Abstract
The initiation of eukaryotic DNA replication is a highly regulated process conserved from yeast to human. The past decade has seen significant advances in understanding how the CMG (Cdc45-MCM-GINS) replicative helicase is loaded onto DNA. However, very little was known on how this complex is removed from chromatin at the end of S phase. Two papers in a recent issue of Science 1,2 show that in yeast and in Xenopus, the CMG complex is unloaded at replication termination sites by an active mechanism involving the polyubiquitylation of Mcm7.
The initiation of DNA replication is a two-step process that is tightly regulated during the cell cycle. In G1, the Mcm2-7 complex is loaded on chromatin at ORC-bound replication origins by Cdc6 and Cdt1 to form the so-called pre-replication complex (pre-RC). Upon entry into S phase, cyclin-dependent and Dbf4-dependent kinases (CDK and DDK, respectively) promote the binding of Cdc45 and GINS to Mcm2-7 to form the CMG replicative helicase (Fig1). This helicase opens the DNA duplex to recruit DNA polymerases and assemble the replisome at the replication fork. From each origin, two replication forks progress bidirectionally until they encounter forks moving in the opposite direction. The termination of DNA replication is a poorly characterized process that involves the disassembly of converging replisomes and the resolution of replication termination intermediates by DNA topoisomerases 3,4. In particular, the CMG ring is unloaded from termination sites upon completion of DNA replication. This process must be tightly controlled, as the replicative helicase can only be assembled at origins and must remain on chromatin until termination. However, the unloading of the CMG complex had remained largely unexplored.
Figure 1.
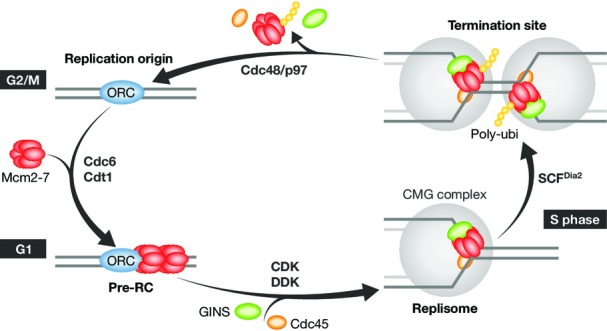
The MCM cycle.
In eukaryotes, the Mcm2-7 complex is loaded on chromatin upon exit from mitosis, exclusively during G1, by the ORC complex, Cdc6 and Cdt1 to form the pre-replicative complex (pre-RC). At the G1/S transition, cyclin- and Dbf4-dependent kinases (CDK and DDK) promote the recruitment of Cdc45 and the GINS complex, which form the CMG replicative helicase together with Mcm2-7. The CMG complex acts in front of the replisome to separate parental DNA strands. Upon termination of DNA replication, Mcm7 is ubiquitylated to promote the disassembly of the CMG helicase by Cdc48/p97. This mechanism is conserved in S. cerevisiae and in Xenopus. In yeast, the ubiquitylation of Mcm7 depends on Dia2, an integral component of the replisome. The ubiquitin ligase responsible for Mcm7 ubiquitylation in metazoans is currently unknown.
The laboratories of Karim Labib and Agnieszka Gambus have recently shed new light on the mechanism by which the CMG replicative helicase is disassembled in budding yeast and Xenopus egg extracts 1,2. Using a combination of in vitro and in vivo approaches, they showed that Mcm7, one of the six subunits of the MCM complex, is specifically polyubiquitylated at the end of S phase. In both organisms, addition of a K48-linked ubiquitin chain to Mcm7 leads to the disassembly of the CMG complex upon termination of DNA replication. In budding yeast, this modification depends on Dia2, an F-box protein that is the substrate adaptor of the SCFDia2 ubiquitin ligase complex. Dia2 has been implicated in the maintenance of fork integrity, and dia2Δ mutants show synthetic lethality/sickness with many mutations that affect DNA replication, recombination, checkpoint and chromatin-remodeling pathways 5. Remarkably, Dia2 interacts with Mrc1 and Ctf4—two components of the replisome progression complex—and travels with the replication fork 6. Yet, it only promotes ubiquitylation of Mcm7 at the end of DNA replication. How cells prevent the premature ubiquitylation of Mcm7 is currently unknown, but an attractive possibility could be that this modification occurs only when two replisomes converge. In dia2Δ mutants, the CMG complex remains on chromatin at the end of S phase and persists in the following G1 and S phases. Re-expression of DIA2 in G1 led to efficient disassembly of CMG complexes.
In Xenopus egg extracts, Mcm7 is also polyubiquitylated upon termination of DNA replication by an unidentified cullin E3 ligase. Importantly, the ubiquitylation of Mcm7 is necessary but not sufficient to disassemble the CMG complex. In yeast and Xenopus, this process depends on the Cdc48/p97 segregase, a factor involved in the displacement of polyubiquitylated proteins from chromatin and their degradation by the proteasome 7. The unloading of CMG by Cdc48/p97 is independent of the activity of the proteasome, and the ubiquitylation status of Mcm7 does not affect the DNA replication program. Altogether, these results indicate that Mcm7 is polyubiquitylated by the yeast SCFDia2 (or its functional homologue in vertebrates) to promote the displacement of Mcm2-7 from the CMG complex by the Cdc48/p97 segregase at termination sites, causing the CMG to fall apart (Fig1).
These findings raise several important questions regarding the mechanism by which cells prevent the premature unloading of the CMG complex, considering the physical proximity between Dia2 and its target Mcm7 within the replisome. This could be a problem when forks encounter unfired pre-RCs, which contain chromatin-bound MCM hexamers and could be mistaken for converging replication forks. In budding yeast, inactive pre-RCs are displaced by Rrm3, a DNA helicase of the Pif1 family 8. Dia2 could, for example, cooperate with Rrm3 to displace inactive MCM complexes ahead of replication forks.
Another important question is how yeast cells cope with the persistence of chromatin-bound CMG complexes in the absence of Dia2. It has been reported that dia2Δ mutants show spontaneous DNA damage during S phase, activate the DNA damage checkpoint, and accumulate in G2/M. Whether this phenotype is due to the persistence of CMG complexes on chromatin or to the accumulation of other replisome components such as Mrc1 and Ctf4 9 is currently unclear. The fact that dia2Δ cells are viable, although very sick, suggests that other mechanisms could remove chromatin-bound CMG complexes. In Xenopus, a candidate to unload the MCM complex is MCM-BP, a factor that is imported to the nucleus in late S phase and binds Mcm7 10. Extracts depleted of MCM-BP replicate normally, but show a delayed dissociation of the Mcm2-7 complex. How Cdc48/p97 and MCM-BP cooperate to unload MCMs is currently unclear, but a likely possibility is that MCM-BP unloads inactive MCM complexes at dormant origins, whereas Cdc48/p97 disassembles MCMs from active CMG complexes. Finally, Xenopus egg extracts treated with the topoisomerase 2 inhibitor ICRF-193 also accumulate MCM on chromatin at the end of S phase 2,10. Since topoisomerase 2 is implicated in the termination of DNA replication 3,4, this effect of ICRF-193 could reflect the persistence of CMG complexes on unreplicated regions.
In conclusion, these two papers describe a very important aspect of MCM biology that has remained unexplored for nearly two decades since the discovery of this complex. Now that the MCM cycle is closed, it appears that the stepwise mechanisms controlling the loading and the unloading of the replicative helicase are equally conserved, complex, and redundant. This is not surprising, considering the importance of initiation and termination of DNA replication for the maintenance of genome integrity and the transmission of genetic information to daughter cells.
References
- Maric M, Maculins T, De Piccoli G, et al. Science. 2014 doi: 10.1126/science.1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, et al. Science. 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- Baxter J, Diffley JFX. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, et al. Mol Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake D, Luke B, Kanellis P, et al. Genetics. 2006;174:1709–1727. doi: 10.1534/genetics.106.057836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi H, Maculins T, Labib K. Curr Biol. 2009;19:1943–1949. doi: 10.1016/j.cub.2009.09.062. [DOI] [PubMed] [Google Scholar]
- Finley D, Ulrich HD, Sommer T, et al. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, et al. Mol Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- Mimura S, Komata M, Kishi T, et al. EMBO J. 2009;28:3693–3705. doi: 10.1038/emboj.2009.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Frappier L, Mechali M. Genes Dev. 2011;25:165–175. doi: 10.1101/gad.614411. [DOI] [PMC free article] [PubMed] [Google Scholar]


