 2005;106:4370–6. doi: 10.1182/blood-2005-04-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann T, et al. A suppopulation of human peripheral blood NK cells that lacks inhibitory receptors for self MHC is developmentally immature. Blood. 2007;110:578–86. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, et al. NK cell education after allogeneic transplantation: dissociation between recovery of cytokine-producing and cytotoxic functions. Blood. 2011;118:2784–92. doi: 10.1182/blood-2011-04-347070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breedveld F, Agarwal S, Yin M, Ren S, Li NF, Shaw TM, et al. Rituximab Pharmacokinetics in Patients With Rheumatoid Arthritis: B-Cell Levels Do Not Correlate With Clinical Response. J Clin Pharmacol. 2007;47:1119–28. doi: 10.1177/0091270007305297. [DOI] [PubMed] [Google Scholar]
- 34.Tran L, Baars JW, Aarden L, Beijnen JH, Huitema ADR. Pharmacokinetics of rituximab in patients with CD20 positive B-cell malignancies. Human Antibodies. 2010;19:7–13. doi: 10.3233/HAB-2010-0215. [DOI] [PubMed] [Google Scholar]
- 35.Maloney DG, Grillo-López AJ, White CA, Bodkin D, Schilder RJ, Neidhart JA, et al. IDEC-C2B8 (Rituximab) Anti-CD20 Monoclonal Antibody Therapy in Patients With Relapsed Low-Grade Non-Hodgkin’s Lymphoma. Blood. 1997;90:2188–95. [PubMed] [Google Scholar]
- 36.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLA-A3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34:1673–9. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
- 37.Yawata M, Yawata N, Draghi M, Partheniou F, Little A-M, Parham P. MHC class I–specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–80. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fauriat C, Andersson S, Björklund AT, Carlsten M, Schaffer M, Björkström NK, et al. Estimation of the Size of the Alloreactive NK Cell Repertoire: Studies in Individuals Homozygous for the Group A KIR Haplotype. J Immunol. 2008;181:6010–9. doi: 10.4049/jimmunol.181.9.6010. [DOI] [PubMed] [Google Scholar]
- 39.Venstrom J, Pittari G, Gooley T, Chewning J, Spellman S, Haagenson M, et al. HLA-C-Dependent Prevention of Leukemia Relapse by Donor Activating KIR2DS1. N Engl J Med. 2012;367:805–16. doi: 10.1056/NEJMoa1200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cook MA, Milligan DW, Fegan CD, Darbyshire PJ, Mahendra P, Craddock CF, et al. The impact of donor KIR and patient HLA-C genotypes on outcome following HLA-identical sibling hematopoietic stem cell transplantation for myeloid leukemia. Blood. 2004;103:1521–6. doi: 10.1182/blood-2003-02-0438. [DOI] [PubMed] [Google Scholar]
- 41.Fischer J, Ottinger H, Ferencik S, Sribar M, Punzel M, Beelen D, et al. Relevance of C1 and C2 Epitopes for Hemopoietic Stem Cell Transplantation: Role of Sequential Acquisition of HLA-C-Specific Inhibitory Killer Ig-Like Receptor. J Immunol. 2007;178:3918–23. doi: 10.4049/jimmunol.178.6.3918. [DOI] [PubMed] [Google Scholar]
- 42.Kim S, Sunwoo J, Yang L, Choi T, Song Y, French A, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105:3053–8. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan WK, Rujkijyanont P, Neale G, Yang J, Bari R, Das Gupta N, et al. Multiplex and Genome-Wide Analyses Reveal Distinctive Properties of KIR+ and CD56+ T Cells in Human Blood. J Immunol. 2013;191:1625–36. doi: 10.4049/jimmunol.1300111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl Ross M, et al. Naive CD4+ T Cell Frequency Varies for Different Epitopes and Predicts Repertoire Diversity and Response Magnitude. Immunity. 2007;27:203–13. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwok WW, Tan V, Gillette L, Littell CT, Soltis MA, LaFond RB, et al. Frequency of Epitope-Specific Naive CD4+ T Cells Correlates with Immunodominance in the Human Memory Repertoire. J Immunol. 2012;188:2537–44. doi: 10.4049/jimmunol.1102190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Obar JJ, Khanna KM, Lefrançois L. Endogenous Naive CD8+ T Cell Precursor Frequency Regulates Primary and Memory Responses to Infection. Immunity. 2008;28:859–69. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotturi MF, Scott I, Wolfe T, Peters B, Sidney J, Cheroutre H, et al. Naive Precursor Frequencies and MHC Binding Rather Than the Degree of Epitope Diversity Shape CD8+ T Cell Immunodominance. J Immunol. 2008;181:2124–33. doi: 10.4049/jimmunol.181.3.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delgado DC, Hank JA, Kolesar J, Lorentzen D, Gan J, Seo S, et al. Genotypes of NK Cell KIR Receptors, Their Ligands, and Fcγ Receptors in the Response of Neuroblastoma Patients to Hu14.18-IL2 Immunotherapy. Cancer Res. 2010;70:9554–61. doi: 10.1158/0008-5472.CAN-10-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams MA, Bevan MJ. Effector and Memory CTL Differentiation. Annu Rev Immunol. 2007;25:171–92. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 50.Sun JC, Lopez-Verges S, Kim CC, DeRisi JL, Lanier LL. NK Cells and Immune “Memory”. J Immunol. 2011;186:1891–7. doi: 10.4049/jimmunol.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–51. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 52.Gill S, Vasey AE, De Souza A, Baker J, Smith AT, Kohrt HE, et al. Rapid development of exhaustion and down-regulation of eomesodermin limit the antitumor activity of adoptively transferred murine natural killer cells. Blood. 2012;119:5758–68. doi: 10.1182/blood-2012-03-415364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Benson DM, Hofmeister CC, Padmanabhan S, Suvannasankha A, Jagannath S, Abonour R, et al. A phase 1 trial of the anti-KIR antibody IPH2101 in patients with relapsed/refractory multiple myeloma. Blood. 2012;120:4324–33. doi: 10.1182/blood-2012-06-438028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benson DM, Bakan CE, Zhang S, Collins SM, Liang J, Srivastava S, et al. IPH2101, a novel anti-inhibitory KIR antibody, and lenalidomide combine to enhance the natural killer cell versus multiple myeloma effect. Blood. 2011;118:6387–91. doi: 10.1182/blood-2011-06-360255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohrt HE, Thielens A, Marabelle A, Sagiv-Barfi I, Sola C, Chanuc F, et al. Anti-KIR antibody enhancement of anti-lymphoma activity of natural killer cells as monotherapy and in combination with anti-CD20 antibodies. Blood. 2014;123:678–86. doi: 10.1182/blood-2013-08-519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gluck WL, Hurst D, Yuen A, Levine AM, Dayton MA, Gockerman JP, et al. Phase I Studies of Interleukin (IL)-2 and Rituximab in B-Cell Non-Hodgkin’s Lymphoma: IL-2 Mediated Natural Killer Cell Expansion Correlations with Clinical Response. Clin Cancer Res. 2004;10:2253–64. doi: 10.1158/1078-0432.ccr-1087-3. [DOI] [PubMed] [Google Scholar]
- 57.Matsuoka K-i, Koreth J, Kim HT, Bascug G, McDonough S, Kawano Y, et al. Low-Dose Interleukin-2 Therapy Restores Regulatory T Cell Homeostasis in Patients with Chronic Graft-Versus-Host Disease. Sci Transl Med. 2013;5:179ra43. doi: 10.1126/scitranslmed.3005265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koreth J, Matsuoka K-i, Kim HT, McDonough SM, Bindra B, Alyea EP, et al. Interleukin-2 and Regulatory T Cells in Graft-versus-Host Disease. N Engl J Med. 2011;365:2055–66. doi: 10.1056/NEJMoa1108188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.