Abstract
Macrophage migration Inhibitory Factor (MIF) is a pro-inflammatory cytokine sustaining the acute response to gram–negative bacteria and a regulatory role for MIF in Cystic Fibrosis has been suggested by the presence of a functional, polymorphic, four-nucleotide repeat in this gene's promoter at position −794, with the 5-repeat allele displaying lower promoter activity. We aimed at assessing the association of this polymorphism with disease severity in a group of Cystic Fibrosis patients homozygous for F508del CFTR gene mutation. Genotype frequencies were determined in 189 Cystic Fibrosis and 134 control subjects; key clinical features of patients were recorded and compared among homozygous 5-allele patients and the other MIF genotypes. Patients homozygous for the 5-repeat allele of MIF promoter displayed a slower rate of lung function decline (p = 0.027) at multivariate survival analysis. Multiple regression analysis on age-normalized respiratory volume showed no association of the homozygous 5-repeat genotype with lung function under stable conditions and no correlation with P.aeruginosa chronic colonization. Therefore, only the Homozygous 5-repeat genotype at MIF −794 is associated with milder disease in F508del Cystic Fibrosis patients.
Introduction
Cystic fibrosis (CF) is the most common, severe, inherited disorder in the Caucasian population. It is caused by mutations in the CF Transmembrane conductance Regulator (CFTR) gene and mainly characterized by bronchopulmonary disease, pancreatic insufficiency and male infertility. Patients with identical CFTR genotypes can display markedly different phenotypic expression [1], [2] and modifier genes were previously described among the factors causing this discrepancy [3], [4]. Macrophage Migration Inhibitory Factor (MIF) is a key pro-inflammatory mediator [5]: it sustains an acute inflammatory response both directly, by inducing cytokines secretion, and indirectly, by overriding the anti-inflammatory activity of glucocorticoids [6]. MIF plays a significant role in immune and inflammatory-based diseases such as asthma [7], rheumatoid arthritis [8], acute respiratory distress syndrome [9] and septic shock [10], [11]. Although MIF is involved in the defence against severe infection, modulation of the high cytokine levels elicited by its action may prevent harmful effects during the inflammatory response. Indeed, lethal sepsis induced in mice by lipopolysaccharide (LPS) or E. coli causes increased mortality in the presence of recombinant MIF [12], while anti–MIF neutralizing antibodies were able to protect mice from lethal endotoxic sepsis induced by bacterial (E. coli) peritonitis [11]. It has also been suggested that neutralizing MIF could lead to improved resistance against P. aeruginosa infection, since clearance of the bacteria following tracheal instillation was improved in MIF-knockout mice [10]. Recently, Baugh et al. [13] identified a functionally significant polymorphism in the human MIF gene, consisting of a four-nucleotide CATT repeat located at position −794 of the MIF promoter (MIF-CATT). In an in vitro model, the 5-CATT repeat showed significantly lower transcriptional activity when compared to the 6-, 7- or 8-CATT repeat alleles. This polymorphism is reported as a TTCA insertion or deletion relative to the 6-repeats genotype in NCBI dbSNP entries rs3063368 and rs36224313 respectively, at the genomic coordinates (UCSC genome browser – hg19) chr22:24235773-24235772. Five percent of healthy subjects are homozygous for the 5-CATT repeat allele. Homozygosity for this allele was significantly associated with milder forms of rheumatoid arthritis, suggesting it may have a protective effect. In CF patients, Plant et al. [14] reported a significant decrease in both P. aeruginosa colonization and pancreatic insufficiency among adult patients carrying at least one 5-CATT MIF allele. Since many studies of modifier genes in CF have yielded conflicting results, it is essential to validate any association in a new, independent population and MIF gene is no exception [4]. This study aimed at clarifying and validating the association between MIF-CATT repeats and disease severity in a more homogeneous cohort of CF patients with homozygous F508del CFTR mutation. Given the biological relationship between MIF and acute inflammation suggested by the above cited literature, we chose as a primary outcome the time to the first acute episode causing forced expiratory volume (FEV1) to fall below the 60% of the predicted value. We also verified the possible relationship between MIF and age-normalized FEV1 and chronic P. aeruginosa colonization under stable conditions.
Materials and Methods
Study population
One hundred and eighty-nine CF patients homozygous for the F508del mutation were recruited from two European centres (Verona: 138, Brussels: 51). All of the patients were able to perform reliable spirometry. A cohort of 134 adult Italian subjects was used as control. Healthy subjects were negative for the most common mutations of the CFTR gene, except for 4 heterozygous subjects (healthy carriers). This is consistent with epidemiological data about carrier frequency in Europe. This study was approved by the Ethics Committee of the University and Hospital Trust of Verona (protocol #24737); informed signed consent for DNA analysis was obtained from participants or from their parents, as required.
Genotyping
DNA was extracted from whole blood using the salting out method, then samples were genotyped for the polymorphism of MIF promoter (varying number of CATT repeats) at -794. DNA was amplified by Polymerase Chain Reaction (PCR) in a GeneAmp PCRsystem 9700® (Applied Biosystem, Foster City, CA, USA) as previously described [13]. Genotyping was performed by the BMR Genomics Sequencing Service (CRIBI, University of Padova, Italy). Results were analysed using GenescanView 1.2 software (CRIBI, University of Padova, Italy).
Clinical data of CF patients
Clinical data for the 189 patients were collected in electronic databases. Main characteristics and MIF genotype of the patients are summarized in table 1 and extensively reported in S1 Table. Anthropometric parameters and forced expiratory volume (FEV1) were normalized using Freeman's [15] and Knudson's [16] equations respectively. Given the accelerated FEV1 decline with age in CF patients, the last value of FEV1 was expressed as FEV1 percentile using CF-specific reference equations [17]. Diabetes was defined by the need for insulin therapy. Chronic P. aeruginosa colonization was reported using a European consensus definition [18]. In brief, chronic colonization was defined as the isolation of at least 3 isolates in a six month period (at minimum 30 days interval) while sporadic colonization referred to the isolation of P. aeruginosa in the bronchial tree in presence or absence of inflammation.
Table 1. Characteristics of 189 Cystic Fibrosis patients homozygous for the F508del mutation recruited from 2 different European centers.
| Brussels | Verona | p-value | |
| Subjects n | 51 | 138 | |
| Female n (%) | 22 (%) | 78 (%) | 0.14 |
| Age, years | 21.5±9.4 | 24.27±9.3 | 0.09 |
| FEV1, Kulich * | 67±24 | 45±31 | <0.0001 |
| BMI z-score | −0.63±1.03 | −0.82±1.28 | 0.36 |
| cc by PA n (%) | 16 (31.4%) | 87 (63.0%) | <0.0002 |
| Diabetes n (%) | 11 (21.6%) | 40 (29.0%) | 0.36 |
| MIF-CATT 5-5 n (%) | 4 (7.8%) | 12 (8.7%) | |
| MIF-CATT 5-6 n (%) | 12 (23.5%) | 59 (42.8%) | |
| MIF-CATT 5-7 n (%) | 7 (13.7%) | 10 (7.2%) | 0.11 |
| MIF-CATT 6-6 n (%) | 20 (39.2%) | 43 (31.2%) | |
| MIF-CATT 6-7 n (%) | 8 (15.7%) | 12 (8.7%) | |
| MIF-CATT 7-7 n (%) | 0 (0%) | 2 (1.4%) |
Continuous data are presented as mean ± SD unless otherwise stated; categorical data are presented as counts and proportions. FEV1: forced expiratory volume in one second; cc by PA: chronic colonization by P. aeruginosa. The most recent FEV1 was used for each patient. MIF-CATT: MIF gene -CATT repeat genotype at position -794
* CF specific percentile according to Kulich et al, Am J Respir Crit Care Med, 2005.
Statistical analysis and study design
Given the involvement of MIF in acute inflammation, we focused on the FEV1 parameter as primary outcome variable, comparing patients with the 5-5 MIF genotype to the rest of the cohort. The relationship between FEV1 and MIF genotype was analysed considering both the time to the first acute episode causing a FEV1 value under 60%, and the “static” FEV1 at last visit (normalised according to Kulich's CF-specific reference equations [17]).
Considering the differences in clinical parameters between patients from the two centres (table 1), the analyses were performed using multivariate techniques. Cox regression was used to analyse the time to first FEV1 under 60% and patient's origin (Verona or Brussels) was included as a covariate together with presence of MIF 5-5 genotype. Chronic colonization by P. aeruginosa and presence of insulin-dependent diabetes were not considered because only a minority of patients displayed these features before first acute episode (28.9% for P.aeruginosa and 5.0% for diabetes). Given the unequal size of 5-5 and X-X groups (patients number ratio = 0.09), we used the R package PowerSurvEpi to calculate the power to detect an effect like the one estimated from our data (Hazard ratio = 0.32) with the number of patients available. The power resulted to be 59%, while the sample size needed to get 80% power would have been 307 patients. Multiple linear regression was used to analyse last visit FEV1, including as covariates MIF 5-5 genotype, patients' origin, chronic colonization by P. aeruginosa and presence of insulin-dependent diabetes; age was not included because FEV1 values were already age-normalized according to Kulich. Variance inflation factor was used to monitor the presence of multicollinearity; its value was below 1.15 for all covariates.
Power analysis for last visit FEV1 was also used to calculate the power to detect a medium effect size (0.5, here corresponding to a difference between means of 15%, which was similar to the estimated difference in our cohort) in an unpaired t-test with our sample size and considering the unequal size of 5-5 (n = 16, mean FEV1 = 50.2, SD = 31.3) and X-X groups (n = 173, mean FEV1 = 64.2, SD = 25.5). The power calculated was 48%, while the sample size needed to get 80% power would have been 382 patients. All analyses were performed using GraphPad Prism version 5.04 for Windows (GraphPad Software, San Diego California USA, www.graphpad.com), MedCalc for Windows, version 14.8.1 (MedCalc Software, Ostend, Belgium) and the R software (R Development Core Team, version 2.9; R Foundation for Statistical Computing, Vienna, Austria; www.R-project.org). Power analysis for multiple linear regression was performed with G*Power [19].
Results
Genotyping of the −794 CATT polymorphic repeats of MIF promoter (MIF-CATT)
In 189 CF patients, frequencies for 5, 6, 7 and 8 MIF-CATT repeats were 31.7%, 57.4%, 10.9% and 0% respectively. Corresponding values for the 135 healthy control subjects were similar (28.5%, 60%, 11.1% and 0.4%). In CF patients, MIF-CATT genotype frequencies were similar among children and adults, excluding a survival bias for 5-5 subjects.
Clinical status of CF patients and MIF-CATT genotype
Patients were categorized according to the −794 CATT polymorphic repeats (MIF-CATT). Anthropometric and clinical data are summarized in Table 1 and 2 , and extensively reported in S1 Table.
Table 2. Clinical data of 187 Cystic Fibrosis patients homozygous for the F508del mutation according to the genotype for the MIF gene -CATT repeat at position −794.
| 5–5 | 5–6 | 5–7 | 6–6 | 6–7 | |
| n | 16 | 71 | 17 | 63 | 20 |
| Age, years (95% CI) | 18.2 (14.5– 21.9) | 23.6 (21.3–25.9) | 27.2 (23.1– 31.3) | 23.7 (21.2–26.1) | 22.3 (18.2–26.5) |
| BMI z-score (95% CI) | −0.51 (−1.10/0.08) | −0.84 (−1.17/−0.52) | −1.09 (−1.63/−0.54) | −0.71 (−0.97/−0.46) | −0.62 (−1.26/0.01) |
| FEV1, Kulich * (95% CI) | 64.2 (50.6–77.8) | 43.9 (36.6–51.2) | 52.4 (37.7–67.08) | 53.8 (45.7–61.8) | 59.3 (44.8–73.8) |
| CC by PA – n (%; 95% CI) | 5 (31.2%; 13.9–55.8%) | 44 (62.0%; 50.3–72.4%) | 11 (64.7%; 41.2–82.8%) | 33 (52.4%; 40.3–64.2%) | 8 (40.0%; 21.8–61.4%) |
| Diabetes – n (%; 95% CI) | 2 (12.5%; 2.2–37.3%) | 22 (31.0%; 21.4–42.5%) | 6 (35.3%; 17.2–58.8%) | 17 (27.0%; 17.5–39.1) | 4 (20.0%; 7.5–42.2%) |
Data are presented as mean and 95% Confidence Interval (95% CI). FEV1: forced expiratory volume in one second; BMI: body mass index; cc by PA: chronic colonization by P. aeruginosa.
* CF specific percentile according to Kulich et al, Am J Respir Crit Care Med, 2005.
Given the involvement of MIF in acute inflammation, we focused on FEV1 as primary outcome variable, comparing patients with the 5-5 MIF-CATT genotype to the rest of the cohort.
The relationship between FEV1 and MIF genotype was analysed considering both the time to the first acute episode causing a FEV1 value under 60% and, secondly, the “static” FEV1 at last visit. Considering the differences in clinical parameters between patients from the two centres and the possible effect of diabetes and P. aeruginosa colonization on pulmonary volume, the following analyses were performed using multivariate techniques.
MIF-CATT 5-5 genotype is associated to a later onset of acute episodes
Cox regression was used to analyse the time to the first acute episode causing FEV1 to fall under 60% predicted. MIF-CATT 5-5 genotype and patient's origin (Verona or Brussels) were included as independent variables. Chronic colonization by P. aeruginosa and presence of insulin-dependent diabetes were not considered because only a minority of patients displayed these features before first acute episode (28.9% for P.aeruginosa and 5.0% for diabetes). Both MIF-CATT 5-5 genotype (Hazard Ratio = 0.327; 95% CI 0.121-0.884) and belonging to the Brussels cohort (Hazard Ratio = 0.514; 95% CI 0.310 to 0.849) resulted to be independent predictors of a later onset of acute episodes ( Table 3 ).
Table 3. Cox regression analysis for age at first acute episode with FEV1 <60% of predicted value on 185 Cystic Fibrosis patients homozygous for the F508del mutation.
| Variable | Value | Hazard Ratio | 95% CI | p-value |
| MIF-CATT genotype | X-X | 1 | - | - |
| 5–5 | 0.325 | 0.120–0.878 | 0.027 | |
| Centre of origin | Verona | 1 | - | - |
| Brussels | 0.510 | 0.309–0.843 | 0.0090 |
Number of patients with acute episode = 119
Number of censored patients = 66
Overall significance p-value = 0.0010
MIF-CATT genotype: MIF gene -CATT repeat genotype at position −794
95% CI = 95% Confidence Interval
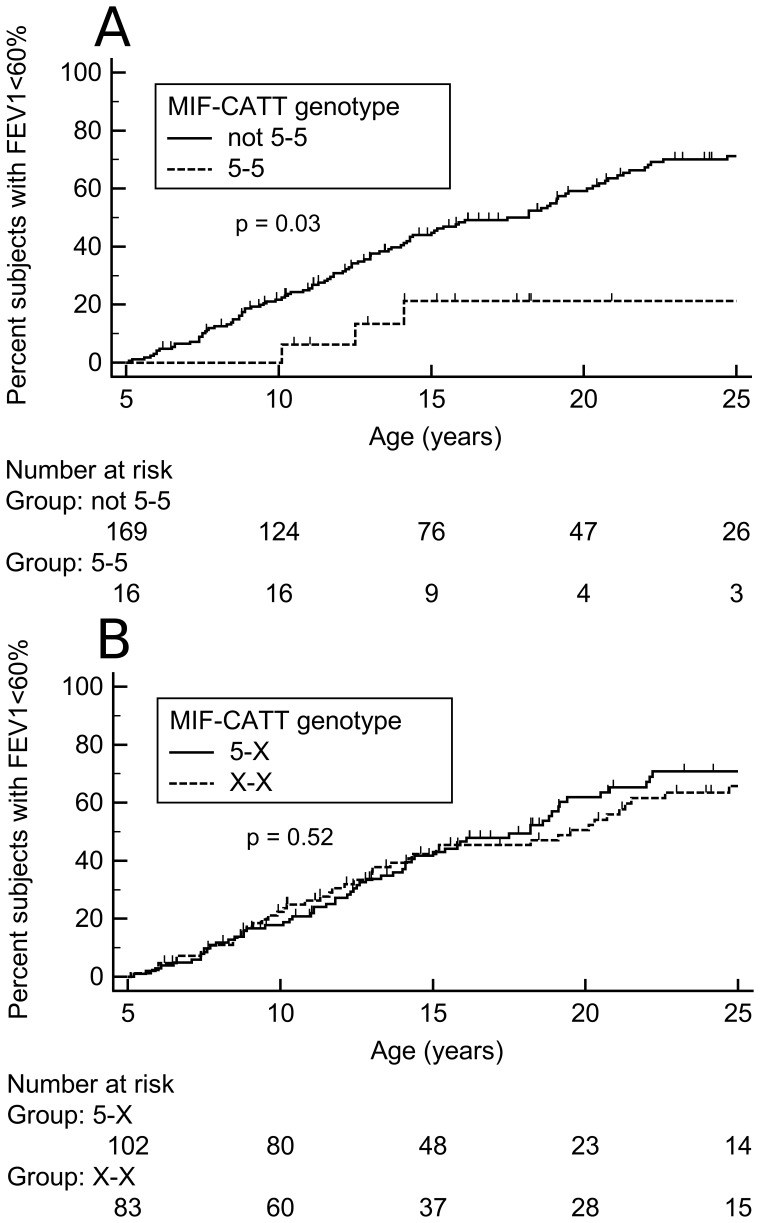
Kaplan-Meier analysis on Fig. 1 shows that age at first acute episode with FEV1 value below 60% predicted was higher in patients with 5–5 genotype compared to all the other genotypes; median time to event was 29.3 years for 5–5 subjects compared to 18.2 for the others (Hazard ratio = 0.35; 95% CI 0.19–0.66; p = 0.03, Fig. 1A ). Applying per-allele analysis, no relevant differences emerged, consistent with a recessive effect of 5-CATT allele ( Fig. 1B ).
Figure 1. Lung function decline in 185 Cystic Fibrosis patients grouped according to MIF -794 CATT genotypes.
Kaplan-Meier plots relative to age at first acute episode with FEV1 <60% of predicted value. (A) Comparison between patients with MIF 5-5 (homozygous 5-CATT repeats) vs. not 5-5 genotype; (B) comparison between patients with at least one 5-CATT allele vs. the others. Ticks indicate censored subjects follow-up times. “Number at risk” at the bottom indicates the number of patients without acute episodes at a given time interval and whose follow- up extends at least that far into the curve.
MIF-CATT 5-5 genotype shows no association with lower pulmonary function and P. aeruginosa chronic colonization under stable conditions
Since recent literature reported that MIF 5-CATT allele also correlates with lower FEV1 under stable conditions and higher prevalence of P. aeruginosa colonization [14], [20], we applied multiple linear regression to analyse age-normalized FEV1 [17] under stable conditions, including MIF-CATT 5-5 genotype, patients' origin, chronic colonization by P. aeruginosa and presence of insulin-dependent diabetes as covariates. The only factor not associated with FEV1 resulted to be MIF-CATT 5-5 genotype ( table 4 ). Variance inflation factor was used to monitor the presence of multicollinearity; its value was below 1.15 for all covariates, indicating no apparent correlation between MIF-CATT genotype and P. aeruginosa chronic colonization.
Table 4. Multiple regression analysis of FEV1 (Kulich)* data on 189 Cystic Fibrosis patients homozygous for the F508del mutation.
| Variable | Coefficient | SE | r-partial | P |
| Constant | 60.96 | - | - | - |
| MIF-CATT genotype = 5-5 | 7.06 | 7.09 | 0.073 | 0.320 |
| Centre of origin = Brussels | 15.15 | 4.58 | 0.24 | 0.001 |
| CC by PA | −16.79 | 4.16 | −0.29 | 0.0001 |
| Diabetes | −19.04 | 4.46 | −0.30 | <0.0001 |
Overall R2 = 0.274; multiple correlation coefficient = 0.524
Overall significance p-value<0.0001;
FEV1: forced expiratory volume in one second; cc by PA: chronic colonization by P. aeruginosa. MIF-CATT genotype: MIF gene -CATT repeat genotype at position -794
*CF specific percentile according to Kulich et al, Am J Respir Crit Care Med, 2005.
Discussion
In this study, 8% of 189 CF patients homozygous for the F508del CFTR mutation carried the 5-5 allele combination for a functional CATT repeat polymorphism in the MIF gene promoter (MIF-CATT), which is expected to be associated with decreased pro-inflammatory activity. Patients carrying this genotype showed a later onset of acute episodes; the rate of lung function decline, as assessed by age at the first FEV1 value below 60% predicted, was lower in this group (table 3). By contrast, no apparent link was found with FEV1 and chronic P. aeruginosa colonization under stable conditions at multivariate regression analysis (table 4). Overall, our data support the role of MIF as a modifier gene of lung disease in CF only to a very limited extent, in contrast to what suggested by Plant et al. [14] and Adamali et al. [20], though the results of both studies differ from ours in several aspects. These authors identified MIF-CATT genotype in 167 adult white CF patients from a single centre, 11% of whom were pancreatic sufficient. Only a fraction of patients (57%) were homozygous for the F508del mutation while 35% were heterozygous for this mutation and the remaining 8% were heterozygous for other CFTR mutations. Patients carrying at least one copy of the 5-repeat MIF-CATT allele were found to have a decreased incidence of P. aeruginosa colonization (defined as the presence of bacteria in the sputum) and a significant reduction in the risk of pancreatic insufficiency.
The results of Plant et al. were partly reproduced (only for FEV and FVC) by Adamali et al. in a recent publication of the same research team [20], with a cohort of 143 patients selected from the same referral centre on the basis of a CF diagnosis and not of a F508del genotype. This work presents the same heterogeneous genetic background as in Plant et al., and the same differences compared to our data. Moreover, an unspecified fraction of the patients from this latter study were also enrolled in the former. Interestingly, the ex-vivo part of the work by Adamali et al. showing differences of MIF levels in plasma and peripheral blood monocytes from CF patients, relies only on individuals with 5-5 and 6-6 MIF genotypes. In these experiments, the 5-5 MIF genotype is confirmed to be the genotype with lowest expression of the protein.
MIF-CATT genotype distribution in patients and controls were comparable in both studies (S2 Table). However, we found that clinical benefit was restricted to later onset of acute episodes in patients homozygous for the 5-repeats MIF-CATT allele.
Aside from these discrepancies, our results are in keeping with the observation by Baugh et al. [13] that in the context of another inflammatory disease (rheumatoid arthritis), only homozygosity for the 5-repeats MIF-CATT allele was protective against the development of severe disease.
Research into CF modifier genes has often yielded conflicting results and numerous challenges have been identified [21]; methodological issues are also likely to be involved. When compared to the earlier study, several strengths of the present work can be stressed. The study population is homogenous at the CF locus, since we focused on a single CFTR genotype. The centres involved have been routinely using reliable CF-specific electronic databases for a substantial period of time. Additional efforts were made to normalize data, using CF-specific FEV1 predicted values and multivariate analysis to accout for centre-dependent variations. Z-score for BMI is now considered a more appropriate index expression of nutrition than percentage of ideal body weight [22] and chronic colonization by P. aeruginosa is clinically more relevant than airway colonization (simply referring to its presence). A large GWAS study on CF patients failed to test MIF as a modifier gene due to the lack of probes for this gene in the used DNA arrays; indeed the whole MIF gene is not covered by the Illumina 610-Quad platform used for genotyping. As for possible SNPs in linkage disequilibrium with MIF-CATT, the study used three different cohorts of patients for a total of 3467 CF patients; a sample size that, as the authors themselves stated, is several-fold smaller than the standard for GWAS studies. This implies that only strongest associations might emerge from the study. Due to the small number of MIF 5-CATT subjects and to the fact that, at least from our data, a moderate effect is suggested only in recessive homozygotes, such an effect would have probably been difficult to detect in any case [23]. Altogether the two previously available studies involved an undefined number of CF patients between 167 and 356. This might still be insufficient [24], given the wide range of FEV1 values and the low prevalence of the homozygous 5-CATT MIF promoter allele, so further work is encouraged. Besides clinical data, biologic plausibility of a candidate modifier gene is essential and the case of MIF gene is a good example. Indeed, it is a key pro-inflammatory mediator that is implicated in the pathogenesis of inflammatory diseases such as asthma, rheumatoid arthritis and acute respiratory distress. It has also been shown to sustain toll-like receptor 4 expression in murine macrophages [25]. Alveolar macrophages are believed to play an important role in regulating the local inflammatory and immune responses in the CF lung [26]–[29]. In addition, both macrophages [30] and their circulating precursors, monocytes [31], have been shown to express functional CFTR, with alveolar macrophages from CFTR -/- mice exhibiting defective killing of internalized bacteria [30]. In Cystic Fibrosis, a vicious circle of inflammation and infection leads to destruction of obstructed airways, and it is notable that, up to now, only 4 medications - all with direct or indirect anti-inflammatory properties - are known to slow FEV1 rate of decline in this disease: high-dose ibuprofen [32], inhaled corticosteroids [20], [33], [34], macrolides [35] and DNase [36]. Conceivably, identification of modifier genes of lung disease in CF should help to provide novel therapeutic targets. In this perspective, neutralizing MIF activity by using antibodies or gene knockout, protected mice against severe sepsis by E. coli [11] and Pseudomonas and also enhanced P. aeruginosa airway clearance [10]. Furthermore, several powerful tautomerase inhibitors, highly selective for human MIF, have recently been identified and Adamali et al. showed promising ex-vivo results regarding their use to tame MIF-sustained immune reaction in CF patients [20], [37], [38]. In conclusion, we have provided additional independent data supporting the role of MIF as a modifier gene of lung disease in CF; the beneficial effect, however, was limited to the homozygous genotype displaying the lowest transcriptional activity. Since the protective MIF genotype is rare, most CF patients could benefit from a targeted approach aimed at reducing the powerful inflammatory response associated with high expression of this gene.
Supporting Information
Full dataset of Cystic Fibrosis patients used in the present study.
(XLS)
Comparison of MIF -794 CATT genotype (a) or 5-CAAT allele (b) frequencies between Plant's [14] and the present study.
(DOC)
Acknowledgments
We thank Patrizia Iansa for clinical data extraction and analysis and the staff at the Laboratory of Molecular Pathology, Azienda Ospedaliera Universitaria Integrata di Verona for storage and processing of DNA samples.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
MO was supported by the ABCFoundation while AM by the Lega Italiana Fibrosi Cistica – Associazione Veneta Onlus. The work is also supported by FFC grant# 06/2010, adopted by Delegazione FFC di V.C.O. Verbania, Associazione Trentina FC Onlus Gruppo di Sostegno FFC in ricordo di Silvia Sommavilla, Consorzio Promotre s.c.r.l., Antonio Guadagnin e Figlio, Alessandra Boccanera and FFC # 26/2011, adopted by Donatori SMS Solidale 2011, Delegazione FFC di Varese, Associazione Trentina FC onlus. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lester LA, Kraut J, Lloyd-Still J, Karrison T, Mott C, et al. (1994) Delta F508 genotype does not predict disease severity in an ethnically diverse cystic fibrosis population. Pediatrics 93:114–118. [PubMed] [Google Scholar]
- 2.Grasemann H (2006) Disease modifier genes in cystic fibrosis. Cystic Fibrosis: European Respiratory Society Journals Ltd. pp. 50−65.
- 3. Davies JC, Griesenbach U, Alton E (2005) Modifier genes in cystic fibrosis. Pediatr Pulmonol 39:383–391. [DOI] [PubMed] [Google Scholar]
- 4. Cutting GR (2010) Modifier genes in Mendelian disorders: the example of cystic fibrosis. Ann N Y Acad Sci 1214:57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cooke G, Armstrong ME, Donnelly SC (2009) Macrophage migration inhibitory factor (MIF), enzymatic activity and the inflammatory response. Biofactors 35:165–168. [DOI] [PubMed] [Google Scholar]
- 6. Calandra T, Bucala R (1995) Macrophage migration inhibitory factor: a counter-regulator of glucocorticoid action and critical mediator of septic shock. J Inflamm 47:39–51. [PubMed] [Google Scholar]
- 7. Rossi AG, Haslett C, Hirani N, Greening AP, Rahman I, et al. (1998) Human circulating eosinophils secrete macrophage migration inhibitory factor (MIF). Potential role in asthma. J Clin Invest 101:2869–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leech M, Metz C, Santos L, Peng T, Holdsworth SR, et al. (1998) Involvement of macrophage migration inhibitory factor in the evolution of rat adjuvant arthritis. Arthritis Rheum 41:910–917. [DOI] [PubMed] [Google Scholar]
- 9. Donnelly SC, Haslett C, Reid PT, Grant IS, Wallace WA, et al. (1997) Regulatory role for macrophage migration inhibitory factor in acute respiratory distress syndrome. Nat Med 3:320–323. [DOI] [PubMed] [Google Scholar]
- 10. Bozza M, Satoskar AR, Lin G, Lu B, Humbles AA, et al. (1999) Targeted disruption of migration inhibitory factor gene reveals its critical role in sepsis. J Exp Med 189:341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Calandra T, Echtenacher B, Roy DL, Pugin J, Metz CN, et al. (2000) Protection from septic shock by neutralization of macrophage migration inhibitory factor. Nat Med 6:164–170. [DOI] [PubMed] [Google Scholar]
- 12. Bernhagen J, Calandra T, Mitchell RA, Martin SB, Tracey KJ, et al. (1993) MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature 365:756–759. [DOI] [PubMed] [Google Scholar]
- 13. Baugh JA, Chitnis S, Donnelly SC, Monteiro J, Lin X, et al. (2002) A functional promoter polymorphism in the macrophage migration inhibitory factor (MIF) gene associated with disease severity in rheumatoid arthritis. Genes Immun 3:170–176. [DOI] [PubMed] [Google Scholar]
- 14. Plant BJ, Gallagher CG, Bucala R, Baugh JA, Chappell S, et al. (2005) Cystic fibrosis, disease severity, and a macrophage migration inhibitory factor polymorphism. Am J Respir Crit Care Med 172:1412–1415. [DOI] [PubMed] [Google Scholar]
- 15. Freeman JV, Cole TJ, Chinn S, Jones PR, White EM, et al. (1995) Cross sectional stature and weight reference curves for the UK, 1990. Arch Dis Child 73:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knudson RJ, Lebowitz MD, Holberg CJ, Burrows B (1983) Changes in the normal maximal expiratory flow-volume curve with growth and aging. Am Rev Respir Dis 127:725–734. [DOI] [PubMed] [Google Scholar]
- 17. Kulich M, Rosenfeld M, Campbell J, Kronmal R, Gibson RL, et al. (2005) Disease-specific reference equations for lung function in patients with cystic fibrosis. Am J Respir Crit Care Med 172:885–891. [DOI] [PubMed] [Google Scholar]
- 18. Doring G, Conway SP, Heijerman HG, Hodson ME, Hoiby N, et al. (2000) Antibiotic therapy against Pseudomonas aeruginosa in cystic fibrosis: a European consensus. Eur Respir J 16:749–767. [DOI] [PubMed] [Google Scholar]
- 19. Faul F, Erdfelder E, Buchner A, Lang AG (2009) Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav Res Methods 41:1149–1160. [DOI] [PubMed] [Google Scholar]
- 20. Adamali H, Armstrong ME, McLaughlin AM, Cooke G, McKone E, et al. (2012) Macrophage migration inhibitory factor enzymatic activity, lung inflammation, and cystic fibrosis. Am J Respir Crit Care Med 186:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boyle MP (2007) Strategies for identifying modifier genes in cystic fibrosis. Proc Am Thorac Soc 4:52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai HJ, Shoff SM (2008) Classification of malnutrition in cystic fibrosis: implications for evaluating and benchmarking clinical practice performance. Am J Clin Nutr 88:161–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, et al. (2011) Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet 43:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Long AD, Langley CH (1999) The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res 9:720–731. [PMC free article] [PubMed] [Google Scholar]
- 25. Roger T, David J, Glauser MP, Calandra T (2001) MIF regulates innate immune responses through modulation of Toll-like receptor 4. Nature 414:920–924. [DOI] [PubMed] [Google Scholar]
- 26. Bonfield TL, Panuska JR, Konstan MW, Hilliard KA, Hilliard JB, et al. (1995) Inflammatory cytokines in cystic fibrosis lungs. Am J Respir Crit Care Med 152:2111–2118. [DOI] [PubMed] [Google Scholar]
- 27. Stotland PK, Radzioch D, Stevenson MM (2000) Mouse models of chronic lung infection with Pseudomonas aeruginosa: models for the study of cystic fibrosis. Pediatr Pulmonol 30:413–424. [DOI] [PubMed] [Google Scholar]
- 28. Meyer M, Huaux F, Gavilanes X, van den Brule S, Lebecque P, et al. (2009) Azithromycin reduces exaggerated cytokine production by M1 alveolar macrophages in cystic fibrosis. Am J Respir Cell Mol Biol 41:590–602. [DOI] [PubMed] [Google Scholar]
- 29. Sorio C, Melotti P (2009) Editorial: The role of macrophages and their scavenger receptors in cystic fibrosis. J Leukoc Biol 86:465–468. [DOI] [PubMed] [Google Scholar]
- 30. Di A, Brown ME, Deriy LV, Li C, Szeto FL, et al. (2006) CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat Cell Biol 8:933–944. [DOI] [PubMed] [Google Scholar]
- 31. Sorio C, Buffelli M, Angiari C, Ettorre M, Johansson J, et al. (2011) Defective CFTR expression and function are detectable in blood monocytes: development of a new blood test for cystic fibrosis. PLoS One 6:e22212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Konstan MW, Schluchter MD, Xue W, Davis PB (2007) Clinical use of Ibuprofen is associated with slower FEV1 decline in children with cystic fibrosis. Am J Respir Crit Care Med 176:1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ren CL, Pasta DJ, Rasouliyan L, Wagener JS, Konstan MW, et al. (2008) Relationship between inhaled corticosteroid therapy and rate of lung function decline in children with cystic fibrosis. J Pediatr 153:746–751. [DOI] [PubMed] [Google Scholar]
- 34. De Boeck K, Vermeulen F, Wanyama S, Thomas M (2011) Inhaled corticosteroids and lower lung function decline in young children with cystic fibrosis. Eur Respir J 37:1091–1095. [DOI] [PubMed] [Google Scholar]
- 35. Hansen CR, Pressler T, Koch C, Hoiby N (2005) Long-term azitromycin treatment of cystic fibrosis patients with chronic Pseudomonas aeruginosa infection; an observational cohort study. J Cyst Fibros 4:35–40. [DOI] [PubMed] [Google Scholar]
- 36. Konstan MW, Wagener JS, Pasta DJ, Millar SJ, Jacobs JR, et al. (2011) Clinical use of dornase alpha is associated with a slower rate of FEV1 decline in cystic fibrosis. Pediatr Pulmonol 46:545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Al-Abed Y, Dabideen D, Aljabari B, Valster A, Messmer D, et al. (2005) ISO-1 binding to the tautomerase active site of MIF inhibits its pro-inflammatory activity and increases survival in severe sepsis. J Biol Chem 280:36541–36544. [DOI] [PubMed] [Google Scholar]
- 38. Dahlgren MK, Garcia AB, Hare AA, Tirado-Rives J, Leng L, et al. (2012) Virtual screening and optimization yield low-nanomolar inhibitors of the tautomerase activity of Plasmodium falciparum macrophage migration inhibitory factor. J Med Chem 55:10148–10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full dataset of Cystic Fibrosis patients used in the present study.
(XLS)
Comparison of MIF -794 CATT genotype (a) or 5-CAAT allele (b) frequencies between Plant's [14] and the present study.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.