Abstract
Integrons, mobile genetic units, capture and incorporate antibiotic resistance gene cassette by site-specific recombination. Class 1 integrons are widespread and associated with dispersion of antibiotic resistance among Gram-negative bacteria. The expression of gene cassette in Class 1 can vary, based on the Pc promoter but seldom from another promoter hiding downstream of Pc, called P2. To probe distribution and prevalence of gene cassette promoter variants, we analyzed 169 S. Choleraesuis and 191 S. Typhimurium isolates from humans and animals, finding 95.27% occurrence of integrin among S. Choleraesuis, 83.25% among S. Typhimurium. PCR-RFLP analysis identified four promoters (PcS+P2, PcWTGN-10+P2, PcH1+P2, and PcWTGN-10+P2-GGG) in said integron-positive isolates; major types in S. Choleraesuis and S. Typhimurium were PcS+P2 and PcWTGN-10+P2, respectively. Likewise, β-galactosidase assay rated promoter strength of variants by transcriptional fusion constructs to show extended -10 promoter (TGn/-10 promoter) in Pc and three-nucleotide insertion (GGG) between -35 and -10 region of P2 improving promoter strength of gene cassette.
Keywords: Salmonella, Class 1 integrons, Gene cassette, Promoter
1. Introduction
Salmonellosis ranks among the most common bacterial infections worldwide [1]. Salmonella species are rod-shaped, aerobic, and Gram-negative bacteria, all major food-borne pathogens in the world [2]. Until 2004, over 2,500 Salmonella serotypes were identified [3]. Among these serovars, Salmonella enterica serovar Choleraesuis and Typhimurium are common non-typhoidal serotypes that pose global concern [4,5]. The USA diagnoses over 4 million cases of Salmonella infection per annum [6], about 500 fatal [2,7]. While mild and self-limited in adults, salmonellosis can require drugs, especially antibiotics, to treat infant, elderly, or immunocomprised patients [8]. Abuse of antibiotics in many locales nowadays spurs development of resistant strains. Studies show ever more multidrug resistance by Salmonella, causing serious public health hazards [5,9,10]. Such mechanisms entail obtaining genes or point mutation in genomes [11], resistance dispersed by [1] clonal expansion of drug-resistant strains or [2] horizontal transfer of determinants. Multidrug resistant genes transmitted between human and animal pathogens [12] mean mobile genetic elements playing a key role in dispersion of drug resistance among bacterial population [13-16].
Plasmids, transposons, and integrons are well known mobile genetic elements that mediate drug resistance genes disseminating via horizontal or vertical transfer [2]. Quantity of integron research has grown recently, with five classes of identified by sequences of integrases. Class 1 is most prevalent and closely linked with multidrug resistance in Gram-negative bacteria [11,17,18]. Typical Class 1 integron consists of intI gene encoding integrase, recombination specific site attI, major promoter Pc, and gene cassettes [18-21]. Over 100 gene cassettes harbored in Class 1 integron have been identified [22]; Pc is thought responsible for expression of gene cassettes [23]. Several Pc variants are described based on strength [24]: PcS for “strong”, PcW for “weak”, PcH1 for Hybrid 1, and PcH2 for Hybrid 2, the last two containing -35 and -10 hexamers of PcW and PcS in opposite combinations. Promoter strengths of PcH1 and PcH2 are intermediate between PcS and PcW. Studies indicate presence of “TGN” extended -10 motif between -35 and -10 hexamers raising transcription efficacy of σ70 promoters in E. coli [24,25]. Occasionally, Pc combines with a second promoter designated P2, located 119 bp downstream of Pc in 10% of Class 1 integrons [24,26,27]. A rare P2 type was described by Tenover [28] and Tae-Eun Kim [29]: three G residue insertion optimizes spacing (17 bp) between potential -35 and -10 hexamer sequences. Strength of four Pc types has been detailed in previous studies [24,30-32], several of which show a great polymorphism among variant Pc-P2 combinations. To evaluate dissemination of intergon-driven drug resistance, this study examined 360 Salmonella isolates for prevalence of Pc variants and strengths of Pc-P2 variant combinations in Taiwan.
2. Materials and methods
2.1. Bacterial strains and culture conditions
Salmonella isolates in this study had been described in previous report [5]. A total of 360 Salmonella isolates (169 S. Choleraesuis and 191 S. Typhimurium) were amassed from human and animal hosts. For serovar identification of Salmonella enterica, antiserum of O and H antigen detection were purchased from Denka Seiken Co., Ltd. in Japan and S&A Reagents Lab Limited in Thailand, respectively. Analysis based on the Kaufmann-White scheme and protocols for serotyping established by the Centers for Disease Control and Prevention in Atlanta, GA [33]. Salmonella isolates were maintained in 25 % frozen glycerol stock and inoculated on Salmonella-Shigella (SS) agar (Difco, USA) at 37°C.
2.2. PCR detection of Class 1 integrons in Salmonella isolates
All Salmonella isolates were probed for integrons by polymerase chain reaction (PCR) and nucleotide sequencing. After culturing bacteria in Luria-Bertani (LB) broth to log-stationary phase at 37 °C with vigorous shaking, genomic template DNA were extracted from isolates, as per manufacturer’s instructions for Tissue & Cell Genomic DNA Purification Kit (Genemark, Taiwan). Specific primers IntegronA and IntegronB [34] (Table 1) screened intI1, Class 1 integrase gene, within bacterial isolates. PCR mixture was in a total volume of 25 μl containing 3 μl genomic DNA as template, 1 μl of each primer (10 μM), 5 μl of 5x PCR Plus Master Mix II solution (Genemark, Taiwan), and 15 μl of distilled water. PCR mixture used T1 Thermocycler (Biometra, USA). Template was initially denatured at 95°C for 5 min followed by 35 cycles at 95°C for 30 sec, 57°C for 30 sec, and 72°C for 30 sec. Final extension for 10 min was done at 72 °C, PCR products confirmed via 1% agarose gel electrophoresis.
| Namea | Sequence (5’-3’) | Product size (bp) | References |
|---|---|---|---|
| IntegronA | GCCTTGCTGTTCTTCTACGG | 558 | [34] |
| IntegronB | GATGCCTGCTTGTTCTACGG | ||
| SC-RGA-F1b | ATTGGATCCGGTGACGCACACCGTGGAAACGGAT | 328 | This study |
| SC-RGA-F2b | ATTGGATCCACCTTGACCGAACGCAGCGGTGGTA | 218 | |
| SC-RGA-Rc | ATTAAGCTTCGAGTTCATATGGCTAACTTTGTTT |
a. IntegronA and IntegronB primers detected Class I integron. SC-RGA-F1 and SC-RGA-R primers for Pc-P2 region in integrons (328 bp); SC-RGA-F2 and SC-RGA-R for only P2 region in integron (218 bp).
b. Nucleotide sequences recognized by BamHI restriction enzyme underlined
c. Nucleotide sequences recognized by HindIII restriction enzyme underlined
2.3. Characterization of gene cassette promoter (Pc-P2) variants in Salmonella isolates by PCR-Restriction fragment length polymorphism (PCR-RFLP)
To ferret out promoter variants of Class 1 integrons in Salmonella intI1-positive isolates, PCR-RFLP method served for analysis: 330-bp fragment of Pc-P2 regions in Class 1 integrons amplified by PCR with SC-RGA-F1 and SC-RGA-R specific primers (Table 1). Preparation of PCR mixture as detailed above proceeded as follows: after initial denaturation (5 min at 95°C), DNA fragment was amplified for 35 cycles of 30 sec at 95°C, annealing at 60°C for 30 sec, and 30 sec for extension at 72°C, with a final extension step at 72°C for 7 min. Before performing RFLP, all PCR products were purified by PCR clean-up kit (Genemark, Taiwan). To screen promoter variants, HincII or AluI restriction enzymes identified Pc variants; BsrGI restriction enzyme was also applied to analyze three nucleotide insertions between -35 and -10 region of P2. Each digestion reaction containing 2 μl of 10X NEBuffer 4, 0.2 μl of 100X BSA, 1 μl of restriction enzyme (10U; New England BioLabs, Inc.), 10 μl of purified PCR products, and added distilled water to 20 μl. Mixture was incubated at 37°C for 6 hrs, after which treatments were analyzed on 1.5 % agarose gel electrophoresis.
2.4. Plasmid constructions for promoter activity assay
After characterizing types of gene cassette promoter, one bacterial strain stood for each type was picked randomly from Salmonella intI1-positive isolates. To study relative strength of a gene cassette promoter, transcriptional fusion with both Pc and P2 were cloned into the promoterless lacZ gene upstream in a reporter vector (pCB267, [35]). Extracting genomic DNA from bacterial strains representing promoter types, we amplified Pc-P2 region by specific SC-RGA-F1 and SC-RGA-R primers (Table 1). To gauge effect of three nucleotide insertions in -35 and -10 region of P2 on promoter strength, only P2 region with or without insertion was amplified by SC-RGA-F2 and SC-RGA-R primer pairs (Table 1). PCR were run for 5 min at 95°C followed by 35 cycles of 30 sec at 95°C, 30 sec at 60°C, 30 sec at 72°C and final extension of 15 min at 72°C. PCR products purified by kit (Genemark, Taiwan) were digested with BamHI and HindIII, then ligated to BamHI- and HindIII-digested pCB267. These constructs could assess strength of promoters.
2.5. β-galactosidase assay of promoter strength
Each recombinant plasmid carrying transcriptional fusion was transformed into E. coli strain DH5α to measure β-galactosidase enzyme activity, assays performed with 0.5-ml aliquots of overnight cultures as described [36, 37]. Bacteria cultured in LB broth containing 100 μg/ml of ampicillin at 37 °C were vigorously shaken overnight, collected by centrifuge, washed with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, and 50 mM β-mercaptoethanol, pH 7.0), and lysed by adding chloroform plus 0.1% SDS. Then 200 μl of 4 mg/ml ortho-nitrophenyl-β-galactoside (ONPG) substrate was added to reactants and incubated at 30°C, with incubation time recorded; 500 μl of 1M Na2CO3 served as stopper of the reaction. Value of OD420 was measured by spectrometer, units of β-galactosidase enzyme activity compared within constructs. Experiments were done at least three times per construct.
2.6. DNA sequencing
All PCR products represented different types of promoter and plasmids used for evaluating promoter strength were purified via PCR clean-up kit or plasmid miniprep purification kit (Genemark, Taiwan) and sequenced from both sides by AmpliTaq-FS DNA polymerase, dye terminator chemistry, and an automatic nucleic acid sequence analyzer (ABI Prism, USA) at the DNA sequence facility at Mission Biotech Co., Ltd. in Taiwan. Specific primers used for cloning and sequencing were synthesized by the same company. Nucleotide sequences were compared with the BLAST network in the GenBank database of National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine [38].
2.7. Statistical analysis
All data of β-galactosidase enzyme activities were calculated at least three times, analyzed by commercial software SPSS Version 16 for Windows (SPSS Company; Chicago, IL). Pearson’s chi-square test derived linkage between independent groups, p value < 0.05 considered significant.
3. Results
3.1. Detection of Class 1 integrons among Salmonella isolates
All isolates in this study were characterized in previous work [5], including the Class 1 integron presence and antimicrobial resistance patterns. A total of 360 isolates, belonging to either S. Typhimurium or S. Choleraesuis, were collected from humans and animals during 1997-2009 in Taiwan. Among S. Choleraesuis isolates, 111 are from pigs and 50 from humans; among S. Typhimurium isolates, 17 are from pigs, 115 from humans, 1 from pigeons, 9 from turtles, 12 from chickens, 1 from snakes, 4 from ducks. Using PCR, we showed 95.27% (161/169) of S. Choleraesuis and 83.25% (159/191) of S. Typhimurium isolates harbor Class 1 integron (Table 2).
| Promoter Type | Variant | -35 Region | Spacing | -10 Region | |||
| Sequence | HincII (GTY/RAC) | No. of nt | N14-TCN (TGN) | Sequence | AluI (AG/CT) | ||
| PC | PcS | TTGACA | + | 17 | TCN | TAAACT | - |
| PcWTGN-10 | TGGACA | - | 17 | TGN | TAAGCT | + | |
| PcH1 | TGGACA | - | 17 | TCN | TAAACT | - | |
| Promoter Type | Variant | -35 Region | Spacing | -10 Region | |||
| Sequence | No. of nt | 3-nt-insertion | BsrGI (T/GTACA) | Sequence | |||
| P2 | P2 | TTGTTA | 14 | - | + | TACAGT | |
| P2-GGG | TTGTTA | 17 | + | - | TACAGT | ||
3.2. Molecular characterization of gene cassettes’ common promoters in intI1-positive Salmonella isolates
Pc is the major promoter located upstream of gene cassettes in Class 1 integron. Occasionally, a second promoter (P2) is located 119 bp downstream of Pc. Based on nucleotide sequence of -35 and -10 Pc regions, it can be classified into strong, hybrid, and weak types [24,27,29,32]. By driving the expression of CAT reporter with Pc variants, relative strengths of PcS and PcW + P2 to PcW were measured, with PcS and PcW+ P2 promoters about 30- and 15-fold higher than PcW, respectively [24,31].
PCR-RFLP characterized Pc-P2 variants. Promoter regions were amplified by primers SC-RGA-F1 and SC-RGA-R (Table 1), then amplicons subjected to enzyme digestion. The -35 region of PcS (TTGACA) and -10 region of PcW (TAAGCT) were digested by HincII and AluI, respectively. Digestion of fragment containing -35 region of PcS with HincII would yield two fragments, 277bp and 51 bp in length; digestion of fragment containing -10 region of PcW with AluI would produce 252bp and 76 bp fragments. Those not fitting this restriction enzyme digestion pattern were subjected to sequence and classified as PcWTGN-10. Three guanine (GGG) insertions between -35 and -10 region of P2 were studied to create a new promoter (P2-GGG) occasionally found in Class 1 integrons [26]. For quicker P2 characterization, PCR amplified fragments were digested by BsrG1. P2-GGG promoter could not be digested, but P2 promoter yielded fragments (188 and 140 bp) after digestion. Not only PCR-RFLP analysis but also DNA sequence was applied to variants, confirming promoters’ molecular characterization. Major Pc-P2 variants identified (Table 2) were PcS+P2, PcWTGN-10+ P2, PcH1+P2, and PcWTGN-10+P2-GGG.. PcS+P2 was the major variant in S. Choleraesuis (143/161, 88.82%) and PcW TGN-10+P2 for S. Typhimurium (131/159, 82.39%). Surprisingly, multiple variants appeared in PcS + P2/PcH1 + P2/PcWTGN-10 + P2 combination in S. Choleraesuis and PcWTGN-10 + P2/PcS + P2/PcWTGN-10 + P2-GGG combination in S. Typhimurium. Of six isolates, two of S. Choleraesuis and four of S. Typhimurium harbored more than one Pc-P2 combination in one strain (Table 3).
| Serotype | Total no. | No. of integrons (%) | Promoter variant (occurrence, %) |
| S. Choleraesuis | 169 | 161 (95.27 %) |
PcS + P2 (143/161, 88.82 %) PcH1 + P2 (16/161, 9.94 %) PcS + P2/ PcH1 + P2/ PcWTGN-10 + P2 (2/161, 1.24 %) |
| S. Typhimurium | 191 | 159 (83.25 %) |
PcWTGN-10 + P2 (131/159, 82.39 %) PcS + P2 (15/159, 9.43 %) PcWTGN-10 + P2-GGG (9/159, 5.66 %) PcWTGN-10 + P2/ PcS + P2/ PcWTGN-10 + P2-GGG (4/159, 5.52 %) |
3.3. Relative strength of gene cassette promoter variants
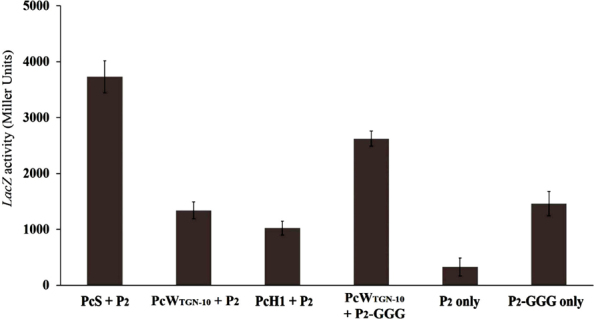
To rate promoter strength, Pc-P2 each variant identified was cloned into a promoterless vector (pCB267) to drive lacZ reporter gene expression. β-galactosidase activities were compared between variants; PcS+P2 proved strongest (Fig. 1). Strength of PcWTGN-10 + P2 was 31% greater than PcH1+P2. In prior studies, strength of hybrid Pc had intermediate activity between PcS and PcW [24]. Promoter activity of PcW rises with PcW carrying TGN-10 motif between -35 and -10 region (PcWTGN-10). Strength of P2-GGG region alone was 4.5-fold that of P2 without insertion. Our data concurred with earlier studies [28, 29]: promoter strength of PcWTGN-10 combined with P2-GGG was about 2-fold that of PcWTGN-10 combined with P2. Therefore, 3-G insertion can enhance promoter strength in general.
4. Discussion
S. Typhimurium DT104, a bacterial strain isolated during the early 1980s in the United Kingdom, is resistant to multiple antibiotics: e.g., ampicillin, chloramphenicol, streptomycin, sulfonamides, tetracycline (ACSSuT). After that, multi-drug resistance to ACSSuT is a common Salmonella trait [5, 39-41]. ACSSuT resistance genes are mostly disseminated through Class 1 integron [5]. In our study, 88.9% of Salmonella isolates harbored Class 1 integron in Taiwan. Comparing two serovars, positive rate of Class I integron in S. Choleraesuis (95.27 %) was higher than in S. Typhimurium (83.25 %)., suggesting Class 1 integron already widespread in Taiwan.
PCR-RFLP analysis identified four Pc-P2 combinations: PcS+P2, PcWTGN-10+P2, PcH1+P2, and PcWTGN-10+P2-GGG., the first two predominant in S. Choleraesuis and S. Typhimurium, respectively. These variants in our study are also the most prevalent forms in silico study [24]. Based on previous research on gene cassettes in Class 1 integron [5], for isolates carrying more than one set of Pc-P2 combination, some only carry one kind of gene cassette. Data portend more than one copy of Class 1 integrons in one strain, albeit with variant promoter combination.
Transcriptional fusion constructs were used to monitor promoter strength among Pc-P2 variants. Our data agreed with prior study that strengths of promoter variants is PcS > PcWTGN-10 > PcH1. Besides, our data showed TGN-10 motif between -35 and -10 region of Pc and three nucleotide insertions (GGG) between -35 and -10 region of P2 boosting both promoter strength and expression of gene cassettes [24]. Sequencing analysis avers that most isolates carrying PcH1+ P2 variant have guanine located 11bp downstream of -10 region of P2, but adenine could also occur. Still, no significant difference appeared between strengths of PcH1 and mutated PcH1 variants, showing single mutation of this site as random and not affecting expression of gene cassettes. In sum, integrons are well-known machinery to spread bacterial genetic elements, especially antibiotic resistance genes. This research proved Pc highly polymorphic; its strength may affect downstream gene cassette expression. Promoter polymorphism might alter levels of bacterial antibiotic resistance in response to environmental stress.
Acknowledgments
This study was funded by Grants CMU102-S-11 from China Medical University and 102-2313-B-005-008-MY2 from the National Science Council, Taiwan.
Declaration of Interest: Authors declare no conflicts of interest for this work.
REFERENCES
- 1].Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2].Ashraf A., Khan EP, Nawaz M. S., Chorng-Ming Cheng, Junaid A. Khan and Christine S. West. Identification and Characterization of Class 1 Integron Resistance Gene Cassettes among Salmonella Strains Isolated from Imported Seafood. Appl Environ Microbiol. 2009;75:1192–6. doi: 10.1128/AEM.02054-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3].Popoff MY, Bockemuhl J, Gheesling LL. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res Mkcrobiol. 2004;155:568–70. doi: 10.1016/j.resmic.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 4].Chiu CH, Su LH, Chu C. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin Microbiol Rev. 2004;17:311–22. doi: 10.1128/CMR.17.2.311-322.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5].Hsu YM, Tang CY, Lin H, Chen YH, Chen YL, Su YH. Comparative study of class 1 integron, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline (ACSSuT) and fluoroquinolone resistance in various Salmonella serovars from humans and animals. Comp Immunol Microbiol Infect Dis. 2013;36:9–16. doi: 10.1016/j.cimid.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 6].Pang T, Bhutta ZA, Finlay BB, Altwegg M. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbio. 1995;3:253–5. doi: 10.1016/S0966-842X(00)88937-4. [DOI] [PubMed] [Google Scholar]
- 7].Zansky S, Wallace B, Schoonmaker-Bopp D, Smith P, Ramsey F, Painter J. From the Centers for Disease Control and Prevention. Outbreak of multi-drug resistant Salmonella Newport—United States, January-April 2002. JAMA. J Am Med Assoc. 2002;288:951–3. [PubMed] [Google Scholar]
- 8].Asperilla MO, Smego RA, Jr., Scott LK. Quinolone antibiotics in the treatment of Salmonella infections. Rev Infect Dis. 1990;12:873–89. doi: 10.1093/clinids/12.5.873. [DOI] [PubMed] [Google Scholar]
- 9].Hsu SC, Chiu TH, Pang JC, Hsuan-Yuan CH, Chang GN, Tsen HY. Characterisation of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int J Antimicrob Agents. 2006;27:383–91. doi: 10.1016/j.ijantimicag.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 10].Hsueh PR, Teng LJ, Tseng SP, Chang CF, Wan JH, Yan JJ. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg Infect Dis. 2004;10:60–8. doi: 10.3201/eid1001.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11].Krauland MG, Marsh JW, Paterson DL, Harrison LH. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg Infect Dis. 2009;15:388–96. doi: 10.3201/eid1503.081131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12].Holmberg SD, Wells JG, Cohen ML. Animal-to-man transmission of antimicrobial-resistant Salmonella: investigations of U.S. outbreaks, 1971-1983. Science. 1984;225:833–5. doi: 10.1126/science.6382605. [DOI] [PubMed] [Google Scholar]
- 13].Lynne AM, Rhodes-Clark BS, Bliven K, Zhao S, Foley SL. Antimicrobial resistance genes associated with Salmonella enterica serovar newport isolates from food animals. Antimicrob Agents Chemother. 2008;52:353–6. doi: 10.1128/AAC.00842-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14].Aarts HJ, Boumedine KS, Nesme X, Cloeckaert A. Molecular tools for the characterisation of antibiotic-resistant bacteria. Vet Res. 2001;32:363–80. doi: 10.1051/vetres:2001130. [DOI] [PubMed] [Google Scholar]
- 15].Poppe C, Martin LC, Gyles CL, Reid-Smith R, Boerlin P, McEwen SA. Acquisition of resistance toe extended- spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in the turkey poult intestinal tract. Appl Environ Microbiol. 2005;71:1184–92. doi: 10.1128/AEM.71.3.1184-1192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16].Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA. Multiple antimicrobial resistance in plague: an emerging public health risk. PloS one. 2007;2:e309. doi: 10.1371/journal.pone.0000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17].Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–66. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 18].Mazel D. Integrons: agents of bacterial evolution. Nat Rev Microbiol. 2006;4:608–20. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 19].Hall RM, Collis CM, Kim MJ, Partridge SR, Recchia GD, Stokes HW. Mobile gene cassettes and integrons in evolution. Ann NY Acad Sci. 1999;870:68–80. doi: 10.1111/j.1749-6632.1999.tb08866.x. [DOI] [PubMed] [Google Scholar]
- 20].Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–83. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 21].Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 22].Fluit AC, Schmitz FJ. Resistance integrons and super- integrons. Clin Microbiol Infect. 2004;10:272–88. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 23].Jove T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 2010;6:e1000793. [DOI] [PMC free article] [PubMed]
- 24].Thomas Jove’ SDR, Francois Denis, Didier Mazel, Marie-Ce’ cile Ploy. Inverse Correlation between Promoter Strength and Excision Activity in Class 1 Integrons. PLoS Genet 2010; 6. [DOI] [PMC free article] [PubMed]
- 25].Burr T, Mitchell J, Kolb A, Minchin S, Busby S. DNA sequence elements located immediately upstream of the -10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res. 2000;28:1864–70. doi: 10.1093/nar/28.9.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26].Collis CM, Hall RM. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–62. doi: 10.1128/AAC.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27].Vinue L, Jove T, Torres C, Ploy MC. Diversity of class 1 integron gene cassette Pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int J Antimicrob Agents. 2011;38:526–9. doi: 10.1016/j.ijantimicag.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 28].Tenover FC, Filpula, D., Phillips, K.L., Plorde, J.J. Cloning and sequencing of a gene encoding an aminoglycoside 6’-N-acetyl-transferase from an R factor of Citrobacter diversus. J Bacteriol 1988; 170. [DOI] [PMC free article] [PubMed]
- 29].Kim TE, Kwon HJ, Cho SH, Kim S, Lee BK, Yoo HS. Molecular differentiation of common promoters in Salmonella class 1 integrons. J Microbiol Methods. 2007;68:453–7. doi: 10.1016/j.mimet.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 30].Brizio A, Conceicao T, Pimentel M, Da Silva G, Duarte A. High-level expression of IMP-5 carbapenemase owing to point mutation in the -35 promoter region of class 1 integron among Pseudomonas aeruginosa clinical isolates. Int J Antimicrob Agents. 2006;27:27–31. doi: 10.1016/j.ijantimicag.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 31].Levesque C, Brassard S, Lapointe J, Roy PH. Diversity and relative strength of tandem promoters for the antibiotic- resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 32].Papagiannitsis CC, Tzouvelekis LS, Miriagou V. Relative strengths of the class 1 integron promoter hybrid 2 and the combinations of strong and hybrid 1 with an active P2 promoter. Antimicrob Agents Chemother. 2009;53:277–80. doi: 10.1128/AAC.00912-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33].Brenner F, McWhorter-Murlin A. Identification and serotyping of Salmonella: Centers for Disease Control and Prevention, National Salmonella Reference Laboratory; 1998.
- 34].Ng LK, Mulvey MR, Martin I, Peters GA, Johnson W. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob Agents Chemother. 1999;43:3018–21. doi: 10.1128/aac.43.12.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35].Schneider K, Beck CF. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42:37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- 36].Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 37].Cagle CA, Shearer JE, Summers AO. Regulation of the integrase and cassette promoters of the class 1 integron by nucleoid-associated proteins. Microbiology. 2011;157:2841–53. doi: 10.1099/mic.0.046987-0. [DOI] [PubMed] [Google Scholar]
- 38].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39].Wall PG, Morgan D, Lamden K, Ryan M, Griffin M, Threlfall EJ. A case control study of infection with an epidemic strain of multiresistant Salmonella typhimurium DT104 in England and Wales. Commun Dis Rep CDR Rev. 1994;4:R130–5. [PubMed] [Google Scholar]
- 40].Yu CY, Chou SJ, Yeh CM, Chao MR, Huang KC, Chang YF. Prevalence and characterization of multidrug-resistant (type ACSSuT) Salmonella enterica serovar Typhimurium strains in isolates from four gosling farms and a hatchery farm. J Clin Microbiol. 2008;46:522–6. doi: 10.1128/JCM.00709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41].Gebreyes WA, Thakur S. Multidrug-resistant Salmonella enterica serovar Muenchen from pigs and humans and potential interserovar transfer of antimicrobial resistance. Antimicrob Agents Chemother. 2005;49:503–11. doi: 10.1128/AAC.49.2.503-511.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]


