Abstract
Cryptochromes are photolyase-like blue/UV-A light receptors that evolved from photolyases. In plants, cryptochromes regulate various aspects of plant growth and development. Despite of their involvement in the control of important plant traits, however, most studies on cryptochromes have focused on lower plants and herbaceous crops, and no data on cryptochrome function are available for forest trees. In this study, we isolated a cryptochrome gene, PeCRY1, from Euphrates poplar (Populus euphratica), and analyzed its structure and function in detail. The deduced PeCRY1 amino acid sequence contained a conserved N-terminal photolyase-homologous region (PHR) domain as well as a C-terminal DQXVP-acidic-STAES (DAS) domain. Secondary and tertiary structure analysis showed that PeCRY1 shares high similarity with AtCRY1 from Arabidopsis thaliana. PeCRY1 expression was upregulated at the mRNA level by light. Using heterologous expression in Arabidopsis, we showed that PeCRY1 overexpression rescued the cry1 mutant phenotype. In addition, PeCRY1 overexpression inhibited hypocotyl elongation, promoted root growth, and enhanced anthocyanin accumulation in wild-type background seedlings grown under blue light. Furthermore, we examined the interaction between PeCRY1 and AtCOP1 using a bimolecular fluorescence complementation (BiFc) assay. Our data provide evidence for the involvement of PeCRY1 in the control of photomorphogenesis in poplar.
Introduction
Light is one of the most important environmental factors for plants as it provides the source of energy to sustain plant life [1]. Light is also a key signal controlling virtually every aspect of plant growth and development [2]. As a consequence, plants have the ability to sense multiple parameters of ambient light signals including light quantity (fluence), quality (wavelength), direction, and duration [1]. Light promotes the developmental transition from skotomorphogenesis to photomorphogenesis in plants through the combinatorial interaction of diverse sensory photoreceptors, which are classified based on the wavelength of light they perceive [3]. Light signals are perceived through at least four distinct families of photoreceptors including red/far-red (600–750 nm) light receptor phytochromes [4], blue/UV-A (320–500 nm) light receptor phototropins [5], cryptochromes [6], [7], ZEITLUPE (ZTL), FLAVIN BINDING, KELCH REPEAT, F-BOX1 (FKF1), and LOV KELCH PROTEIN2 (LKP2) and the UV-B (280–320 nm) light receptor UVR8 [8].
Cryptochromes are photolyase-like blue/UV-A light receptors that evolved from photolyases, a class of blue light-activated microbial DNA repair enzymes [9], [10], but rather than repairing DNA, cryptochromes regulate numerous aspects of growth and development in a wide range of organisms from bacteria to humans [11], [12]. In plants, cryptochromes mediate blue light-dependent inhibition of hypocotyl elongation, deetiolation responses, control of vegetative growth, flowering initiation, anthocyanin accumulation, regulation of gene expression, and the maintenance of plant endogenous rhythms [7].
Cryptochrome was first identified in Arabidopsis thaliana during the molecular characterization of the Arabidopsis mutant hy4, which exhibited elongated hypocotyls when grown in blue light [13]. The hy4 mutant was subsequently renamed cry1, defining the CRY1 gene encoding cryptochrome 1 [14], which mediates deetiolation in response to blue light. Subsequently, a second member of the Arabidopsis cryptochrome family, CRY2, which primarily regulates photoperiodic flowering [15], was identified by screening Arabidopsis cDNA libraries with CRY1 cDNA probes [16], [17]. A third CRY family member, CRY-DASH, from Arabidopsis has been characterized [18] and exhibits single-stranded DNA-specific photolyase activity [19]. Since the discovery of the first cryptochrome in Arabidopsis, this type of photoreceptor has been found widely in organisms ranging from bacteria to humans [11], [12]. In addition to Arabidopsis, cryptochromes have been studied in various photosynthetic species including algae [20], moss [21], fern [22], tomato [23], [24], rapeseed [25], pea [26], rice [27], [28], and apple [29], [30].
All cryptochromes are composed of two major domains, the N-terminal photolyase-homologous region (PHR) domain of approximately 500 residues and the CRY C-terminal extension (CCE) domain of various lengths. The PHR domain is required to bind the two chromophores flavin adenine dinucleotide (FAD) [31]–[33] and 5,10-methenyltetrahydrofolate (MTHF) [19], [34], whereas CCE is a cryptochrome effector domain [35] that governs the signaling activity of photoactivated cryptochromes.
The CRY1-PHR of Arabidopsis contains two subdomains similar to photolyase and CRY-DASH: an N-terminal α/β subdomain (residues 13–139) connected via a loop to the C-terminal α-helical subdomain (residues 217–495) [36]. The α/β subdomain has a five-stranded, parallel β-sheet flanked by four α-helices and a 310 helix (a less common α-helix with 3.0 residues per turn instead of 3.6 residues per turn) resembling a dinucleotide-binding domain. The CCE domains of plant cryptochromes are intrinsically unstructured [9] with little sequence similarity, and the cryptochromes of different plant species are distinguished mainly by their CCE domains [35]. However, plant cryptochromes from different species share a common DQXVP–acidic- STAESSS (DAS) sequence motif in their CCE domains.
Previous studies have indicated that Arabidopsis CRY1 and CRY2 were expressed ubiquitously in all cells and organ types examined [14], [15], [37]. The cryptochrome genes of some plant species exhibit expression patterns in different tissues similar to those in Arabidopsis and are expressed at higher levels during the early stages of development. As blue light receptors, plant CRY mRNA levels are regulated dramatically by light. For example, the expression of both Arabidopsis CRY1 and CRY2 is regulated by the circadian clock with peaks during the light phase and troughs during the dark phase [37].
In addition to CRY gene regulation at the transcript level, CRY protein levels are also regulated by light. Arabidopsis CRY2 protein is light-labile, but CRY1 protein is light-stable. Most plant CRY proteins are nuclear-localized; both Arabidopsis CRY1 and CRY2 accumulate in the nucleus, but AtCRY1 can also be detected in the cytosol of seedlings grown under either dark or light conditions [38]. Nuclear CRY1 protein has been shown to be responsible for blue light inhibition of hypocotyl elongation, whereas cytosolically localized CRY1 mediates blue light stimulation of cotyledon expansion and root elongation [38]. In contrast, CRY2 appears to complete its posttranslational life cycle in the nucleus [39].
Although the signal transduction mechanism of plant cryptochromes is not fully understood, cryptochromes are generally thought to interact with signaling proteins to regulate gene expression [1], [11], [12]. Results of DNA microarray analyses suggested that the expression of approximately 5–25% of the genes in the Arabidopsis genome changes in response to blue light with most of the changes mediated by CRY1 and CRY2 [40]–[43]. In Arabidopsis, both CRY1 and CRY2 were shown to interact directly with the constitutive photomorphogenic 1 (COP1) protein and inhibit its E3 ubiquitin ligase activity to prevent the degradation of COP1 substrates such as the long hypocotyl in far-red 1 (HFR1) [44] and CONSTANS (CO) proteins [45]. Therefore, the CRY proteins stabilize transcription factors that can regulate gene expression and promote photomorphogenesis in Arabidopsis.
Despite their involvement in the control of important plant traits such as plant height, root length, and flowering time, most studies on cryptochromes have focused only on lower plant species and herbaceous crops [30], [46]. The cryptochromes of forest tree species have not been extensively characterized. In this study, we isolated the PeCRY1 gene encoding a blue light receptor from the desert species Populus euphratica, analyzed its structure and relationship with CRY genes from other species, and substantiated the role of PeCRY1 in regulating plant height, root length, and anthocyanin accumulation. Our results indicate that PeCRY1 plays an important role in the regulation of growth and development of Euphrates poplar.
Results
Cloning of a Full-Length PeCRY1 cDNA
To isolate a full-length cDNA sequence of the Euphrates poplar cryptochrome gene PeCRY1, expressed sequence tag (EST) clones with similarity to Arabidopsis cryptochrome 1 were identified by analysis of Euphrates poplar dbEST sequences in the National Center for Biotechnology Information (NCBI) database. A 581-bp PeCRY1 fragment was isolated from Euphrates poplar leaves and 5′/3′-RACE extension methods were used to obtain the missing PeCRY1 sequences. The EST sequence (AJ768957) and two fragments were then combined based on analysis using DNAman software (Lynnon Corp., Pointe-Claire, QB, Canada) to obtain a full-length PeCRY1 sequence. The full-length PeCRY1 cDNA sequence contained a 2,046-bp open reading frame (ORF). The PeCRY1 ORF encoded a protein of 681 amino acids with a calculated mass of 76.9 kDa as predicted using DNAstar software. The deduced protein was basic with an isoelectric point (pI) of 5.64 as predicted using the DNAman software.
PeCRY1 Amino Acid Sequence Analysis
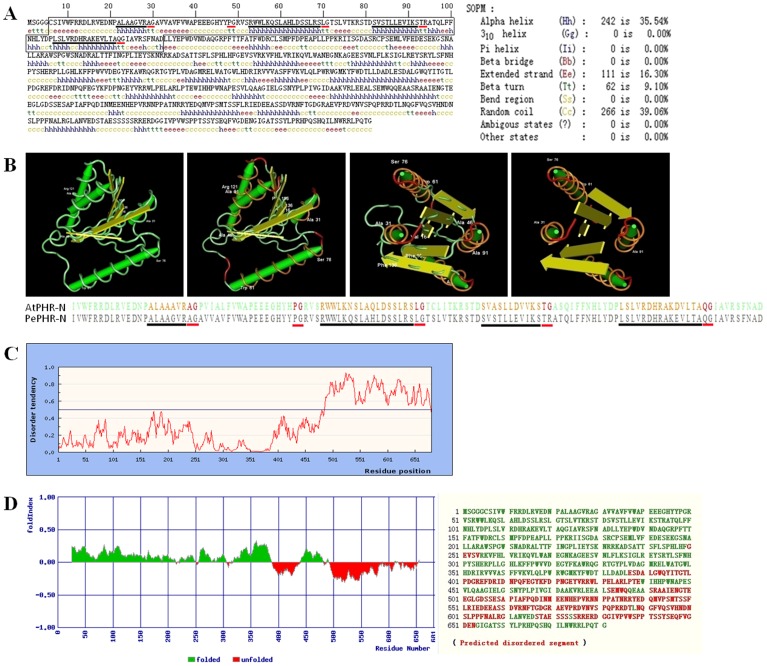
Sequence analysis using the CD-search program to explore the NCBI database (http://structure.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) revealed that the PeCRY1 amino acid sequence contained a well-conserved N-terminal PHR domain, which is required for binding of the chromophore flavin adenine dinucleotide (FAD) [25] and a C-terminal CCE region containing a DAS domain [35] (Fig. 1A).
Figure 1. PeCRY1 protein sequence analysis.
(A) Structural domains of the PeCRY1 protein. Analysis of protein sequences in the National Center for Biotechnology Information (NCBI) database was performed using the CD-search software. (B) Amino acid sequence alignment of cryptochrome proteins from Arabidopsis, winter rape, apple, tomato, European aspen, and Euphrates poplar. The alignment was constructed using DNAman version 5.2.2 software. Identical residues are highlighted by black boxes. Red lines under the sequences indicate the TGYP, WRWK, and LLDAD motifs. Black lines above the sequences indicate the DQMVP-E/D-STAESS (DAS) domain located in the C-terminal region. Residues that interact with FAD and MTHF are indicated by black rectangles and black dots, respectively. A nuclear localization signal (NLS) predicted using NLStradamus software is indicated by red stars. (C) DAS domain sequence logos. The sequence alignment of the domains was generated using ClustalX, and conserved motif logos were created using the WebLogo program (http://weblogo.threeplusone.com/).
AtCRY1 associates with two cofactors, the catalytic cofactor (FAD) and a light-harvesting cofactor (MTHF), in the same manner as Type I photolyases [31], [47]. Twelve of 13 amino acids in AtCRY1 predicted to interact with FAD were conserved in PeCRY1 with the exception of the serine at position 359, which was replaced by an alanine (Fig. 1B). In addition, six of seven identical amino acid residues (His-52 was replaced by Gln) known to interact with MTHF were also conserved in PeCRY1 (Fig. 1B). In addition, the TGYP motif, which is conserved in all Type I photolyases and forms a part of the FAD-binding domain [47]; the WRWG motif, which is well conserved among photolyases and cryptochromes [14], [48]; and the LLDAD motif, which is a conserved region of the FAD-binding pocket of cryptochromes [31], [49], were also present in PeCRY1 (Fig. 1B).
Three important motifs, known collectively as the DAS domain (Fig. 1B), including the DQXVP motif of unknown function, an acidic region (represented by E and D), and the STAESS motif implicated in the interaction with phytochrome A (phyA) (Fig. 1B, C) [50], were present in the PeCRY1C-terminal region and conserved, although the overall similarity was low.
Structural Analysis of the PeCRY1 Amino Acid Sequence
The PeCRY1 secondary structure was solved using the self-optimized prediction method (SOPM, http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopm.html) [51]. PeCRY1 consists of α-helices (242 aa, 35.54%), β-turns (62 aa, 9.10%), extended strands (111 aa, 16.30%), and random coil (266 aa, 39.06%) regions (Fig. 2A). The α-helices and β-turns were distributed randomly throughout the PeCRY1 polypeptide (Fig. 2A).
Figure 2. Structural analysis of the PeCRY1 protein.
(A) The secondary structure of PeCRY1 solved by the self-optimized prediction method (SPOM). The black box indicates the PeCRY1 PHR-N domain corresponding to that of AtCRY1. Black and red lines beneath the sequence indicate the α-helices and the β-turns in the PHR-N domain. (B) Comparison of the predicted three-dimensional structures of PeCRY1 and AtCRY1 using Cn3D software. Orange and red segments in the images correspond to the sequences marked by black and red lines, respectively, in (A). (C) Local disorder tendency of the PeCRY1 CCE domain based on an estimated-amino-acid-pairwise-energy-content analysis using IUPred software. (D) The fold disordering character of PeCRY1 predicted using FoldIndex software.
The α/β subdomain of CRY1-PHR in Arabidopsis has five-stranded, parallel β-sheets flanked by four α-helices and a 310 helix, which collectively resemble a dinucleotide-binding domain. Although a 310 helix was not present in PeCRY1, four α-helices were identified in the same region as in AtCRY1 using the SOPM method (Fig. 2A). Moreover, we identified five β-turns in the α/β subdomain (Fig. 2A), similar to AtCRY1 using the Cn3D macromolecular structure viewer software (PDB: 1U3C_A), suggesting that these β-turns might form parallel β-sheets in this region and be flanked by the four α-helices as in AtCRY1 (Fig. 2B).
CRY proteins from different plant species are distinguished mainly by C-terminal extensions [29]. Previous studies suggested that the CCE domain is important for cryptochrome function in plants [14] and is intrinsically unstructured [12]. Analysis using circular dichroism and nuclear magnetic resonance (NMR) demonstrated that the CCE domains of Arabidopsis CRY1 and human CRY2 are unstructured [12]. We confirmed the intrinsically unstructured nature of the PeCRY1 CCE domain based on an analysis of estimated amino acid pairwise energy content using IUPred software (http://iupred.enzim.hu/) (Fig. 2C). In addition, we investigated the fold disordering character of PeCRY1 using FoldIndex software (http://bip.weizmann.ac.il/fldbin/findex) [52]. We identified seven disordered regions in the PeCRY1 sequence with the longest disordered region (97 aa) located in the CCE domain with a total of 218 disordered amino acid residues (Fig. 2D).
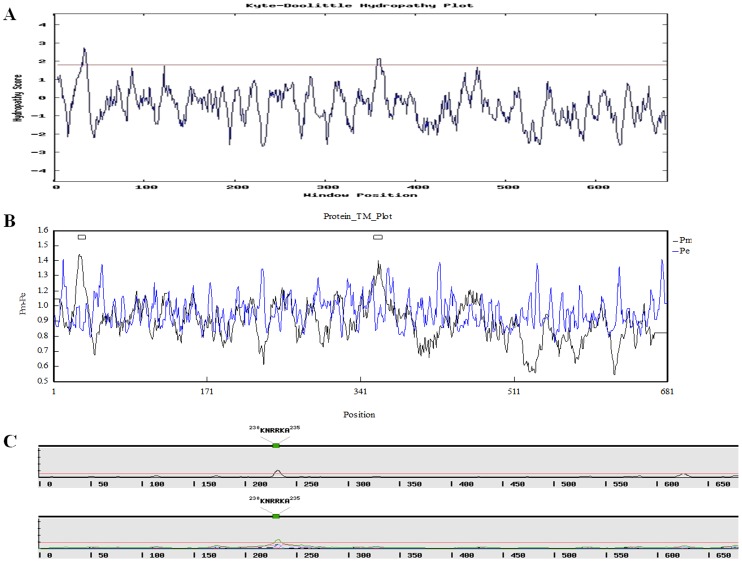
We also investigated the hydrophilicity/hydrophobicity of PeCRY1 using the Kyte and Doolittle method (http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm) [53]. The majority of PeCRY1 amino acids were hydrophilic, and almost all of the C-terminal region amino acids were hydrophilic, indicating that PeCRY1 is a hydrophilic protein (Fig. 3A). In addition, two PeCRY1 transmembrane domains with scores above 1.8 were also predicted using the Kyte and Doolittle method (Fig. 3A). To confirm this result, we examined the PeCRY1 transmembrane domains using DNAman software, which identified two possible transmembrane domains in PeCRY1, the first located at amino acids 23–40 and the second at amino acids 351–369 (Fig. 3B), in agreement with the results obtained using the Kyte and Doolittle method.
Figure 3. Hydrophilicity/hydrophobicity analysis and PeCRY1 transmembrane domain prediction.
(A) Hydrophilicity/hydrophobicity analysis of PeCRY1 performed using the Kyte and Doolittle method. A score of 1.8 is indicated by the red line. (B) PeCRY1 transmembrane domain prediction using DNAman version 5.2.2 software with default options. Predicted transmembrane regions are indicated by boxes above the profile. (C) PeCRY1 nuclear localization signal (NLS) prediction using a simple hidden Markov model (HMM) and NLStradamus software. The analysis was performed using both a 2-state HMM dynamic model (above) and a 4-state HMM static model (below).
Most plant CRY proteins are thought to be nuclear localized proteins. Both Arabidopsis CRY1 and CRY2 accumulate in the nucleus [35]. Using NLStradamus software (http://www.moseslab.csb.utoronto.ca/NLStradamus/), a simple hidden Markov model (HMM) for nuclear localization signal prediction, we identified a nuclear localization signal (NLS) sequence “KNRRKA” in PeCRY1 at amino acids 230–235 (Fig. 3C), indicating that PeCRY1 is a nuclear localized protein. Analysis with the NLStradamus software using both 2-state HMM dynamic and 4-state HMM static models gave the same results.
Relationship of PeCRY1 to Other Cryptochromes
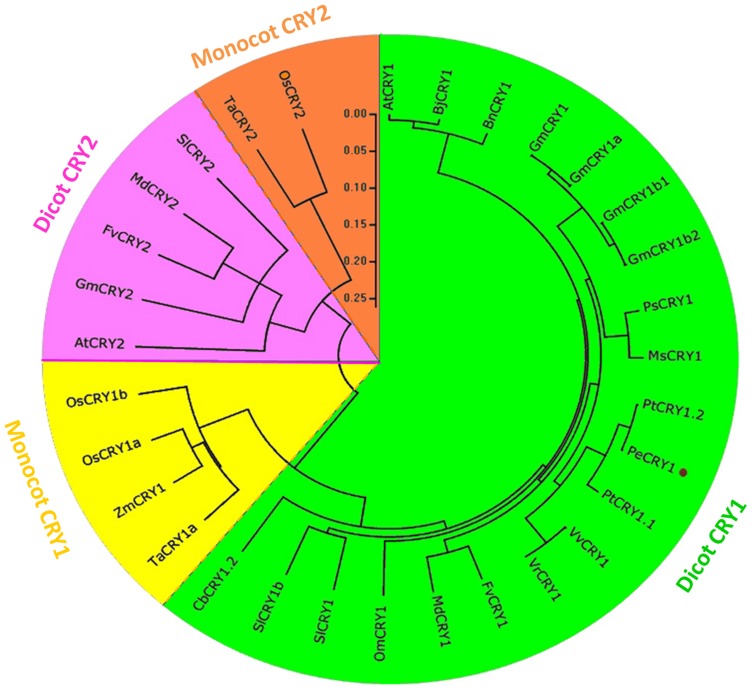
To analyze the phylogenetic relationship between PeCRY1 and the CRY proteins from other plant species, we performed a phylogenetic analysis of 30 plant cryptochromes representing 14 diverse species using the Mega4.1 program and the Clustal method (Fig. 4). The phylogenetic tree indicated obvious boundaries between the dicotyledonous and monocotyledonous CRY1 proteins. PeCRY1 grouped in the dicot CRY1 clade and was most closely related to the PtCRY1.1 (JN235115.1) and PtCRY1.2 (JN235116.1) CRY proteins of European aspen and the VvCRY1 (EU188919.1) and VrCRY1 (ABX80391.1) CRY proteins of grape, all of which clustered in the same clade. In contrast, PeCRY1 was most distantly related to the CRY1 proteins of monocotyledonous species, including wheat TaCRY1a (ABX58028.1), maize ZmCRY1 (AFW71952.1), and rice OsCRY1a (BAB70686.1) and OsCRY1b (BAB70688.2). In addition, the phylogenetic tree indicated that the plant CRY1 and CRY2 proteins formed in different groups with clear boundaries between dicotyledonous and monocotyledonous species (Fig. 4), demonstrating the consistency of plant evolution.
Figure 4. Phylogram of plant cryptochromes.
CRY amino acid sequences from 14 diverse plant species were obtained from the NCBI database. The alignment was constructed using ClustalX and the phylogenetic tree was constructed using the neighbor-joining method of MEGA version 4.1 software. Each node corresponds to a number indicating the bootstrap value for 1000 replicates. The scale bar represents 0.05 substitutions per sequence position. PeCRY1 is denoted by a red dot. At, Arabidopsis thaliana; Bj, Brassica juncea; Bn, Brassica napus; Gm, Glycine max; Ps, Pisum sativum; Ms, Medicago sativa; Pt, Populus tremula; Pe, Populus euphratica; Vv, Vitis vinifera; Vr, Vitis riparia; Fv, Fragaria vesca; Md, Malus domestica; Om, Orobanche minor; Sl, Solanum lycopersicum; Cb, Chrysanthemum boreale; Ta, Triticum aestivum; Zm, Zea mays; Os, Oryza sativa.
PeCRY1 Expression Analysis
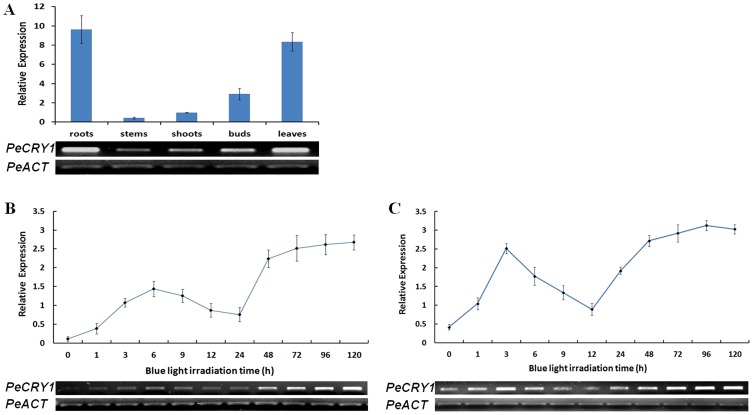
Previous studies showed that cryptochrome genes such as AtCRY1 were expressed ubiquitously in all cell types and organs examined [14], [15], [37] and that they regulated different aspects of plant growth and development [7]. To examine whether PeCRY1 transcript levels were tissue-specific, we performed semiquantitative and real-time quantitative reverse transcription (RT)-PCR analysis using total RNAs obtained from different tissues including roots, stems, shoots, buds, and leaves. PeCRY1 was expressed in all of the tissues we examined and the expression levels varied among the tissues (Fig. 5A). PeCRY1 expression was highest in the roots and leaves, followed by the buds and shoots with only weak expression detected in the stems.
Figure 5. Analysis of PeCRY1 expression using semiquantitative and real-time quantitative RT-PCR.
(A) PeCRY1 transcript levels in different tissues. (B) and (C) PeCRY1 transcript levels in cultured seedlings or callus with different durations of blue light irradiation. Poplar actin (PeACT) expression was used as an internal control.
As blue light receptors, the expression of cryptochrome genes such as AtCRY1 and AtCRY2 is regulated by blue light [35]. To study the effect of blue light on PeCRY1 expression, cultured Euphrates poplar seedlings were grown in darkness for 3 days and then exposed to blue light for 0–120 h. Upon irradiation of the seedlings with blue light, PeCRY1 exhibited low basal expression followed by a clear increase in transcript levels with extended irradiation time (Fig. 5B). The PeCRY1 transcript levels reached a peak at 6 h, then decreased to a minimum at 24 h. Subsequently, transcript levels increased markedly to a peak at 72 h and remained stable through 120 h. Although the seedlings were treated with continuous blue light, the PeCRY1 expression pattern exhibited a circadian rhythm during the first 24 h (Fig. 5B), similar to the observation in previous studies that CRY transcript levels exhibit an oscillation period of almost 24 h [26], [37]. To confirm the expression pattern of PeCRY1 under blue light, we also analyzed PeCRY1 expression using Euphrates poplar callus under the same light conditions with results similar to those obtained from the seedlings. However, the relative PeCRY1 background expression was higher and responded more rapidly to blue light in the callus cells than in the cultured seedlings with a expression peak at 3 h, and the transcript rhythm cycle was within 12 h (Fig. 5C).
Functional Complementation of the Arabidopsis cry1 Mutant by PeCRY1
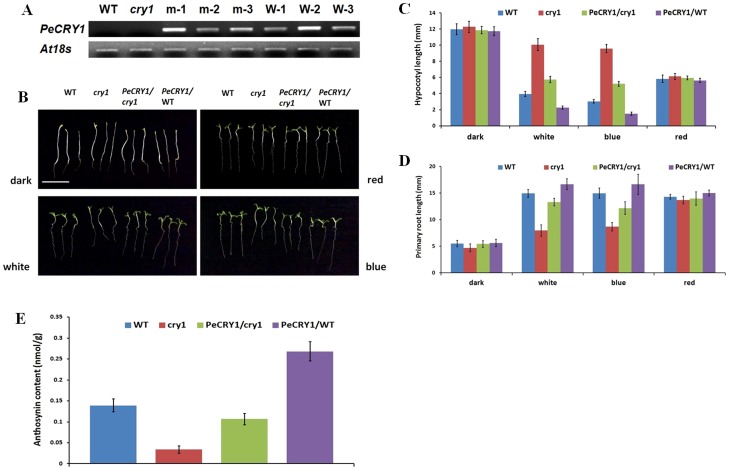
To investigate the function of PeCRY1 in plants, we conducted a functional complementation assay using an Arabidopsis cry1 mutant. PeCRY1 was introduced into a pRI vector (pRI 101-AN) with expression driven by a CaMV-35S promoter followed by a 58-bp AtADH 5′UTR enhancer (35S::PeCRY1) and transformed into wild-type (WT; Columbia ecotype) and cry1-mutant Arabidopsis backgrounds using the floral dip method [54]. After repeated selection on kanamycin and PCR screening for the presence of the transgene, at least three transformants of each background were obtained. Transgene expression levels were determined by semiquantitative RT-PCR to verify successful transformation (Fig. 6A). Two transgenic lines, m-1 and W-2, with high levels of PeCRY1 expression were chosen to compare the phenotypes of the WT, cry1 mutant, cry1 mutant transformed with PeCRY1, and PeCRY1 overexpressing lines.
Figure 6. Phenotypes of wild-type (WT), cry1-mutant, PeCRY1-transgenic cry1-mutant, and PeCRY1-transgenic WT plants.
(A) PeCRY1 transcript levels in WT, cry1 mutant, and transgenic lines. m-1, m-2, and m-3, three cry1 mutant lines transformed with PeCRY1; W-1, W-2, and W-3, three WT lines transformed with PeCRY1. (B) Phenotypes of WT, cry1-mutant, and the two transgenic lines (m-1 and W-2) grown under dark, white, blue, and red light. Scale bar represents 1 cm. (C) and (D) Hypocotyl and primary root lengths of WT, cry1-mutant, and the two transgenic lines grown under dark, white, blue, and red light. (E) Anthocyanin content of WT, cry1-mutant, and the two transgenic lines grown under blue light.
PeCRY1 Inhibits Hypocotyl Elongation in Arabidopsis
Previous research indicated that AtCRY1 plays a prominent role in the inhibition of hypocotyl elongation [14] and that many other plant cryptochrome genes also inhibit hypocotyl elongation [23], [24], [29], [30]. To determine if PeCRY1 inhibits hypocotyl elongation, we measured the hypocotyl lengths of the four types of Arabidopsis lines. When the seedlings were grown in complete darkness, the hypocotyl lengths did not differ significantly among the four genotypes (Fig. 6B, C). However, when grown under white light, the cry1 mutant lines exhibited reduced inhibition of hypocotyl elongation, which was restored by transformation of the cry1 mutant with PeCRY1. The PeCRY1-overexpressing lines exhibited the shortest hypocotyl lengths of the three genotypes (Fig. 6B, C). To examine whether the inhibitory effect was specific to blue light, seedlings of the four genotypes were grown under continuous blue or red light. Under blue light, the hypocotyls of cry1 mutant and WT plants transformed with PeCRY1 were shorter than those of the untransformed cry1 mutant or WT plants. The hypocotyl lengths were not significantly different among the four Arabidopsis types when grown under red light, indicating that the inhibitory effects of PeCRY1 were much less dramatic under red light (Fig. 6B, C). These results demonstrated that PeCRY1 exhibited the same function as AtCRY1 in the inhibition of hypocotyl elongation in plants.
PeCRY1 Promotes Root Elongation in Arabidopsis
Similar to the differences in hypocotyl elongation, primary root lengths were clearly different among the four genotypes when grown under light with the root lengths of the cry1 mutants being significantly shorter than those of WT and PeCRY1 transgenic plants (Fig. 6B, D). AtCRY1 has been shown to promote root elongation under blue light conditions, while cry1 mutant seedlings exhibit decreased root elongation [2]. In this study, we showed that PeCRY1 also promoted root elongation. Transformation with PeCRY1 complemented the reduced root elongation phenotype of the cry1 mutant and WT plants transformed with PeCRY1 had the longest roots. These results demonstrated that PeCRY1 can regulate plant root elongation and that this regulation is more sensitive to blue light, similar to AtCRY1.
PeCRY1 Increases Anthocyanin Accumulation in Arabidopsis
Previous studies showed that anthocyanin accumulation decreased markedly in the Arabidopsis cry1 mutant and increased significantly in Arabidopsis seedlings overexpressing AtCRY1 [14]. Many other plant cryptochrome genes, such as MdCRY1, SlCRY1, and BnCRY1, promote anthocyanin accumulation as shown by increased anthocyanin levels in Arabidopsis seedlings transformed with these genes. Transformation with PeCRY1 also resulted in significantly increased anthocyanin accumulation in both cry1 mutant and WT seedlings (Fig. 6E). These results suggest that PeCRY1 promotes anthocyanin accumulation in plants grown under blue light.
PeCRY1 Interacts with AtCOP1 and AtSPA1
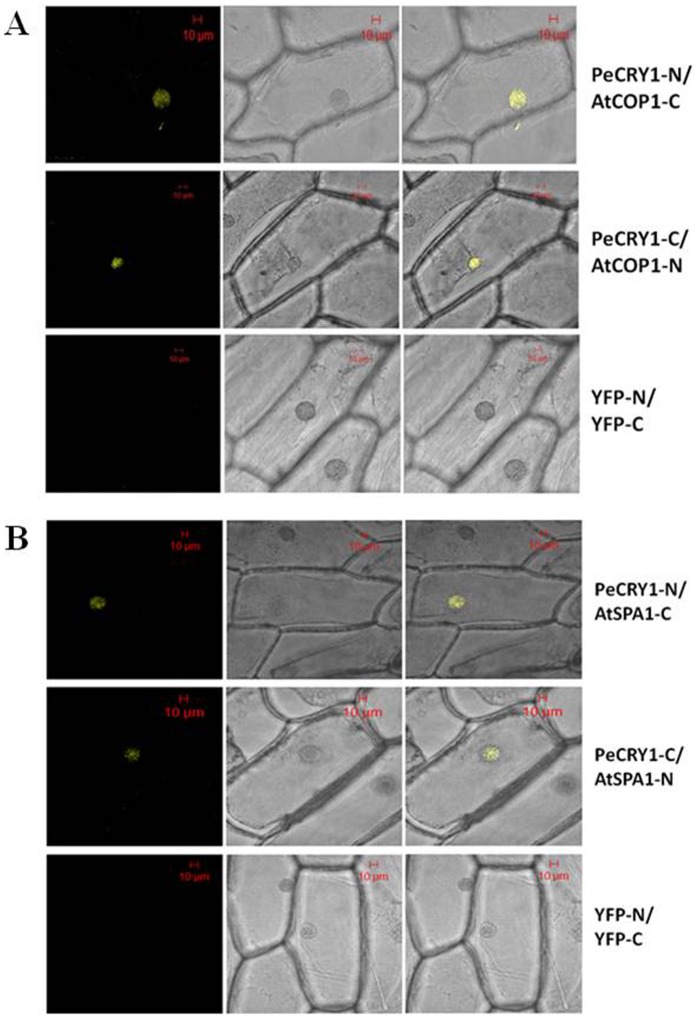
Although the signal transduction mechanism of plant cryptochromes is not fully understood, cryptochromes are generally considered to regulate plant growth and development through interactions with signaling proteins that regulate the expression of downstream genes [1], [11], [12]. In Arabidopsis, both CRY1 and CRY2 act largely through direct interaction with COP1 [55], [56]. Based on the highly similar structures and functions of PeCRY1 and AtCRY1, we performed bimolecular fluorescence complementation (BiFC) assays to determine whether PeCRY1 interacts with AtCOP1 and whether PeCRY1 function is conserved between poplar and Arabidopsis.
Full-length PeCRY1 and AtCOP1 cDNAs were fused to the N-terminal and C-terminal halves, respectively, of yellow fluorescent protein (YFP). The fusion proteins were then introduced transiently into onion epidermal cells. Yellow fluorescence was clearly detectable when the two proteins were co-transformed, suggesting that PeCRY1 and AtCOP1 interact in vivo in plant cells (Fig. 7A). Besides, it has also been hypothesized that CRY1 suppress the activity of COP1 by interacting with SPA1. We found PeCRY1 also interacts with AtSPA1 just as in Arabidopsis using BiFc assays (Fig. 7B). These results provided additional evidence suggesting that PeCRY1 is a poplar counterpart of AtCRY1 and plays a role in the molecular mechanism of the light signal transduction pathway in poplar. Moreover, these results demonstrated that PeCRY1 is a nuclear-localized protein as predicted (Fig. 3C).
Figure 7. PeCRY1 interacts with AtCOP1 and AtSPA1 respectively in bimolecular fluorescence complementation (BiFc) assays.
Discussion
The blue light receptor cryptochromes play vital roles in plant growth and development. Studies on different species over the last 20 years indicate that cryptochromes are probably the most widely spread photoreceptors in nature and play various biological functions across the three major evolutionary lineages, bacteria, plants, and animals [35]. In this study, we cloned the cryptochrome gene PeCRY1 from Euphrates poplar and demonstrated its function as a blue light receptor similar to that of AtCRY1 in Arabidopsis.
PeCRY1 Sequence Analysis
CRY apoprotein contains two characteristic domains: the N-terminal PHR and C-terminal CCE domains [57]. In this study, we showed that PeCRY1 also contains these conserved domains as well as the FAD-binding and C-terminal DAS domains (Fig. 1). In addition, specific amino acids and the three amino acid motifs (TGYP, WRWG, and LLDAD) located in the N-terminal PHR domain and crucial for binding with FAD or MTHF were also present in the PeCRY1 sequence (Fig. 1B). The presence of these highly conserved domains indicated that the function of PeCRY1 should be similar to other CRY proteins, especially AtCRY1.
In contrast to the conserved N-terminal PHR domain, cryptochrome C-terminal CCE domains share little sequence similarity with each other, which provides the primary means of distinguishing the cryptochromes of different plant species [35]. Based on amino acid sequence analysis using IUPred and FoldIndex software, we indentified an unstructured C-terminal region in PeCRY1 located between amino acids 480 and 650, similar to the AtCRY1 protein (Fig. 2B, C). Because unstructured regions are found frequently in signaling proteins, they have been hypothesized to be important for protein–protein interactions because they confer structural plasticity for interactions with multiple partners, more favorable energy costs for high specificity/low affinity binding of the partner proteins, and accessible posttranslational modification sites often recognized not only by signaling partners, but also by regulatory proteins [58], [59]. The CCE domain of CRY1 in Arabidopsis is necessary for its interaction with COP1 [55] and the interaction is light-dependent. Based on the high similarity of predicted secondary and tertiary structures between PeCRY1 and AtCRY1, we propose that PeCRY1 may be a blue light photoreceptor in Euphrates poplar that undergoes a light-induced conformational change to transmit the light signal through interaction with other proteins such as COP1 to regulate plant growth and development. The results of the BiFc assay showing that PeCRY1 can interact with AtCOP1 support this hypothesis and raise the possibility that one or more proteins like COP1 may interact with PeCRY1 in Euphrates poplar to regulate plant growth under light conditions. The identification of other proteins that can interact with PeCRY1 and how such protein–protein interactions might transmit light signals awaits further study.
Analysis of PeCRY1 Expression
Expression of CRY genes is regulated by endogenous circadian rhythms, light quality, and day length. As in Arabidopsis, cryptochrome mRNA levels are regulated by light in tomato [60], pea [26], [61], Brassica [25], and apple [29], [30]. CRY genes in different plant species respond differently to light induction. In garden pea, blue light inhibits CRY gene expression [26], but enhances CRY1 expression in Brassica napus [25]. In Arabidopsis, the expression of cryptochrome genes was induced by blue light and exhibited an oscillation period of almost 24 h [26], [37]. In our light treatment experiments, we showed that PeCRY1 expression is induced by blue light and that the circadian rhythm of its expression is similar to that of AtCRY1 (Fig. 5B). In addition, analysis of PeCRY1 expression in different Euphrates poplar tissues indicated that it was expressed constitutively (Fig. 5A) similar to AtCRY1 expression in Arabidopsis [14], [15], [37] and in accordance with the various functions of cryptochrome genes in different tissues [35]. These results demonstrate the role of PeCRY1 in photoresponses and provide preliminary evidence that PeCRY1 may be a blue/UV-A light receptor in Euphrates poplar.
Studies using green fluorescent protein (GFP)/phytochrome fusion proteins have demonstrated that both phyA and phyB are expressed in Arabidopsis root tissues [62] as are phototropins [63] and cryptochromes [37]. In addition, both red and blue light were effective in promoting significantly increased primary root length compared to roots of dark-grown control plants [2]. In this study, we showed that PeCRY1 was expressed at a high level in Euphrates poplar root tissue and promoted primary root elongation (Fig. 6B, D). How the expression of a photoreceptor such as PeCRY1 can be induced in underground tissues without direct light is an interesting question. One possible explanation is that some light may penetrate soil to activate the three major plant photosensory receptors [64]. However, the functions of both AtCRY1 and AtCRY2 in regulating primary root elongation have been shown to be dependent on the transport of auxin hormone from the aerial part of the plant to the root tissues [2]. Studies of the last 20 years in different plant species also indicated that cryptochrome genes can regulate the synthesis, transport, or concentration of hormones such as auxin [65], [66], gibberellic acid (GA) [41], and ethylene [40]. However, if root tissue growth is regulated solely by hormones transmitted from aerial tissues, the reason for the high level of PeCRY1 expression in root tissue remains unclear.
PHYA and PHYB proteins in root tissues have been reported to be involved in the transmission of some hormone signals from the shoot [67]. In addition, previous studies indicated that the expression of the cryptochrome gene MdCRY2 in apple may be regulated by the hormone abscisic acid (ABA) [29]. Based on these results, we propose that in addition to their growth-regulating functions, the hormones that are regulated by cryptochrome genes and transmitted from aerial to underground tissues may promote the expression of crucial genes in roots such as PeCRY1. For example, we found that PeCRY1 expression was regulated by indole acetic acid (IAA) and ABA (data not shown). Once the genes are thus induced in the roots, they can perform their functions in the same manner as in the aerial tissues, but in the absence of light. This hypothesis also leads to other questions such as whether cryptochrome expression can also be induced by hormones in the aerial tissues and whether genes expressed in underground tissues can regulate the expression of genes in the aerial tissues in the same manner. Based on the high level of PeCRY1 expression in root tissue, we can deduce that PeCRY1 has important functions in root development such as regulating primary and lateral root elongation and the proportion of primary and lateral roots, which are important in tree growth and development.
Functional Complementation of the Arabidopsis cry1 Mutant
Analysis of the effects of PeCRY1 on inhibition of hypocotyl elongation, promotion of root elongation, and anthocyanin accumulation indicated that PeCRY1 could complement the Arabidopsis cry1 mutant phenotype (Fig. 6). These results, coupled with the expression analysis, suggest that PeCRY1 is a counterpart of AtCRY1. In addition, PeCRY1 has the two opposite functions of promoting root elongation, but inhibiting hypocotyl elongation, just as AtCRY1 does in Arabidopsis.
Previous studies suggested that reduced hypocotyl length resulting from blue light perception is caused primarily by a reduction in cell length and not by a reduction in cell number [68]. In addition, the effect of cryptochrome photoreceptors on root growth can be explained, to a large extent, by changes in cell length, as opposed to a change in the number of cell divisions [2]. We can conclude that at the cellular level, cryptochrome photoreceptors regulate hypocotyl and root elongation by the same mechanism. Why the same mechanism leads to opposite results remains unclear.
Auxin regulates cell elongation in numerous plant tissues, and polar auxin transport inhibitors have long been known to affect elongation growth and tropisms in roots and shoots [2], [69]. Given that cryptochrome can regulate the synthesis, transport, and concentration of hormones, we believe that cryptochrome can regulate the extent of cell elongation in different tissues by adjusting the concentration of auxin with low auxin concentration promoting growth and high auxin concentration inhibiting growth. Additional studies are needed to test this hypothesis and to fully clarify the correlation between blue light transmission and hormone signal transduction mechanisms.
Materials and Methods
Ethics Statement
No specific permits were required for the described field studies. The location is not privately-owned or protected in any way, and the field studies did not involve endangered or protected species.
Plant Material And Growth Conditions
Samples of roots, stems, shoots, buds and leaves were collected from adult trees of euphrates poplar (Populus euphratica) in Xinjiang province, China. The poplar forest locates along the Tarim River in western China (41.0526°N; 86.2289°E).
Euphrates poplar seedlings were grown on MS medium with 0.4 mM IBA, and grown in controlled environment cabinets under 16 h light/8 h dark conditions at 23°C unless stated otherwise.
Euphrates poplar callus were grown on MS medium with 0.5 mM 2,4-D and 1.5 mM 6-BA, and grown in controlled environment cabinets in the dark at 23°C unless stated otherwise.
Columbia lines of Arabidopsis thaliana were used as the wild type (WT). Seeds of Columbia, cry1 -mutant and transgenic lines were sown on MS medium, cold-treated for 3 days at 4°C, and then transferred to controlled environment cabinets under long days (16 h light/8 h dark) conditions at 22°C.
Experiments involving dark or different light treatments were performed in a controlled environment chamber using the blue (5 µmol m−2 s−1), red (5 µmol m−2 s−1) or white (150 µmol m−2 s−1) tubes.
Isolation of Full-Length cDNA of PeCRY1 by Rapid Amplification of cDNA Ends (RACE)
An EST sequence (AJ768957) with 581 bp in length encoding PeCRY1 was isolated though RT-PCR using the degenerate primer pair Y1EST-F (5′-TCTTGGTTGGCAATACATAACC-3′) and Y1EST-R (5′- TTGGTTGTGGACTGACATT-3′). To obtain the full-length gene, 5′- and 3′-RACE was used. Total RNA was isolated from euphrates poplar leaves using the Trizol (Invitrogen, Carlsbad, CA, USA) method according to the manufacturer's instructions. The 5′-RACE primers Y1-5-R1 (5′-GCACGTATTCTCCATTTGGGTC-3′) and Y1-5-R2 (5′-GGTTATGTATTGCCAACCAAG -3′), and the 3′-RACE primers Y1-3-F1 (5′-GACGAAGAAGCTTCTTCAG-3′) and Y1-3-F2 (5′-GATGTAAATGTCAGTCCAC-3′) were designed on the basis of the PeCRY1 EST sequence. The PCR products of expected sizes were purified, cloned into the pMD18-T vector (Takara, Otsu, Japan), and sequenced. Then, the putative 3′-and 5′-RACE cDNAs and the EST sequence were over-lapped with DNAMAN to form a cDNA contig, which was used to determine the putative initiating translation codon (ATG) and open reading frame (ORF). To obtain a full-length cDNA of PeCRY1, a pair of full-length primers PeCRY1-F (5′-ATGTCAGGAGGTGGGTGTAG-3′) and PeCRY1-R (5′-TTACCCGGTTTGGGGTAGCC-3′) were designed according to the contig. The full length of PeCRY1 was then obtained by RT-PCR using the full-length primers.
Expression Analysis
RNA was extracted using TRizol Reagent (Invitrogen, Carlsbad, CA, USA), then reverse transcribed using the PrimeScript First Strand cDNA Synthesis Kit (Takara, Otsu, Japan), following the manufacturer's instructions. 10 µl of cDNA was diluted to a final volume of 100 µl with water.
Semiquantitative RT-PCR was carried out in 25-µl reactions with 5 ng diluted cDNA template. The PCR profile was 94°C for 3 min, 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s, with a 5 min extension at 72°C. The primers used were bdlY1-F (5′-CTGCTACAAATCGCCGCTAC-3′) and bdlY1-R (5′-CATCACCAACAAACTGCTCTG-3′). PeACT cDNA amplification was used as the external control, and primers used were PeACT-F (5′-GTCCTCTTCCAGCCATCTC-3′) and PeACT-R (5′-TTCGGTCAGCAATACCAGG-3′). PCR products were electrophoresed on a 1.5% agarose gel and viewed under UV light after standard staining with ethidium bromide.
For real-time quantitative RT-PCR analysis, the specific primer used were DLY1-F (5′-GTCCCTTCACAACCTTTGCT-3′) and DLY1-R (5′-GCATCACCCGAAATAATCCT -3′). Poplar actin (PeACT) gene was used as loading controls. Fluorescence-quantitative PCR reactions were repeated three times.
Generation of PeCRY1-Transgenic Arabidopsis Plants
To generate Arabidopsis PeCRY1 overexpressing lines, the full length of PeCRY1 was amplified by PCR using the primers PeCRY1-F and PeCRY1-R. The amplified cDNA was cloned into the expression vector pRI (pRI 101-AN) under the control of the cauliflower mosaic virus (CaMV)-35S promoter. The plasmids were then transformed into WT and cry1 mutant Arabidopsis separately, mediated by Agrobacterium GV3101 using the floral dip method (Clough and Bent 1998).
Hypocotyl And Root Measurements
For hypocotyl and root growth experiments, hypocotyl and root lengths of at least 30 Arabidopsis seedlings grown in appointed conditions were measured. Experiments were performed in at least three independent biological replicates.
Measurement of the Total Anthocyanin Concentration
Total anthocyanin was extracted according to methanol–HCl method, in which the samples were extracted independently overnight in 5 ml methanol and 1% (v/v) HCl with extraction at room temperature. The absorbance of each extract was measured at 530, 620 and 650 nm with a spectrophotometer (UV-1600, Shimadzu, Kyoto, Japan). The relative anthocyanin content was determined by the formula OD = (A530 - A620) - 0.1(A650 - A620). One unit of anthocyanin content was expressed as a change of 0.1 OD (unit ×103/g FW).
Bimolecular Fluorescence Complementation (BiFC)
The full-length PeCRY1 and AtCOP1 cDNAs were cloned into pYFP-N (1–155) and pYFP-C (156–239) vectors and sequenced. Onion epidermal cells were transiently transformed using the Agrobacteriumin fection method with different combinations of these constructs. YFP-dependent fluorescence was detected 24 h after transfection using a confocal laser scanning microscope (Carl Zeiss; LSM 510 Meta).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This work is supported by the Special Fund for Forest Scientific Research in the Public Welfare (201404102), NSF/IOS-0923975, Changjiang Scholars Award, and “Thousand-person Plan” Award (http://www.1000plan.org/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230. [DOI] [PubMed] [Google Scholar]
- 2. Canamero RC, Bakrim N, Bouly JP, Garay A, Dudkin EE, et al. (2006) Cryptochrome photoreceptors cry1 and cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana . Planta 224:995–1003. [DOI] [PubMed] [Google Scholar]
- 3. Sullivan JA, Deng XW (2003) From seed to seed: the role of photoreceptors in Arabidopsis development. Developmental Biology 260:289–297. [DOI] [PubMed] [Google Scholar]
- 4. Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, et al. (1995) Phytochromes: photosensory perception and signal transduction. Science 268:675–680. [DOI] [PubMed] [Google Scholar]
- 5. Briggs WR, Huala E (1999) Blue-light photoreceptors in higher plants. Annu Rev Cell Dev Biol 15:33–62. [DOI] [PubMed] [Google Scholar]
- 6. Cashmore AR, Jarillo JA, Wu YJ, Liu D (1999) Cryptochromes: Blue light receptors for plants and animals. Science 284:760–765. [DOI] [PubMed] [Google Scholar]
- 7. Lin C (2002) Blue light receptors and signal transduction. Plant Cell 14 suppl.:S207–S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rizzini L, Favory JJ, Cloix C, Faggionato D, O'Hara A, et al. (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332:103–106. [DOI] [PubMed] [Google Scholar]
- 9. Partch C, Sancar A (2005a) Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem Photobiol 81:1291–1304. [DOI] [PubMed] [Google Scholar]
- 10. Banerjee R, Batschauer A (2005) Plant blue-light receptors. Planta 220:498–502. [DOI] [PubMed] [Google Scholar]
- 11. Cashmore AR (2003) Cryptochromes: enabling plants and animals to determine circadian time. Cell 114:537–543. [PubMed] [Google Scholar]
- 12. Partch CL, Sancar A (2005b) Cryptochromes and circadian photoreception in animals. Methods Enzymol 393:726–745. [DOI] [PubMed] [Google Scholar]
- 13. Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh Z Pfl anzenphysiol Bd. 100:147–160. [Google Scholar]
- 14. Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366:162–166. [DOI] [PubMed] [Google Scholar]
- 15. Lin C, Yang H, Guo H, Mockler T, Chen J, et al. (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci U S A 95:2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoffman PD, Batschauer A, Hays JB (1996) PHH1, a novel gene from Arabidopsis thaliana that encodes a protein similar to plant blue-light photoreceptors and microbial photolyases. Mol Gen Genet 253:259–265. [DOI] [PubMed] [Google Scholar]
- 17. Lin C, Ahmad M, Chan J, Cashmore AR (1996b) CRY2, a second member of the Arabidopsis cryptochrome gene family. Plant Physiol. 110:1047.8819875 [Google Scholar]
- 18. Kleine T, Lockhart P, Batschauer A (2003) An Arabidopsis protein closely related to Synechocystis cryptochrome is targeted to organelles. Plant J 35:93–103. [DOI] [PubMed] [Google Scholar]
- 19. Selby CP, Sancar A (2006) A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci U S A 103:17696–17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Immeln D, Schlesinger R, Heberle J, Kottke T (2007) Blue light induces radical formation and autophosphorylation in the light-sensitive domain of Chlamydomonas cryptochrome. J Biol Chem 282:21720–21728. [DOI] [PubMed] [Google Scholar]
- 21. Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome Light Signals Control Development to Suppress Auxin Sensitivity in the Moss Physcomitrella patens. Plant Cell 14:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imaizumi T, Kanegae T, Wada M (2000) Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern adiantum capillusveneris. Plant Cell 12:81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ninu L, Ahmad M, Miarelli C, Cashmore AR, Giuliano G (1999) Cryptochrome 1 controls tomato development in response to blue light. Plant J 18:551–556. [DOI] [PubMed] [Google Scholar]
- 24. Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, et al. (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol 137:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chatterjee M, Sharma P, Khurana JP (2006) Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol 141:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Platten JD, Foo E, Elliott RC, Hecht V, Reid JB, et al. (2005a) Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol 139:1472–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Matsumoto N, Hirano T, Iwasaki T, Yamamoto N (2003) Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol 133:1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang YC, Gong SF, Li QH, Sang Y, Yang HQ (2006) Functional and signaling mechanism analysis of rice CRYPTOCHROME 1. Plant J 46:971–983. [DOI] [PubMed] [Google Scholar]
- 29. Li YY, Mao K, Zhao C, Zhao XY, Zhang RF, et al. (2013a) Molecular cloning and functional analysis of a blue light receptor gene MdCRY2 from apple (Malus domestica). Plant Cell Rep 32:555–566. [DOI] [PubMed] [Google Scholar]
- 30. Li YY, Mao K, Zhao C, Zhang RF, Zhao XY, et al. (2013b) Molecular cloning of cryptochrome 1 from apple and its functional characterization in Arabidopsis. Plant Physiol Bioch 67:169–177. [DOI] [PubMed] [Google Scholar]
- 31. Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, et al. (1995b) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269:968–970. [DOI] [PubMed] [Google Scholar]
- 32. Banerjee R, Schleicher E, Meier S, Munoz Viana R, Pokorny R, et al. (2007) The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J Biol Chem 282:14916–14922. [DOI] [PubMed] [Google Scholar]
- 33. Bouly JP, Schleicher E, Dionisio-Sese M, Vandenbussche F, Van Der Straeten D, et al. (2007) Cryptochrome blue light photoreceptors are activated through interconversion of flavin redox states. J Biol Chem 282:9383–9391. [DOI] [PubMed] [Google Scholar]
- 34. Song SH, Dick B, Penzkofer A, Pokorny R, Batschauer A, et al. (2006) Absorption and fluorescence spectroscopic characterization of cryptochrome 3 from Arabidopsis thaliana . J Photochem Photobiol B 85:1–16. [DOI] [PubMed] [Google Scholar]
- 35.Yu X, Liu H, John K, Lin C (2010) The Cryptochrome Blue Light Receptors. The Arabidopsis Book e0135. 10.1199/tab.0135. [DOI] [PMC free article] [PubMed]
- 36. Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, et al. (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana . Proc Natl Acad Sci U S A 101:12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toth R, Kevei EE, Hall A, Millar AJ, Nagy F, et al. (2001) Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis . Plant Physiol 127:1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu G, Spalding EP (2007) Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc Natl Acad Sci U S A 104:18813–18818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, et al. (2007b) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19:3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma L, Li J, Qu L, Hager J, Chen Z, et al. (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13:2589–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Folta KM, Pontin MA, Karlin-Neumann G, Bottini R, Spalding EP (2003) Genomic and physiological studies of early cryptochrome 1 action demonstrate roles for auxin and gibberellin in the control of hypocotyl growth by blue light. Plant J 36:203–214. [DOI] [PubMed] [Google Scholar]
- 42. Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis . Proc Natl Acad Sci U S A 101:2223–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sellaro R, Hoecker U, Yanovsky M, Chory J, Casal JJ (2009) Synergism of red and blue light in the control of Arabidopsis gene expression and development. Curr Biol 19:1216–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J, Lin R, Sullivan J, Hoecker U, Liu B, et al. (2005) Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17:804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, et al. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. Embo J 27:1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma P, Chatterjee M, Burman N, Khurana JP (2013) Cryptochrome 1 regulates growth and development in Brassica through alteration in the expression of genes involved in light, phytohormone and stress signaling. Plant, Cell and Environment 10.1111/pce.12212 [DOI] [PubMed]
- 47. Malhotra K, Kim ST, Batschauer A, Dawut L, Sancar A (1995) Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34:6892–6899. [DOI] [PubMed] [Google Scholar]
- 48. Kanai S, Kikuno R, Toh H, Ryo H, Todo T (1997) Molecular evolution of the photolyase-blue-light photoreceptor family. J Mol Evol 45:535–548. [DOI] [PubMed] [Google Scholar]
- 49. Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, et al. (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana . Proc Natl Acad Sci U S A 101:12142–12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ahmad M, Jarillo JA, Smirnova O, Cashmore AR (1998b) Cryptochrome blue-light photoreceptors of Arabidopsis implicated in phototropism. Nature 392:720–723. [DOI] [PubMed] [Google Scholar]
- 51. Geourjon C, Deléage G (1994) SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng 7:157–164. [DOI] [PubMed] [Google Scholar]
- 52. Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg E, Man O, et al. (2005) FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 21:3435–3438. [DOI] [PubMed] [Google Scholar]
- 53. Kyte J, Doolittle R (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. [DOI] [PubMed] [Google Scholar]
- 54. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16:735–743. [DOI] [PubMed] [Google Scholar]
- 55. Yang HQ, Tang RH, Cashmore AR (2001) The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13:2573–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–158. [DOI] [PubMed] [Google Scholar]
- 57. Liu H, Liu B, Zhao C, Pepper M, Lin C (2011) The action mechanisms of plant cryptochromes. Trends Plant Sci 16(12):684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6:197–208. [DOI] [PubMed] [Google Scholar]
- 59. Gsponer J, Futschik ME, Teichmann SA, Babu MM (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322:1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Facella P, Lopez L, Carbone F, Galbraith DW, Giuliano G, et al. (2008) Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS ONE 3:e2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Platten JD, Foo E, Foucher F, Hecht V, Reid JB, et al. (2005b) The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol 59:683–696. [DOI] [PubMed] [Google Scholar]
- 62. Somers DE, Quail PH (1995) Phytochrome-mediated light regulation of PHYA- and PHYB-GUS transgenes in Arabidopsis thaliana seedlings. Plant Physiol 107:523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14:1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mandoli DF, Ford GA, Waldron LJ, Nemson JA, Briggs WR (1990) Some spectral properties of several soil types: implications for photomorphogenesis. Plant Cell Environ 13:287–294. [Google Scholar]
- 65. Morelli G, Ruberti I (2002) Light and shade in the photocontrol of Arabidopsis growth. Trends in Plant Science 7:399–404. [DOI] [PubMed] [Google Scholar]
- 66. Sorce C, Picciarelli P, Calistri G, Lercari B, Ceccarelli N (2008) The involvement of indole-3-acetic acid in the control of stem elongation in dark- and light-grown pea (Pisum sativum) seedlings. J Plant Physiol 165:482–489. [DOI] [PubMed] [Google Scholar]
- 67. Corell MJ, Kiss JZ (2005) The roles of phytochromes in elongation and gravitropism of roots. Plant Cell Physiol 46:317–323. [DOI] [PubMed] [Google Scholar]
- 68. Gendreau E, Traas J, Desnos T, Grandjean O, Caboche M, et al. (1997) Cellular basis of hypocotyl growth in Arabidopsis thaliana . Plant Physiol 114:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fujita H, Syono K (1996) Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol 37:1094–1101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.