Abstract
RNA ligation can regulate RNA function by altering RNA sequence, structure and coding potential. For example, the function of XBP1 in mediating the unfolded protein response requires RNA ligation, as does the maturation of some tRNAs. Here, we describe a novel in vivo model in Caenorhabditis elegans for the conserved RNA ligase RtcB and show that RtcB ligates the xbp-1 mRNA during the IRE-1 branch of the unfolded protein response. Without RtcB, protein stress results in the accumulation of unligated xbp-1 mRNA fragments, defects in the unfolded protein response, and decreased lifespan. RtcB also ligates endogenous pre-tRNA halves, and RtcB mutants have defects in growth and lifespan that can be bypassed by expression of pre-spliced tRNAs. In addition, animals that lack RtcB have defects that are independent of tRNA maturation and the unfolded protein response. Thus, RNA ligation by RtcB is required for the function of multiple endogenous target RNAs including both xbp-1 and tRNAs. RtcB is uniquely capable of performing these ligation functions, and RNA ligation by RtcB mediates multiple essential processes in vivo.
Subject Categories Protein Biosynthesis & Quality Control; RNA Biology
Keywords: Caenorhabditis elegans, HSPC117, RtcB, tRNA, xbp-1
Introduction
RNA ligation can provide organisms with a powerful mechanism for regulating RNA function by altering RNA sequence, structure, and coding potential. Metazoan cell extracts contain RNA ligase activity, demonstrating that RNA ligation does occur in vivo in metazoans 1–3. Further, in metazoans, the function of two different types of RNA molecule is predicted to depend on RNA ligation. First, in order to mediate protein translation, metazoan intron-containing tRNAs require ligation after removal of their intron by the splicing endonuclease complex 4. Second, the function of the XBP mRNA—encoding a transcription factor that mediates the unfolded protein response—requires ligation after cleavage by the IRE1 endonuclease 5,6. Until recently, however, the molecular identity of the ligase was not known in metazoans, as the RNA ligases identified in other phyla (such as LigT in bacteria, AtRNL in Arabidopsis thaliana, and TRL1 in yeast) are absent from known metazoan genomes 7–9. The initial biochemical characterization showed that the metazoan RNA ligase differs from the yeast and plant enzymes by utilizing specific RNA termini and by ligating RNA by an orthodox phosphodiester without the 2′ phosphate involvement 1–3.
The recent cloning and molecular characterization of RtcB/HPC117, an RNA ligase that is highly conserved from bacteria to metazoans, raises the possibility that this RNA ligase mediates multiple aspects of RNA ligation in metazoans. Indeed, knockdown of RtcB expression in cultured metazoan cells or cell extracts reduces ligation of artificial RNA substrates and of an overexpressed tRNA reporter gene 10. However, the ability of RtcB to mediate ligation of endogenous tRNA and mRNA substrates, and whether it is the primary RNA ligase that mediates these functions in metazoans, is not known. Analysis of mutant animals that lack RtcB would shed light on the in vivo functions of RtcB in particular and on RNA ligation in general. Here, working in the model organism Caenorhabditis elegans, we describe the first metazoan loss-of-function model for RtcB, show that RtcB is essential for the ligation of multiple endogenous substrates in metazoans, demonstrate that ligation of the xbp-1 mRNA is essential for activation of the unfolded protein response, and analyze the function of metazoan RNA ligation at the genetic, cellular, and organismal level.
Results
We developed a genetic loss-of-function model for the RNA ligase RtcB in the metazoan C. elegans. RtcB is a highly conserved protein with a unique structure that is present in archaea, bacteria, and animals, but largely absent in plants and fungi 11. Caenorhabditis elegans has a single gene encoding RtcB, which we named rtcb-1. Caenorhabditis elegans RtcB/RTCB-1 is 73% identical to human RtcB/HSPC117 and 30% identical to E. coli RtcB (Supplementary Fig S1). The deletion allele rtcb-1(gk451) removes the start codon and first exon and is a putative null (Supplementary Fig S2). We found that rtcb-1(gk451) homozygotes can be recovered from balanced heterozygous animals and that these homozygous mutant animals grow to mature adults (although these adults are sterile). The phenotypes of rtcb-1(gk451) homozygotes are efficiently rescued by a single-copy wild-type transgene inserted at a different genomic location, demonstrating that all rtcb-1(gk451) phenotypes are specifically due to loss of RtcB (Supplementary Fig S2). Thus, these homozygous RtcB mutant animals enable the in vivo analysis of the effects of loss of RtcB function.
Ligation of intron-containing tRNAs is defective in RtcB mutants. In bacteria, archaea, and eukaryotes, some precursor tRNA transcripts contain introns 12–14. For example, in C. elegans, 31 of 605 tRNA genes contain such introns; in humans, the number is 32 of 506 14. Although tRNA introns are conceptually similar to introns in pre-mRNA, they are not processed by the spliceosome but rather removed in a two-step enzymatic process involving cleavage and subsequent ligation 15–17. In C. elegans, the 31 intron-containing tRNAs address a total of four codons, and for Leu(CAA) and Tyr(GUA), all genomic copies of the tRNAs contain introns (7/7 and 19/19 copies, respectively) 14. We extracted RNA from age-matched wild-type and RtcB null animals and performed Northern blot analysis for tRNALeu(CAA) (Fig1) and tRNATyr(GUA) (Supplementary Fig S3). Unligated tRNA halves were apparent in RtcB mutants but not in wild-type animals (Fig1A). These data indicate that tRNA ligation is defective in RtcB mutants and demonstrate that RtcB ligates endogenous tRNAs. However, RtcB mutants still contained some spliced tRNAs, although only about 50% as much as wild-type (Fig1B and E). These spliced tRNAs could be produced by an RtcB-independent process; alternatively, they could be maternally contributed or produced in the embryo by maternally contributed RtcB. To determine whether larval RtcB animals still contain tRNA ligase activity, we used a 3′ trailer probe to visualize transient intermediates that are ligated but retain the 3′ trailer (and possibly the 5′ leader). These short-lived species were detected in wild-type animals, demonstrating that active tRNA ligation maintains a pool of intermediates in which the 3′ trailer has not yet been trimmed (Fig1C). By contrast, these transient species were not apparent in RtcB mutants, indicating that tRNA ligation is reduced below detectable levels. Similar phenomena were observed for the intron-containing tRNATyr(GUA) (Supplementary Fig S3), but not for other tRNAs without introns (Fig1 and Supplementary Fig S3). These data indicate that larval-stage RtcB mutants lack tRNA ligation activity and support the idea that residual tRNAs in RtcB mutants are the result of maternal contribution and the known stability of tRNA molecules 18. Unexpectedly, we also found that RtcB mutants contained elevated levels of uncleaved, intron-containing pre-tRNALeu(CAA), suggesting that lack of ligation can influence pre-tRNA transcription, cleavage, or both (Fig1D and Supplementary Fig S3). Taken together, these data show that RtcB is the RNA ligase that mediates processing of endogenous intron-containing pre-tRNAs.
Figure 1.
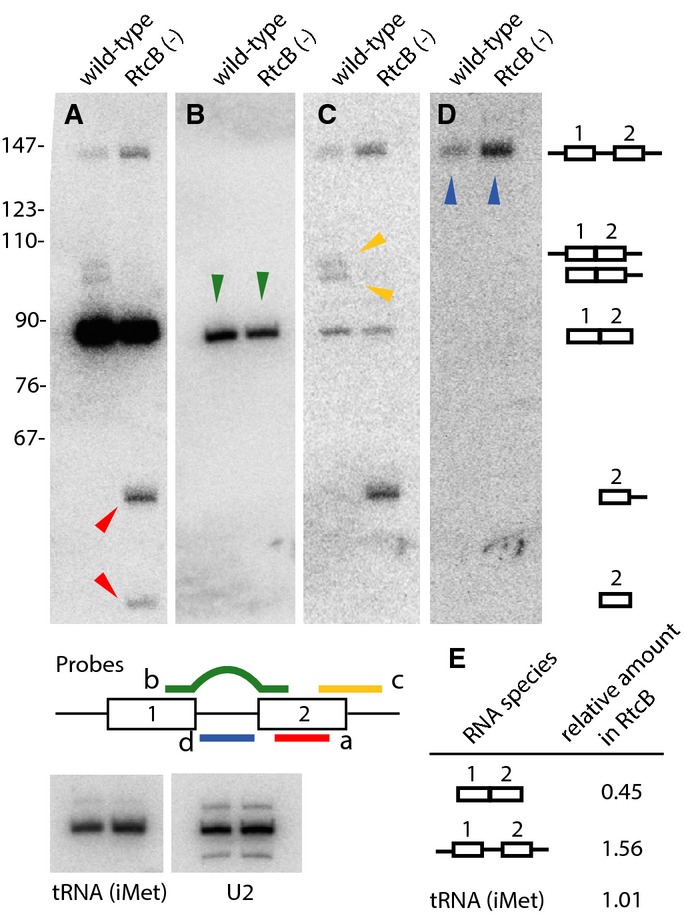
RtcB is required for ligation of endogenous intron-containing pre-tRNAs
Northern blots of RNA extracted from wild-type and RtcB mutant worms, probed for tRNALeu(CAA).
A tRNA halves accumulate in RtcB mutants (red arrowheads).
B Levels of spliced mature tRNALeu(CAA) are reduced in RtcB mutants (green arrowheads).
C Untrimmed, spliced pre-tRNALeu(CAA) intermediates are observed in wild-type but not RtcB mutants (yellow arrowheads).
D Unspliced pre-tRNALeu(CAA) accumulates in RtcB mutants (blue arrowheads).
E Quantitative analysis of the RNA species probed in (B) and (D), showing the relative amount in RtcB versus wild-type controls.
RtcB is required for activation of the IRE1/XBP1 branch of the unfolded protein response (UPR). Processing of the XBP1 mRNA during the IRE1 branch of the UPR occurs via a two-step cleavage and ligation reaction, similar to processing of intron-containing tRNAs 19. However, the RNA ligase TRL1 that ligates Hac1, the functional ortholog of XBP1, during the yeast UPR is not found in metazoans, and the identity of the ligase or ligases that mediate the metazoan UPR is not known 20,21. We assessed XBP1/xbp-1 function in RtcB mutants using a Phsp-4::GFP reporter that is dependent on the IRE1/XBP1 branch of the UPR 5,6. ER stress induced by tunicamycin resulted in upregulation of GFP in control RtcB heterozygous animals, but expression was eliminated in homozygous RtcB mutants (Fig2A). Expression of rtcb-1 cDNA specifically in the intestine restored GFP only in that tissue, indicating that RtcB is required cell-autonomously for XBP-1 function in vivo.
Figure 2.
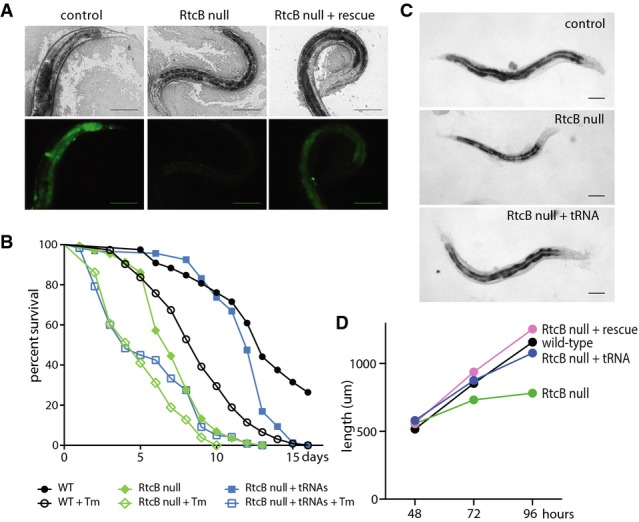
RtcB is required for survival under conditions of ER stress
A Brightfield (top panels) and fluorescence (bottom panels) images of animals expressing the Phsp-4::GFP reporter. Animals were exposed to tunicamycin before imaging. Control animals (RtcB/+) exhibit strong GFP expression in multiple tissues; RtcB mutants fail to express GFP (faint signal is gut autofluorescence); expression of RtcB in the intestine of RtcB mutants rescues GFP expression specifically in intestinal cells. Scale bars, 100 μm.
B Survival curves for wild-type animals (black), RtcB mutants (green), and RtcB mutants expressing pre-spliced tRNAs (blue). Filled symbols indicate normal growth conditions, and unfilled symbols indicate growth on tunicamycin. y-axis indicates the percentage of worms that survived. x-axis indicates the number of days after L4 stage.
C Age-matched wild-type, RtcB mutant, and RtcB mutant worms expressing pre-spliced tRNAs. Scale bars, 100 μm.
D Average length of wild-type (black), RtcB mutants (green), genomic rescued RtcB mutants (pink), and RtcB mutants expressing pre-spliced tRNAs (blue) at 48, 72, and 96 h after eggs were laid. N = 20 per genotype.
Loss of RtcB reduces lifespan under conditions of constant ER stress. As previously reported, wild-type animals had shortened life spans when grown on tunicamycin (Fig2B) 22. RtcB mutants grown on tunicamycin had even shorter life spans, suggesting that loss of XBP-1 function in RtcB mutants results in increased sensitivity to ER stress. However, RtcB animals had reduced life spans even in the absence of tunicamycin. We reasoned that the RtcB life span phenotype in the absence of protein stress might be due to defects in tRNA ligation (Fig1). Reduced tRNA abundance can cause growth deficits due to defects in translation 23, and RtcB mutants grow at a slower rate than wild-type (Fig2C and D). Further, 98.86% of predicted C. elegans coding sequences have at least one RtcB-dependent (Leu(CAA) or Tyr(GUA)) codon, 1.72% of predicted coding sequences have 3 or more consecutive RtcB-dependent codons, and 27 predicted coding sequences have 4 or 5 consecutive RtcB-dependent codons (Supplementary Table S1). To distinguish between potential tRNA and UPR functions in life span, we generated transgenic animals expressing intron-less, artificial tRNA genes for the two RtcB-dependent pre-tRNA species, tRNALeu(CAA) and tRNATyr(GUA). Expression of pre-spliced tRNAs was sufficient to rescue the life span of RtcB mutants under unstressed conditions (Fig2B) and also rescued their growth deficits (Fig2C and D). By contrast, expression of pre-spliced tRNAs had no effect on the extremely short life spans of RtcB mutants under ER stress conditions. Additionally, pre-spliced tRNA RtcB mutant animals were unable to activate the Phsp-4::GFP UPR reporter (Supplementary Fig S4). These data demonstrate that diminished growth and life span of RtcB mutants are due to lack of mature intron-containing tRNAs, likely leading to reduced translation of many mRNAs. These tRNA effects are separate from the requirement for RtcB in XBP-1 function and survival under ER stress.
RtcB ligase activity is required for XBP-1 ligation and activation of the UPR. In C. elegans, xbp-1 mRNA (xbp-1U) is processed by cleavage and ligation during the UPR to remove a 23-nucleotide intron and generate a shortened transcript (xbp-1S) 5,6. We induced the UPR in wild-type and rtcb-1 mutants by treatment with tunicamycin and performed RT–PCR for xbp-1, using primers flanking the intron to detect both xbp-1U and xbp-1S in the same RT–PCR reaction 5,6 (Fig3A). Consistent with previous results 5,6, xbp-1U was detected in wild-type worms treated with tunicamycin. However, there was no detectable xbp-1S in rtcb-1 mutants. Thus, RtcB is essential for the generation of xbp-1S during the metazoan UPR. To determine whether the RNA ligase activity of RtcB is necessary for UPR activation, we mutated the conserved histidine at residue 428 to alanine, which blocks ligase activity by disrupting a metal ion-binding site 15. Wild-type and H428A RtcB were tagged with mRFP and expressed in the intestine of RtcB mutants. Both versions of RtcB were expressed at similar levels in the absence or presence of tunicamycin (Fig3B). The wild-type tagged protein restored the ability of the intestinal cells to activate Phsp-4::GFP in response to tunicamycin (Fig3C), similar to expression of untagged RtcB (Fig2). However, cells expressing catalytically dead H428A RtcB failed to activate Phsp-4::GFP in response to tunicamycin (Fig3C). Tagged RtcB was diffusely localized in the nucleus and in the cytoplasm, consistent with its function in tRNA and mRNA ligation (Fig3D). Next, we performed RNAseq on normal and ER-stressed control and RtcB mutant animals (all expressing pre-spliced tRNAs to reduce secondary effects). At the xbp-1 locus, read coverage on the 3′ side of the UPR-dependent intron was similar in all samples (Fig4A). By contrast, read coverage on the 5′ side was specifically reduced in RtcB mutants treated with tunicamycin. Since RNA samples were poly-A-selected, recovery of the 5′ side of xbp-1 after UPR-induced cleavage depends on ligation to the 3′ side, which contains the poly-A tail. Thus, as with tRNA splicing, loss of RtcB results in a failure to ligate target mRNA molecules and in the accumulation of mRNA fragments. Further, while ER stress resulted in many gene expression changes in both RtcB mutants and control animals, these changes were largely non-overlapping. Of the 121 genes that were upregulated (> 1.75×) in control animals in response to tunicamycin, only 12 were upregulated in tunicamycin-treated RtcB mutants (Fig4B and Supplementary Table S2). Similarly, of the 135 genes that were upregulated in RtcB mutants in response to tunicamycin, only 6 were upregulated in controls (Fig4C and Supplementary Table S3). These data support the idea that in RtcB mutants, ER stress can no longer act via xbp-1 to regulate gene expression. We conclude that RtcB is the ligase that acts on the xbp-1 mRNA during the IRE1 branch of the UPR and that C. elegans does not express any other ligase that can perform this function.
Figure 3.
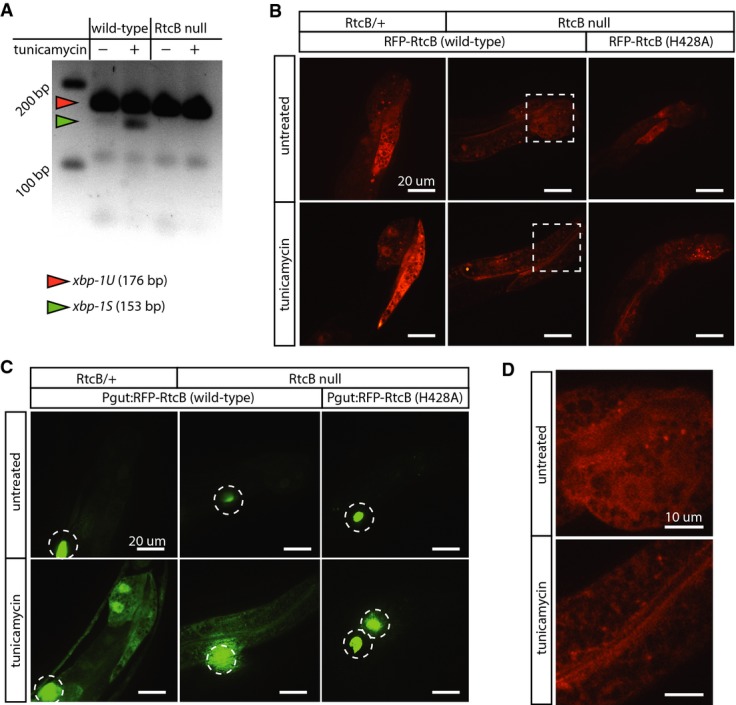
RtcB ligates the xbp-1 mRNA
A RT–PCR using primers that detect both xbp-1U and xbp-1S. In wild-type, xbp-1S is observed after 5 h of tunicamycin treatment. In RtcB nulls, xbp-1S is not detected.
B Expression of RFP-tagged wild-type RtcB and RFP-tagged inactive RtcB (H428A) in the intestine of RtcB/+ controls or RtcB mutant animals. Both wild-type and inactive RtcB are robustly expressed. Dashed boxes are detailed in (D).
C Same animals as (B), visualizing GFP expression from the Phsp-4::GFP reporter under normal conditions (top panels) and after induction of ER stress (bottom panels). Expression of inactive RtcB (H428A) fails to rescue GFP induction in RtcB null mutants. Dotted circles indicate GFP expression in coelomocytes from the injection marker. Scale bars, 20 μm.
D RFP-tagged RtcB is diffusely localized in the cytoplasm and nucleus. Images correspond to dotted squares in panel (B). Scale bars, 10 μm.
Figure 4.
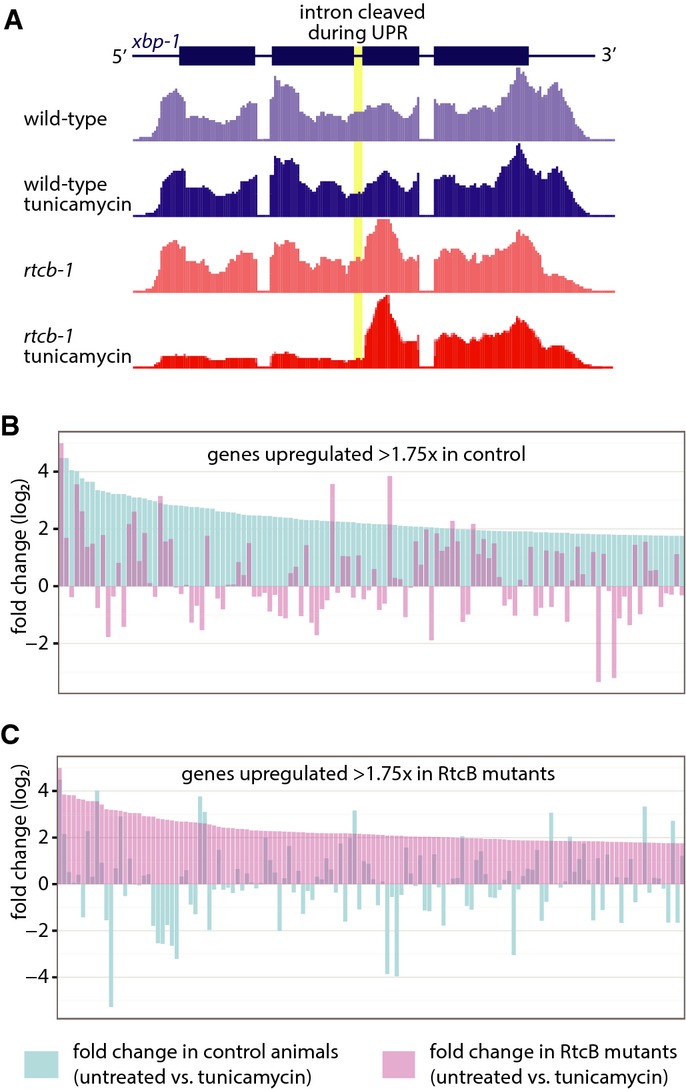
RtcB is required for the unfolded protein response
A RNAseq read coverage at the xbp-1 locus in poly-A-selected, tunicamycin-treated, RtcB null worms.
B Fold change per gene for genes that exhibit a ≥ 1.75 fold change in control worms, comparing untreated to tunicamycin-treated animals. Genes are ranked by fold change in control, shown in blue; pink bars indicate the fold change in RtcB mutants.
C Same analysis as (B), except genes are selected that exhibit a ≥ 1.75 fold change in RtcB mutants. Genes are ranked by fold change in RtcB mutants, shown in pink; blue bars indicate the fold change in control animals.
Data information: Data and gene names are listed in Supplementary Tables S2 and S3
RtcB mutant animals have additional, highly penetrant phenotypes that are independent of deficits in tRNA ligation or the unfolded protein response. First, RtcB mutants are defective for vulval development and exhibit a severe protruding vulva (Pvul) phenotype (Supplementary Fig S5). The Pvul phenotype is independent of RtcB's role in tRNA biogenesis, since engineered RtcB mutants expressing pre-spliced tRNAs still exhibit the Pvul phenotype, even though they are rescued for life span and growth. The Pvul phenotype is also independent of RtcB's essential function in the UPR, since other mutations that abrogate the UPR (ire-1 and xbp-1) do not result in any vulval defects. Second, germline proliferation occurs in RtcB mutants, but germ cells fail to mature into oocytes and the animals are completely sterile (Supplementary Fig S6). Again, this phenotype is not rescued by expression of pre-spliced tRNAs, nor is it phenocopied by ire-1 and xbp-1. Thus, lack of RtcB causes highly penetrant changes in vulval development and fertility that are not correlated with its functions in tRNA biogenesis or the UPR. Together, these data suggest that metazoan RtcB may have biological functions and molecular targets in addition to tRNA and xbp-1 ligation.
Discussion
Our results establish that RNA ligation by RtcB is a key modifier of genetic information, generating increased RNA complexity relative to the genome and supporting essential biological functions. For both xbp-1 mRNA and for tRNA ligation, our data not only demonstrate that RtcB can ligate these targets, but also show that it is absolutely required for these functions, thus suggesting that RtcB is the only RNA ligase in C. elegans that can mediate xbp-1 and tRNA processing. These findings agree with and complement the recent work identifying RtcB as a component of the mammalian UPR 24. Our analysis also suggests that RtcB performs essential functions in vulval development and fertility, independently of tRNA and xbp-1 substrates. It is possible that one primary function of RtcB is to limit tRNA fragments. Such fragments have been shown to accumulate due to stress and are thought to inhibit protein translation 16,17,25. A mouse model that accumulates such fragments displays neurodegeneration and lethality 26. Additionally, RNAi of RtcB enhances aggregation of α-synuclein in a C. elegans model of Parkinson's disease 27. Thus, RtcB may be required in vivo to prevent deleterious effects associated with the accumulation of tRNA fragments. Alternatively, RtcB may have other RNA ligation targets in addition to intron-containing pre-tRNAs and the xbp-1 mRNA. Multiple nucleases can produce RNA fragments with 5′–OH and 2′,3′-cyclic phosphate ends, including tRNA splicing endonucleases and IRE1 15,28. Such fragments represent potential targets for RtcB ligation. By analogy to tRNA and xbp-1 mRNA processing, RtcB-mediated ligation could produce novel RNA species, with functions or coding potential unavailable via conventional processing. A potential site of such RNA processing is RNA granules, which in one case have been shown to contain RtcB 29. The C. elegans RtcB model described here will be useful for future studies of other in vivo functions of RtcB.
Materials and Methods
Genomic rescue
The rtcb-1 (F16A11.2) genomic locus (see Supplementary Fig S2) was amplified from N2 wild-type genomic DNA with primers containing unique restriction digest sites (Forward (SbfI): ccaacctgcaggCATGATTCTGTCTTATTTCGTACC; Reverse (SpeI): ccaaactagtGAAGCTAATTGCGAGCATGTC). The amplicon was digested and ligated into the pCFJ151 vector using standard procedures. The resulting plasmid (pTE048) was injected with other MosSCI reagents into the EG6699 strain as described [30,31]. The resulting single-copy insertion (wpSi17) was verified by PCR, restriction digest, and sequencing. Rescued rtcb-1 homozygotes were isolated from a cross of wpSi17 into the rtcb-1 balanced heterozygote background and were verified by PCR of the rtcb-1 deletion.
Northern blots
RNA extraction: C. elegans RNA for Northern analysis was prepared from 2,000 L4 wild-type or rtcb-1 null worms. Worms were washed twice with M9 buffer, resuspended in TRIzol (Invitrogen), frozen in liquid nitrogen, thawed at 37°C, vortexed briefly, the freeze–thaw repeated five additional times, and then incubated at room temperature for 10 min. RNA was isolated from the aqueous phase following manufacturer's instructions.
Northern blots: Approximately 2 μg of total RNA was separated in 8% polyacrylamide–8.3 M urea gels and transferred to Hybond-N membranes (Amersham) in 0.5× TBE at 150 mA for 16 h. The blots were hybridized with [32P]-labeled oligonucleotide probes as described 32. Oligonucleotides probes for tRNALeu(CAA) were 5′-GAACACCCAGTCGACCTCGAACTCGAGGCAAGCAAT-3′ to detect intron-containing pre-tRNAs, 5′-GAGTACCAGAACTTGAGTC-3′ to detect spliced tRNAs, 5′-CACGCACCCATACGAGTACCAG-3′ for 3′ halves, and 5′-AAAA(G/T)ATCTGCACGAAGTGGG-3′ to detect 3′ trailer-containing tRNAs. Probes for tRNATyr(GTA) were 5′-GATACCTGCTGACTCTACAG-3′ for intron-containing pre-tRNAs, 5′-GTGACCTAAGGATCTACAGTC-3′ for spliced tRNAs, 5′-GAACCAGTGACCTAAGGA-3′ for 3′ halves, and 5′-AAAAGAAATCCGTCGAACCG-3′ to detect 3′ trailer-containing tRNAs. Other probes were 5′-CAGATACTACACTTTGATCTTAGC-3′ for U2 snRNA and 5′-TCCACCGACCTCTGGGTTATG-3′ for tRNA(iMet). Hybridized signals were quantified using a Typhoon Imager System and normalized to U2 snRNA.
UPR fluorescent reporter assay
Worms at the L4/young adult stage were placed on NGM plates with 5 μg/ml tunicamycin for approximately 24 h. RtcB homozygotes (rtcb-1(gk451);zcIs4[hsp-4::GFP]) were isolated from balanced heterozygotes (rtcb-1(gk451)/hT2;zcIs4[hsp-4::GFP]), which served as a control. Transgenes expressing rtcb-1 cDNA behind the intestine-specific spl-1 promoter were injected into balanced heterozygotes. Multiple animals were examined for UPR expression in two transgenic lines, which gave similar results (wpEx185[Pspl-1::rtcb-1::unc-54UTR + Prgs-1::mCherry] and wpEx186[Pspl-1::rtcb-1::unc-54UTR + Prgs-1::mCherry]). Animals were mounted on agarose pads and imaged on a Nikon Eclipse 80i compound microscope with a Nikon 20X LWD (0.40 Ph1 DL) objective. Images were acquired with a Hamamatsu ORCA-05-G camera using NIS Elements BR software and subsequently processed with ImageJ.
Protein tagging experiments
Transgenes expressing TagRFP::rtcb-1 cDNA under the intestinal specific spl-1 promoter were injected into balanced heterozygotes. Multiple transgenic lines expressing wpEx230 [Pspl-1::TagRFP::rtcb-1::unc-54UTR + Punc-122::GFP] were examined for GFP expression under hsp-4 promoter, which reports the presence of the unfolded protein response. Animals were mounted on 3% agarose pads and imaged on an UltraVIEW VoX (PerkinElmer) spinning disk confocal microscope and subsequently processed with Volocity (Improvision).
xbp-1 RT–PCR assay
Approximately 20 L4-stage N2 wild-type or rtcb-1(gk451) animals were placed onto seeded NGM plates with or without 5 μg/ml tunicamycin at 20°C for 5 h. Worms were recovered, washed multiple times in water, and frozen at −80°C. Samples were then freeze–thawed three times, Buffer RLT Plus with β-mercaptoethanol was added, and samples were passed through an 18-gauge needle approximately 10 times. RNA was extracted using the RNeasy Plus Mini kit (Qiagen), eluting the final RNA in 40 μl RNase-free water. First-strand cDNA synthesis was carried out using Affinity Script Reverse Transcriptase. Forward (tgcatctaccagaacgtcgt) and reverse (cggagttggttgctgatgtt) primers flanking the intron were used for PCR using Phusion polymerase. Products were resolved on a 2.2% agarose gel.
Growth and vulval development assay
Gravid hermaphrodites of each genotype were placed on plates, laid eggs for 4 h, and were then removed to create a synchronous F1 cohort. At 48, 72, and 96 h after egg laying, F1 progeny were mounted on agarose pads and imaged on a Nikon Eclipse 80i compound microscope with a Nikon Plan Fluor 4×/0.13 objective for whole worm length and a Nikon Plan Apo 60×/1.4 Oil DIC objective for vulval development. Images were acquired with a Hamamatsu ORCA-05-G camera using NIS Elements BR software and processed with ImageJ. For each genotype, a minimum of 20 worms were measured and evaluated for vulval development. Worms were discarded after imaging, but all three time points were obtained from the same cohort.
Pre-spliced tRNAs
The pre-spliced tRNA construct was generated by synthesizing a single DNA fragment (Genscript) containing two tRNA genes: chrI.trna6-Tyr (GTA) (6135832–6135915) and chrIII.trna27-Leu(CAA) (4362495–4362614) tRNAs without their introns (AGTTCGCAGAT and CGTAACGCTTACCTCAAGTTCGAGGTCTACTGGGTG, respectively). The 200 bp upstream and 100 bp downstream were also included for each gene. The DNA fragment was cloned into the pCFJ151 vector. The resulting construct was injected at 30 ng/μl with 5 ng/μl of Pmyo-3::GFP as a coinjection marker. Three independent lines were generated, with similar phenotypes. tRNAs and their introns were identified using the GtRNAdb4.
Analysis of RtcB-dependent codons
Coding sequences were obtained from WormBase using WormMart, including all live coding sequences from the WBG220 genome release. Codon counts were made using a custom Python script (available on request).
ER stress survival assays
Assays were conducted as described in a previous study 33. Wild-type, rtcb-1(gk451), and transgenic rtcb-1(gk451) animals expressing pre-spliced tRNAs were grown on E. coli OP50 without tunicamycin until L4 stage. Wild-type or homozygous mutant worms at L4 stage were individually picked and placed on NGM plates with 0.5% DMSO alone or NGM plates containing 0.5% DMSO and 50 μg/ml tunicamycin. The animals were fed with OP50 bacteria during the assay. Viability was scored every day. Death was scored by failure to respond to touching. All assays were performed at 20°C with at least 50 worms and were repeated to obtain at least three independent measurements for each strain.
RNAseq
2,000 L4 worms were gathered for the following four samples: (i) wild-type with pre-spliced tRNAs, (ii) wild-type with pre-spliced tRNAs treated with tunicamycin, (iii) RtcB null with pre-spliced tRNAs, and (iv) RtcB null with pre-spliced tRNAs treated with tunicamycin. Tunicamycin treatment involved 4 h on NGM plates containing 50 μg/ml tunicamycin. Total RNA was extracted using TRIzol as described above and sent to the Yale Center for Genome Analysis for library construction and high-throughput sequencing. RNA was poly-A-selected, and pair-end sequencing was performed on the Illumina HiSeq 2000 platform using 75 bp reads. Data from the RNA-seq experiment are available from NCBI GEO under accession number GSE61733.
Acknowledgments
We thank Javier Martinez, Theresa Henkel, and Jennifer Jurkin for their generous assistance with the manuscript. We thank Markus Englert and Dieter Soll for helpful discussions. Strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40OD010440). SGK and TJE were supported by NIH training Grant T32GM007223. MJE is an NSF Graduate Research Fellow. SP is supported by NIH training Grant T32GM008730. This work was supported by a Damon Runyon-Rachleff Innovation award and the American Cancer Society (RSG-13-216-01-DMC) to JRH, NIH Grants R01GM073863 and R01GM048410 to SLW, and NIH Grants R01NS066082 and R56NS081217 to MH.
Author contributions
SGK, TJE, SMH, MJE, SP, JRH, SLW, and MH conceived or designed the experiments. SGK, TJE, SMH, MJE, BIM, and SP performed the experiments. SGK, TJE, SMH, MJE, SP, JRH, SLW, and MH analyzed the data. SGK and MH wrote the manuscript.
Conflict of interest
The author declares that they have no conflict of interest.
Supporting Information
for this article is available online: http://embor.embopress.org
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Legends
Review Process File
References
- Filipowicz W, Konarska M, Gross HJ, Shatkin AJ. RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 1983;11:1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Laski FA, Fire AZ, RajBhandary UL, Sharp PA. Characterization of tRNA precursor splicing in mammalian extracts. J Biol Chem. 1983;258:11974–11980. [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell. 2004;117:311–321. doi: 10.1016/s0092-8674(04)00342-3. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Silber R, Malathi VG, Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc Natl Acad Sci USA. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lührmann R, Söll D, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- Popow J, Schleiffer A, Martinez J. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci. 2012;69:2657–2670. doi: 10.1007/s00018-012-0944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G, Beckmann JS, Johnson PF, Fuhrman SA, Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978;14:221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Kaine BP, Gupta R, Woese CR. Putative introns in tRNA genes of prokaryotes. Proc Natl Acad Sci USA. 1983;80:3309–3312. doi: 10.1073/pnas.80.11.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucl Acids Res. 2009;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR, III, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu G-F, Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwagwu M, Nana M. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J Embryol Exp Morphol. 1980;56:253–267. [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J Biol Chem. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H. Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid Redox Signal. 2007;9:2323–2334. doi: 10.1089/ars.2007.1800. [DOI] [PubMed] [Google Scholar]
- Qabazard B, Ahmed S, Li L, Arlt VM, Moore PK, Stürzenbaum SR. Caenorhabditis elegans aging is modulated by hydrogen sulfide and the sulfhydrylase/cysteine synthase cysl-2. PLoS ONE. 2013;8:e80135. doi: 10.1371/journal.pone.0080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Liang F-X, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell. 2014;55:758–770. doi: 10.1016/j.molcel.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia M, Krokowski D, Guan B-J, Ivanov P, Parisien M, Hu G, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, Hanada R, Orthofer M, Cronin SJ, Komnenovic V, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc Natl Acad Sci USA. 2008;105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dörfler S, Walter P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J. 1999;18:3119–3132. doi: 10.1093/emboj/18.11.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen Jorgensen. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen Jorgensen. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Yario TA, Steitz JA. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA. 1995;1:644–656. [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell. 2013;153:153–1447. doi: 10.1016/j.cell.2013.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Legends
Review Process File


