Abstract
MicroRNAs (miRNAs) are important regulators of mouse brain development. However, their precise roles in this context remain to be elucidated. Through screening of expression profiles from a miRNA microarray and experimental analysis, we show here that miR-15b controls several aspects of cortical neurogenesis. miR-15b inhibits cortical neural progenitor cell (NPC) proliferation and promotes cell-cycle exit and neuronal differentiation. Additionally, miR-15b expression decreases the number of apical progenitors and increases basal progenitors in the VZ/SVZ. We also show that miR-15b binds to the 3′ UTR of TET3, which plays crucial roles during embryonic development by enhancing DNA demethylation. TET3 promotes cyclin D1 expression, and miR-15b reduces TET3 expression and 5hmC levels. Notably, TET3 expression rescues miR-15b-induced impaired NPC proliferation and increased cell-cycle exit in vivo. Our results not only reveal a link between miRNAs, TET, and DNA demethylation but also demonstrate critical roles for miR-15b and TET3 in maintaining the NPC pool during early neocortical development.
Subject Categories Neuroscience; Chromatin, Epigenetics, Genomics & Functional Genomics; RNA Biology
Keywords: miR-15b, neocortical development, neural progenitors, proliferation, TET3
Introduction
The mammalian neocortex is a complex structure composed of six layers of neurons. All these neurons are born from neural progenitor cells (NPCs), although they have differences in their positions and roles in the developing brain. The generation of neurons is strictly programmed and occurs in an inside-out manner 1. The development of the cortex is a complex process, and there is tight control of all the stages, including self-renewal, survival, cell fate determination, differentiation, and migration. Each process happens in a spatially and temporally specific manner and is dependent on the precise regulation of intrinsic and extrinsic signals. Among these signals, transcriptional regulation is an essential element. MicroRNA (miRNA) as a post-transcriptional regulator is likely to play such an important role during neocortical development 2.
miRNAs as endogenous non-coding RNAs can play important roles in physiological progresses such as development by repressing gene expression at the post-transcriptional level 3–5. As important regulatory molecules, miRNAs not only coordinate gene expression during neocortical development but are also important in brain function and dysfunction 2. The miRNA expression profiles during neocortical development have been reviewed 2,6. Many miRNAs are correlated with specific stages of cortical development. Therefore, it is possible to regulate specific aspects of cortical development by expressing miRNAs to coordinate gene expression.
Epigenetic pathways play essential roles in stem cell development. DNA methylation has been linked to both normal neurodevelopment and neurological diseases 7. Recently identified DNA modifications, including 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC), have provided a new perspective on the previously unobserved plasticity in 5mC-dependent regulatory processes. Since the initial discovery of their enzymatic activity 8,9, proteins of the Ten–eleven translocation (TET) family have become a focus of substantial interest. The three TET proteins (TET1, TET2, and TET3) oxidize 5mC to 5hmC, 5fC, and 5caC, respectively 9–12. TET proteins have prominent roles in multiple biological processes, including epigenetic modulation, embryonic development, stem cell renewal, and cancer 11–16. Among mammalian tissues, the brain expresses the highest levels of 5hmC. Higher 5hmC levels and lower DNA methylation levels are clearly correlated with higher levels of gene expression and a more open state of chromatin in neural cells 17. The function of TET proteins and their regulation in brain development remain largely unknown, although some essential roles of TET family during brain development have been reported 18,19.
Here, we report the crucial roles of miR-15b identified from total miRNAs during early neocortical development. We found a dramatic expression pattern of miR-15b during cortical development. miR-15b regulates various aspects of early neocortical development. We observed miR-15b directly targets TET3 and represses TET3 and 5hmC expression levels. Importantly, TET3 rescues the miR-15b-induced proliferation defects, and the miR-15b/TET3 interaction is involved in NPC proliferation.
Results and Discussion
miR-15b regulates cell distribution and cortical NPC proliferation
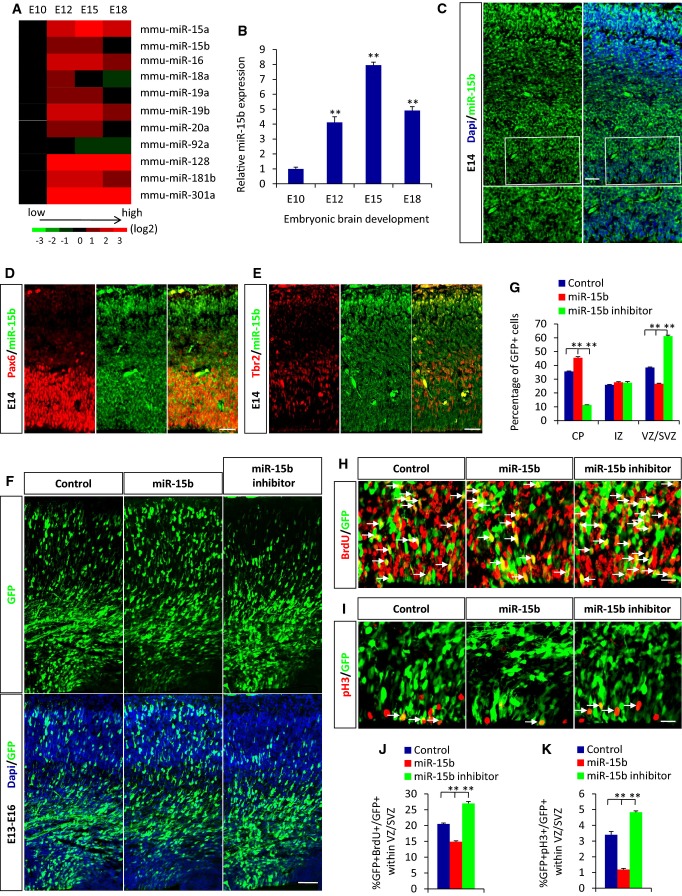
To characterize the roles of miRNAs during the cortical development, we first analyzed the expression pattern of miRNAs in mouse forebrain from embryonic day 10 to embryonic day 18 (E10–E18) with a miRNA array. We selected 11 miRNAs based on dramatic changes in expression during cortical development (Fig1A) from the array data and validated their expression patterns by quantitative real-time PCR (qRT–PCR). Among these miRNAs, miR-15b was identified to display the most significant changes in expression levels during early neocortical development. First, we found that the expression level of miR-15b increased with embryonic development and peaked on E15, followed by a decline until E18 (Fig1B). We next examined the expression of miR-15b using in situ hybridization (ISH). The results displayed higher expression of miR-15b in the cortical plate (CP), subventricular zone (SVZ), and ventricular zone (VZ) in the developing cortex but lower in the intermediate zone (IZ) (Fig1C). To further confirm the expression of miR15b in NPCs, we performed the double immunostaining experiments. We noted that miR-15b more strongly expressed in Pax6-positive apical progenitors, whereas it was also expressed in Tbr2-positive basal progenitors (Fig1D and E). Because neurogenesis begins at around E12, peaks at around E15, and finishes at around E18 in the murine cortex 20,21, our results suggested that miR-15b may play an important part in NPC proliferation and neurogenesis. To identify the roles of miR-15b, the following studies were performed: We electroporated E13 embryonic brains with either a DNA plasmid encoding miR-15b or the miR-15b inhibitor, which is chemically modified anti-sense oligonucleotide designed to compete for binding and protecting the target without changing the abundance of the physiological miRNA. The electroporated brains were analyzed 3 days later, at E16. We found a significant change in the number of GFP-positive cells in the three cortex zones. miR-15b overexpression caused an obvious increase in GFP-positive cells in the CP and corresponding significant decreases in the SVZ and VZ, whereas the delivery of the miR-15b inhibitor caused the contrary phenotype (Fig1F and G). Together, these results indicate that miR-15b regulates cell distribution during early neocortical development.
Figure 1.
miR-15b regulates cell distribution and cortical NPC proliferation
A miRNA expression profile during mouse brain development as determined by miRNA microarray analysis. Relative expression levels for the 11 miRNAs that showed significant changes are shown in four columns corresponding to the four time points. Relative signal intensities are indicated in different colors. Red represents a high relative expression level, black represents no change, and green represents a low relative expression level.
B Relative miRNA-15b expression from embryonic day 10 to embryonic day 18 (E10–E18) was determined by qRT–PCR.
C The expression of miR-15b in the embryonic cortex. The E14 brain sections were hybridized with miR-15b FISH detection probe. Boxed areas are shown at a higher magnification for miR-15b expression in VZ/SVZ. SVZ, subventricular zone; VZ, ventricular zone.
D, E miR-15b is largely expressed in apical progenitors (D) and also expressed in basal progenitors (E) in embryonic cortex. The E14 brain sections were immunostained with anti-Pax6 or anti-Tbr2 antibody followed by miR-15b in situ hybridization.
F, G miR-15b regulates cell position in the cortex. Control, miR-15b expression plasmid, or miR-15b inhibitor was electroporated into embryonic brains at E13 and harvested at E16. CP, cortical plate; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone.
H–K miR-15b regulates BrdU (H) and pH3 (I) incorporation in vivo. Embryonic brains were electroporated at E13 and harvested at E16. BrdU (100 mg/kg) was injected into mice 2 h before sacrifice. Arrows indicate GFP-BrdU or GFP-pH3 double-positive cells. The percentages of GFP-BrdU (J) and GFP-pH3 (K) double-positive cells are shown.
Data information: Scale bars, 20 μm (C–E, H and I) and 50 μm (F). Values are mean ± SEM; n = 3; one-way ANOVAs with Tukey's test for post hoc multiple comparisons; **P < 0.01.
To test whether the changed proportion of cells in the cortex was due to an altered neuronal migration, we performed a BrdU birth-dating experiment. The results showed that the percentage of GFP-BrdU double-positive cells in each region of the cortex changed following change of miR-15b expression (Supplementary Fig S1A–F). This demonstrated that neuronal migration is a cause (at least in part) to the changed proportion of cells in the cortex following miR-15b alteration. The contribution of neuronal migration is less compared with the cell fate change of neural progenitor from the results of statistical analysis.
The fact that fewer GFP-positive cells remained in the VZ/SVZ following miR-15b overexpression suggested the possible defects in the maintenance of cell proliferation. To examine the effects of miR-15b expression on the overall proliferative capacity, E13 embryonic brains were electroporated with miR-15b plasmid or inhibitor, followed by bromodeoxyuridine (BrdU) injection to label S phase dividing cells 2 h before sacrifice at E16. The quantitative results of BrdU labeling in the GFP-positive population showed a significant decrease after miR-15b overexpression (Fig1H and J), indicating an overall decrease in cell proliferation. Meanwhile, at E16, the electroporated brains were also stained with the mitotic marker phosphohistone H3 (pH3), and a significant decrease in mitotic activity was observed from the staining results (Fig1I and K). Conversely, there was a significant increase in cell proliferation and mitotic index in brain treated with the miR-15b inhibitor. To further confirm the effects of miR-15b on the proliferation of NPCs, we performed in vitro NPC culture experiments. Primary neural progenitor cells were infected with control or miR-15b lentivirus or transfected with control or miR-15b inhibitor, and then cultured for 3 days under proliferation conditions. The quantitative real-time polymerase chain reaction results revealed that overexpression of miR-15b decreased the expression in primary NPCs of Ki67 (Supplementary Fig S2A), a proliferation marker that labels cells in all active phases of the cell cycle 22. Following miR-15b inhibition, increased expression of Ki67 was observed (Supplementary Fig S2B). The in vitro immunostaining results for Ki67 and incorporation of ethynyldeoxyuridine (EdU, a proliferation marker also labeling S phase dividing cells) also showed decreased cell proliferation activity (Supplementary Fig S2F, G, I and J). Collectively, these data in vivo and in vitro illustrate that miR-15b regulates the proliferative capacity of cortical neural progenitor cells.
miR-15b regulates cell-cycle exit and neuronal differentiation
Neural progenitor cells are a group of cells that are actively cycling and give rise not only to immature neurons but also to more intermediate progenitors. Because miR-15b decreases the proliferative activity of NPCs and increases the number of GFP-positive cells in the CP, we investigated whether this coincided with changes in cell-cycle exit and premature neuronal differentiation. To test this possibility, we first performed an analysis of cell-cycle exit. E13 embryonic brains were electroporated, BrdU (100 mg/kg) was injected into mice 48 h after electroporation, and then, brains were harvested at E16. The result demonstrated that miR-15b overexpression significantly increased the cell-cycle exit index compared with controls (Fig2A and C). Meanwhile, miR-15b overexpression led to a significant decrease in the 24-h BrdU-labeling index relative to controls (Fig2A and B). This suggested that miR-15b overexpression decreased the proportion of time that cells have spent in S phase, that is, G1 is now becoming longer. To confirm the possibility that increased cell-cycle exit causes premature neuronal differentiation, we next examined the overlap of the GFP-positive cell population with β-III-tubulin (Tuj1), a marker of differentiated neurons, at E16. We found significant increase in Tuj1 incorporation after miR-15b overexpression compared with control (Fig2D and E). To study whether miR-15b regulates embryonic neuronal differentiation in vitro, primary neural progenitor cells were infected with miR-15b lentivirus or transfected with miR-15b inhibitor, respectively. The treated cells were induced to differentiate for 3 days. Western blotting revealed that expression levels of the neuronal marker Tuj1 increased when miR-15b was overexpressed (Supplementary Fig S2C–E). The immunostaining results showed similar results: miR-15b significantly promoted neuronal differentiation, with increased percentages of GFP-Tuj1 double-positive cells at day 3 of differentiation (Supplementary Fig S2H and K). These results indicate that miR-15b accelerates neuronal differentiation of neural progenitor cells. Conversely, miR-15b inhibition caused decreases in cell-cycle exit index and neuronal differentiation. Collectively, these results illustrate that changes in miR-15b expression result in concomitant changes in cell-cycle exit and neuronal differentiation.
Figure 2.
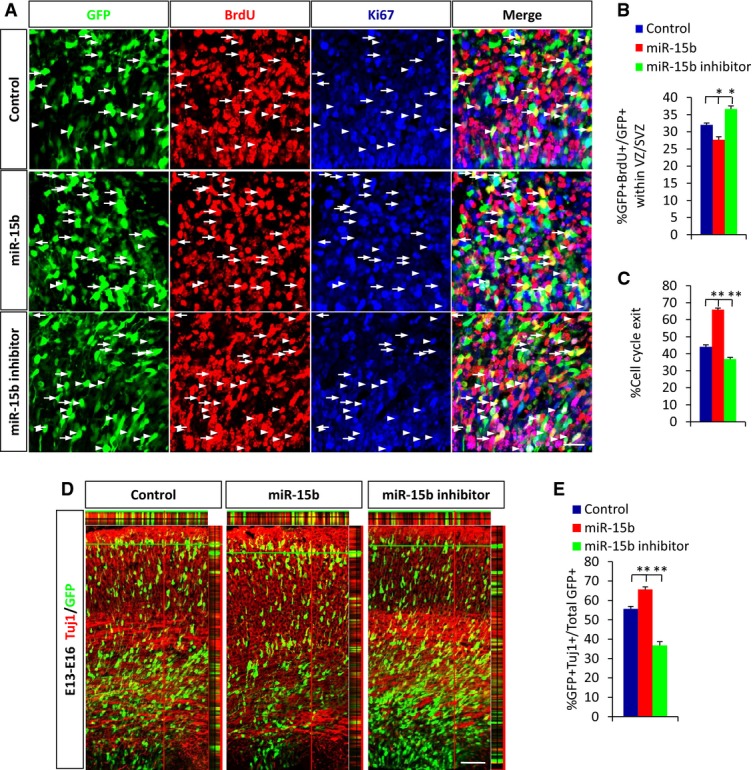
miR-15b regulates cell-cycle exit and neuronal differentiation
A Images of E16 brain sections immunostained for Ki67 and BrdU. Control, miR-15b expression plasmid, or miR15b inhibitor was electroporated into E13 embryonic brains. BrdU (100 mg/kg) was injected into mice 48 h after electroporation, and then, brains were harvested at E16. Arrows indicate GFP+BrdU+Ki67− cells. Arrowheads indicate GFP+BrdU+Ki67+ cells.
B, C Quantification of 24-h BrdU-labeling index (B) and cell-cycle exit index (C) (the percentage of the GFP+BrdU+Ki67− cells relative to the GFP+BrdU+ cells) is shown.
D, E miR-15b regulates cortical neuronal differentiation. The E16 brain sections were stained with anti-Tuj1 antibody. Colocalization is represented by yellow with z-stack. The percentage of GFP-Tuj1 double-positive cells is shown.
Data information: Scale bars, 20 μm (A) and 50 μm (D). Values are mean ± SEM; n = 3; one-way ANOVAs with Tukey's test for post hoc multiple comparisons; *P < 0.05; **P < 0.01.
miR-15b regulates the neural progenitor pool
Proper cortical development requires the appropriate numbers and types of cells derived from the progenitor pools. There are two types of neurogenic progenitor cells in the developing cerebral cortex, apical progenitors, and basal progenitors. Apical progenitors are capable of self-renewal and are responsible for the maintenance of the progenitor pool during neurogenesis. Basal progenitors, also called intermediate neuronal progenitors, undergo symmetric division to generate neurons 23–26. The imbalance of NPC proliferation and differentiation and the changed cell-cycle exit index after miR-15b expression variation implied that miR-15b may play an important role in maintaining the neural progenitor pool. To test this possibility directly, electroporation was performed at E13 to overexpress or inhibit miR. Then, we collected the electroporated brains and examined the makeup of the progenitor pool by immunohistochemistry for Pax6, the apical progenitor marker, and Tbr2, the basal progenitor marker, at E16. The quantitative results showed that miR-15b overexpression led to a decreased proportion of apical progenitors (Fig3A and B) and an increased proportion of basal progenitors (Fig3C and D). The change we observed may well uncover a change in apical progenitors toward a bias to produce basal progenitors and neurons by asymmetric divisions. Our data capture a time during which apical progenitors in the progenitor pool have undergone the changes from self-renewing divisions to basal progenitors undergoing neuronal generation divisions, resulting in a transient alteration in the NPCs' proliferation and neurogenesis. The opposite change was observed in the proportions of apical and basal progenitors following miR-15b inhibition. Thus, miR-15b expression levels regulate the makeup of the progenitor pool, providing a possible explanation for the changed cell proliferation, cell-cycle exit, and neuronal differentiation observed following miR-15b overexpression or inhibition.
Figure 3.
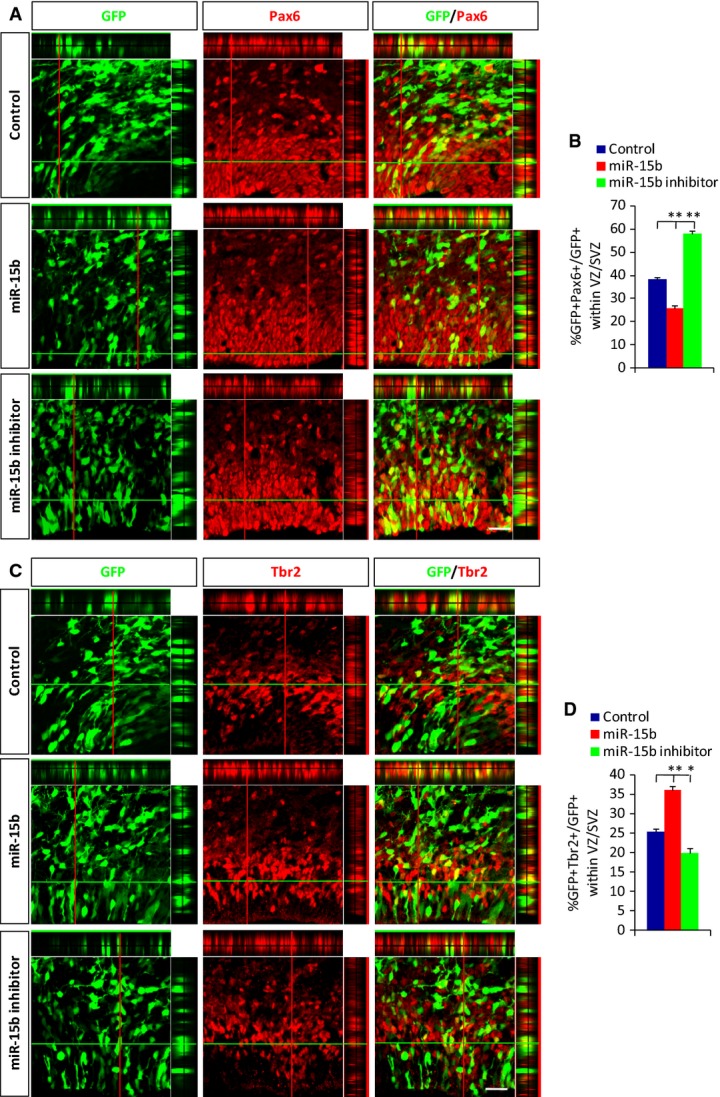
miR-15b regulates the neural progenitor pool
A–D miR-15b regulates the proportion of Pax6-positive progenitor cells (A) and Tbr2-positive progenitor cells (C). The percentages of GFP-Pax6 (B) and GFP-Tbr2 (D) double-positive cells are shown. Embryonic brains were electroporated at E13, harvested at E16, and immunostained with anti-Pax6 or anti-Tbr2 antibody. Co-localization is represented by yellow with z-stack. Scale bars, 20 μm (A and C). Values are mean ± SEM; n = 3; one-way ANOVAs with Tukey's test for post hoc multiple comparisons; *P < 0.05; **P < 0.01.
miR-15b directly targets TET3 and then regulates TET3 and 5hmC levels
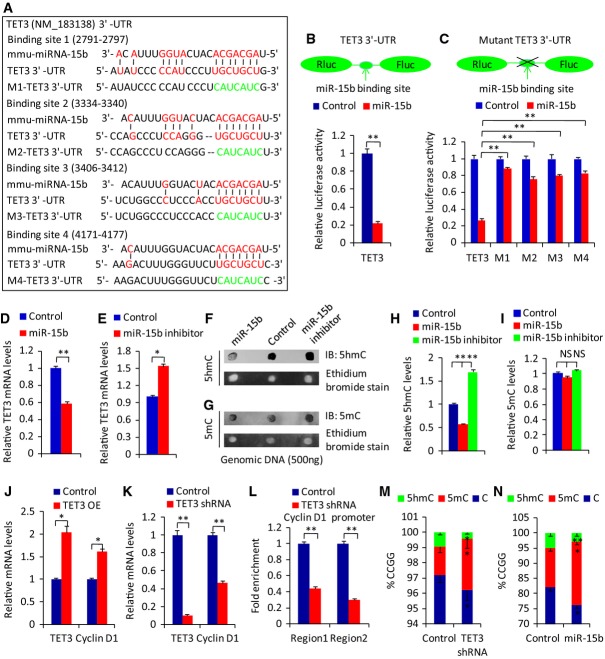
We next addressed the molecular mechanism by which miR-15b regulates neurogenesis during early cortical development. Individual miRNAs can regulate target mRNAs at the post-transcriptional level 3. The target genes of a miRNA show relatively long and conserved 3′ UTRs 27. To investigate the downstream targets of miR-15b, we searched for potential mRNA targets of miR-15b with TargetScan algorithms 28. Among the predicted candidates, TET3 has four putative target binding sites for miR-15b in its 3′ UTR (Fig4A), and TET3 has a long and conserved 3′ UTR. TET protein is known to be important in brain development and function 18,19. To directly test whether TET3 is the target of miR-15b, the 3′ UTR of TET3 was cloned in a luciferase reporter vector, and the reporter construct was cotransfected along with miR-15b into the human cell line HEK293T. The luciferase expression effectively decreased after the co-expression of miR-15b in constructs with TET3 3′ UTR (Fig4B). To test whether the predicted miR-15b target site in the TET3 3′ UTR is critical for repression of TET3 expression by miR-15b, four mutations in the seed sequence of the predicted miR-15b binding sites were introduced into the TET3 3′ UTR. The mutation of the miR-15b target site within the TET3 3′ UTR abolished the inhibitory effects of miR-15b on luciferase expression (Fig4C), suggesting that TET3 is directly regulated by miR-15b.
Figure 4.
miR-15b directly targets TET3 and then regulates TET3 and 5hmC levels
A The putative target binding sites of miR-15b in 3′ UTR of TET3. The red sequence indicates the binding sequence of miR-15b in TET3 3′ UTR. The green sequence indicates the mutated seed sequence in TET3 3′ UTR.
B Relative luciferase activity was suppressed by co-transfection of HEK293T cells with miR-15b and a reporter construct containing the 3′ UTR of TET3.
C Relative luciferase activity in HEK293T cells co-transfected with miR-15b and reporter constructs containing the mutant 3′ UTR of TET3.
D, E The relative TET3 mRNA levels were determined by qRT–PCR 3 days after miR-15b overexpression via lentivirus infection (D) or miR-15b inhibitor transfection (E) in mouse neural progenitor cells under proliferation conditions.
F–I The relative 5hmC (F) and 5mC (G) levels were determined by dot blot. Quantification of 5hmC (H) and 5mC (I) levels is shown. NPCs were infected with miR-15b overexpression lentivirus or transfected with miR-15b inhibitor, and the cells were collected for dot blot.
J, K The correlated change of relative cyclin D1 mRNA levels was determined by qRT–PCR 3 days after TET3 overexpression (J) or inhibition (K) via lentivirus infection in mouse neural progenitor cells under proliferation conditions.
L TET3 binds to cyclin D1 promoter. The enrichment of cyclin D1 was determined by ChIP assay 3 days after TET3 inhibition via lentivirus infection in mouse neural progenitor cells under proliferation conditions.
M, N The correlated changes of methylation and hydroxylation at the cyclin D1 loci after TET3 inhibition (M) and miR-15b overexpression (N) as determined by GlucMS-qPCR assay.
Data information: Values are mean ± SEM; n = 3; two-tailed Student's t-tests (B, D, E, J–N), and one-way ANOVAs with Tukey's test for post hoc multiple comparisons (C, H, I); *P < 0.05; **P < 0.01; NS, non-significant.
Gi2ven that TET3 is a direct target of miR-15b, we tested whether miR-15b regulates TET3 expression levels in primary NPCs. Indeed, we found that expression of the endogenous TET3 was repressed by miR-15b overexpression and enhanced by miR-15b inhibition (Fig4D and E). The response of TET3 expression to miR-15b regulation is correlated with the number of miR-15b target sites within the TET3 3′ UTR. Because TET proteins can convert 5mC to 5hmC 9,29, we investigated whether miR-15b affects 5hmC levels. Dot blot analysis revealed that miR-15b overexpression significantly reduced the levels of 5hmC, whereas miR-15b inhibition increased the levels of 5hmC (Fig4F and H). However, global 5mC levels were not affected by miR-15b overexpression or inhibition (Fig4G and I), consistent with previous reports 30,31. The level of 5hmC in DNA from brain cortex is approximately 1% of all cytosines or 20–25% of total 5mC 32, so the global 5mC levels would not be dramatically changed with the change of 5hmC. Meanwhile, the immunostaining experiments in vitro and in vivo showed similar results (Supplementary Fig S3A–C). Our study established a correlation between the expression levels of miR-15b and TET3, as well as 5hmC. Taken together, these data demonstrate that miR-15b can directly target TET3, thereby regulating TET3 levels and 5hmC expression.
miR-15b/TET3 is involved in the proliferation of cortical NPCs
In addition to being a target of miR-15b, TET3 can also act as a transcription regulator to control gene expression 18,30. However, no roles for TET proteins have been identified in proliferation genes. Cyclin D1 has been shown to have important function in NPCs proliferation and differentiation 33. We therefore tested whether TET3 was able to regulate cyclin D1 expression in NPCs. Indeed, we found that overexpression of TET3 led to significantly increased expression of cyclin D1 (Fig4J). We also found that the expression of endogenous cyclin D1 was drastically downregulated in NPCs when TET3 was knocked down (Fig4K). We further tested whether TET3 bound to the cyclin D1 promoter and caused the epigenetic change of cyclin D1 loci. Chromatin immunoprecipitation (ChIP) assay showed that the enrichment of the cyclin D1 promoter decreased when TET3 knockdown and TET3 bound to the promoter of cyclin D1 (Fig4L). Glucosylated hydroxymethyl-sensitive qPCR (GlucMS-qPCR) assay showed the correlated changes of methylation and hydroxylation at cyclin D1 loci after TET3 knockdown (Fig4M) or miR-15b overexpression (Fig4N). TET3 inhibition or miR-15b overexpression caused the decrease of 5mC hydroxylation. Collectively, these in vitro results suggest that miR-15b and TET3 may be involved in co-regulating NPC proliferation through epigenetic control of cyclin D1 expression. Recent work reported the link between cyclin D1 expression and miRNA networks 34; our result showed that miR-15b regulated cyclin D1 expression through epigenetic control.
To demonstrate whether miR-15b/TET3 is involved in NPC proliferation in vivo, we first performed in utero electroporation for overexpression or knockdown of TET3 at E13, followed by BrdU injection 2 h before sacrifice on E16. We observed that TET3 overexpression resulted in more cells remaining within the VZ/SVZ compared with control (Supplementary Fig S4A and B). We also observed that there was a significant increase in cell proliferation and the mitotic index as assessed by BrdU or pH3 in the GFP-positive population with TET3 overexpression (Supplementary Fig S4C–F). Furthermore, the opposite phenotypes of cell distribution and proliferation were shown with TET3 knockdown. A few reports have revealed key roles for TET3 in the early developing central nervous system. TET3-depleted Xenopus embryos exhibit striking abnormalities in neural development, featuring reduced expression of neural genes and resulting in small heads 18. These phenotypes were partly rescued by a catalytically inactive TET3. It is unknown whether the perinatal lethality caused by TET3 depletion 30 is due to a neurological development defect. Combined with these reports, our results reveal a critical role for TET3 in NPC proliferation during early neocortical development.
Given these results, the possibility appeared that TET3 may rescue cell proliferation defects caused by miR-15b overexpression in the developing cortex. To test this possibility, we co-electroporated TET3 plasmids together with miR-15b plasmids into E13 brains. The brains were harvested at E16. Notably, we observed that the cell distribution phenotype caused by miR-15b overexpression in the cortex was partly rescued by TET3 overexpression (Fig5A and B). Additionally, we found that TET3 was able to completely rescue the NPC proliferation defects caused by miR-15b overexpression, as evaluated by BrdU incorporation (Fig5C and D) and pH3 incorporation (Fig5E and F). The cell-cycle exit analysis showed that TET3 rescued the effects of increased miR-15b expression on NPC proliferation and cell-cycle exit (Supplementary Fig S5A–C). This suggests that the interaction between miR-15b and TET3 is crucial for maintaining the neural progenitor pool during early corticogenesis. Collectively, these results further indicate that miR-15b regulates neural progenitor proliferation by targeting TET3.
Figure 5.
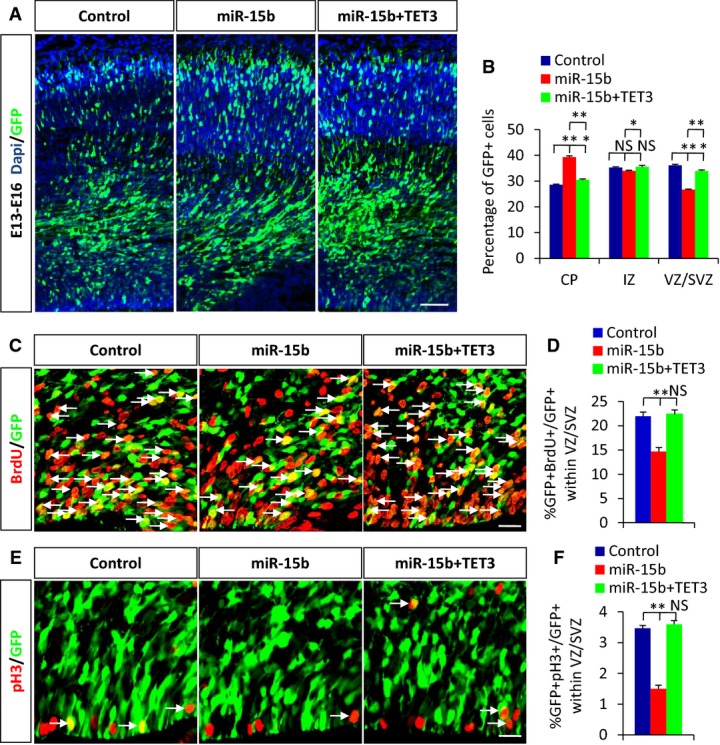
miR-15b/TET3 is involved in the proliferation of cortical NPCs
A, B The changed cell position in each region of cortex caused by miR-15b overexpression is partly rescued by TET3 overexpression.
C–F Decreased BrdU (C, D) or pH3 (E, F) incorporation caused by miR-15b overexpression is rescued by TET3 overexpression. Arrows indicate double-positive cells for GFP and BrdU or pH3.
Data information: Scale bars, 50 μm (A) and 20 μm (C and E). Values are mean ± SEM; n = 3; one-way ANOVAs with Tukey's test for post hoc multiple comparisons; *P < 0.05; **P < 0.01; NS, non-significant.
In summary, we clearly demonstrate that miR-15b regulates various aspects of cortical development and is involved in the expansion and differentiation of NPCs. Our data show that miR-15b regulates the makeup of the progenitor pool, providing a possible explanation for the changed cell proliferation, cell-cycle exit, and neuronal differentiation. We reveal that miR-15b directly targets TET3 and represses the expression of TET3 and 5hmC. Importantly, miR-15b cooperates with TET3 to regulate NPC proliferation. Together, our results not only reveal an important regulatory mechanism of miR-15b/TET3 interaction involved in the amplification of NPCs but also uncover a new relationship between miRNA, TET, and DNA demethylation during early neocortical development.
Materials and Methods
Detailed information can be found in Supplementary Materials and Methods.
Animals
Pregnant ICR mice were purchased from Vital River Laboratories. All animal studies were performed in accordance with experimental protocols and approved by Animal Care and Use Committees at the Institute of Zoology, Chinese Academy of Sciences.
In utero electroporation
In utero electroporation was carried out as described previously 35. Detailed information of in utero electroporation can be found in Supplementary Materials and Methods.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2014CB964903 and 2015CB964500 JJ), the National Science Foundation of China (31371477 JJ), the Strategic Priority Research Program (XDA01020301 JJ), and China Postdoctoral Science Foundation Fellowships (XL and HJ).
Author contributions
XL and HJ designed parts of the study, performed the research, analyzed the data, and wrote the manuscript; YL and XL performed the research; JJ designed parts of the study, supported the finance, and approved the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
for this article is available online: http://embor.embopress.org
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Materials and Methods
Review Process File
References
- Mcconnell SK. Constructing the cerebral-cortex – neurogenesis and fate determination. Neuron. 1995;15:761–768. doi: 10.1016/0896-6273(95)90168-x. [DOI] [PubMed] [Google Scholar]
- Barca-Mayo O, De Pietri Tonelli D. Convergent microRNA actions coordinate neocortical development. Cell Mol Life Sci. 2014;71:2975–2995. doi: 10.1007/s00018-014-1576-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung HL, Zhang K, Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Lyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development. 2012;139:1895–1902. doi: 10.1242/dev.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell. 2011;9:193–204. doi: 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2012;2:568–579. doi: 10.1016/j.celrep.2012.08.030. [DOI] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, Hu Y, Hu D, Zhao B, Cerovina T, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Woodworth MB, Custo Greig L, Kriegstein AR, Macklis JD. SnapShot: cortical development. Cell. 2012;151:918–918.e1. doi: 10.1016/j.cell.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Contribution of intermediate progenitor cells to cortical histogenesis. Arch Neurol. 2007;64:639–642. doi: 10.1001/archneur.64.5.639. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- Fu X, Jin L, Wang X, Luo A, Hu J, Zheng X, Tsark WM, Riggs AD, Ku HT, Huang W. MicroRNA-26a targets ten eleven translocation enzymes and is regulated during pancreatic cell differentiation. Proc Natl Acad Sci USA. 2013;110:17892–17897. doi: 10.1073/pnas.1317397110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SG, Jiang Y, Qiu R, Rauch TA, Wang Y, Schackert G, Krex D, Lu Q, Pfeifer GP. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res. 2011;71:7360–7365. doi: 10.1158/0008-5472.CAN-11-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Ghosh T, Aprea J, Nardelli J, Engel H, Selinger C, Mombereau C, Lemonnier T, Moutkine I, Schwendimann L, Dori M, et al. MicroRNAs establish robustness and adaptability of a critical gene network to regulate progenitor fate decisions during cortical neurogenesis. Cell Rep. 2014;7:1779–1788. doi: 10.1016/j.celrep.2014.05.029. [DOI] [PubMed] [Google Scholar]
- Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1:1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Materials and Methods
Review Process File