Abstract
Next-generation sequencing technologies have greatly lowered the cost of whole-genome sequencing (WGS) and related approaches. Thus, comprehensive sequencing for diagnostic purposes may clear this financial hurdle in the near future. The report by Bainbridge and colleagues in this issue of Science Translational Medicine illustrates the diagnostic power of WGS. In this Perspective, we discuss whether and how genome sequencing might become routine for clinical diagnosis.
The molecular bases of almost one half of the 6,727 disorders with suspected Mendelian inheritance have been defined within the past generation (1). Mendelian disorders account for ~17% of pediatric hospitalizations and an even greater proportion of health care costs (2–4). An immediate benefit of disease-gene discovery is the capability of definitive diagnosis in affected individuals and risk assessment for relatives. Decades of experience have established the utility of Mendelian diagnosis by serial additive Sanger sequencing of candidate genes, yielding guidelines for clinical laboratory procedures, variant interpretation, and the ethics of testing (5–8). However, despite this progress the availability of clinical testing for most Mendelian disorders is hampered by economics: The rarity of many conditions makes them unattractive targets for commercialization; for others, testing is handicapped by exclusive patent-ownership practices (9). As a result, public knowledge of mutation spectrum, genotype-phenotype relationships, and allele frequencies is rudimentary for most Mendelian disorders. In turn, low rates of ascertainment and delayed diagnosis hamper treatment innovations. Of those for which molecular tests are available, many exhibit locus and/or clinical heterogeneity, engendering lengthy and costly differential-diagnostic odysseys. Not infrequently, the cost of diagnosis exceeds $10,000 per patient (10).
Thus, it is with great excitement that scientists and clinicians greet recent reports of Mendelian disease diagnosis by whole-genome sequencing (WGS), such as that by Bainbridge and colleagues in this issue of Science Translational Medicine (11). In this Perspective, we discuss whether, when, and whereby WGS and related approaches might become routine for clinical diagnosis.
Driving these diagnostic developments are next-generation sequencing (NGS) technologies, which have lowered the cost of individual genome sequencing 1700-fold (12). Simultaneously, for the ~20% of genes that are patented, the legal status of exclusive rights to clinical testing is being broadly assailed (9). Additional economies are afforded by sequencing only the exome (exome-seq; the 2% of the genome represented by coding regions, or exons), the “Mendelianome” (coding regions of 2,993 known disease genes), or targeted disease panels using NGS (13).These trends promise to remove the financial hurdle to routine comprehensive sequencing for diagnostic purposes in the near future. But when? And how?
NEXT-GENERATION METHODS, ROUTINE USE?
Sequencing of single candidate disease-driving genes is effective for diagnosis if distinctive clinical features are present and minimal locus heterogeneity exists. However, specific diseases that feature gross locus heterogeneity (multiple genes may be causal) or broad presentations (such as mitochondrial dysfunction, congenital disorders of glycosylation, sensorineural deafness, and intellectual disability) are intractable with conventional capillary sequencing. This bottleneck results from the inability to prioritize individual genes among many candidates for diagnostic testing. NGS approaches circumvent this bottleneck because all candidate genes may be interrogated simultaneously.
Although the utility of WGS and exome-seq for disease-gene discovery is well established, the power of such technologies for routine clinical diagnosis and therapeutic guidance is just beginning to be realized (10, 11, 14–25). A prototypical example of diagnostic WGS in a setting of gross locus heterogeneity is Charcot-Marie-Tooth disease (CMT), in which heterogeneity exists in both the inheritance pattern and genetic loci (causal mutations have been identified in 37 genes to date). Research-grade WGS of the proband (the affected family member through whom the family is ascertained) identified compound heterozygous exonic variants in a gene previously reported to be mutated in CMT type 4C [Online Mendelian Inheritance in Man database number 601596 (1)] (10). These variants were subsequently verified by Sanger sequencing and were shown to track with the disease in three additional siblings, establishing a molecular diagnosis for the family.
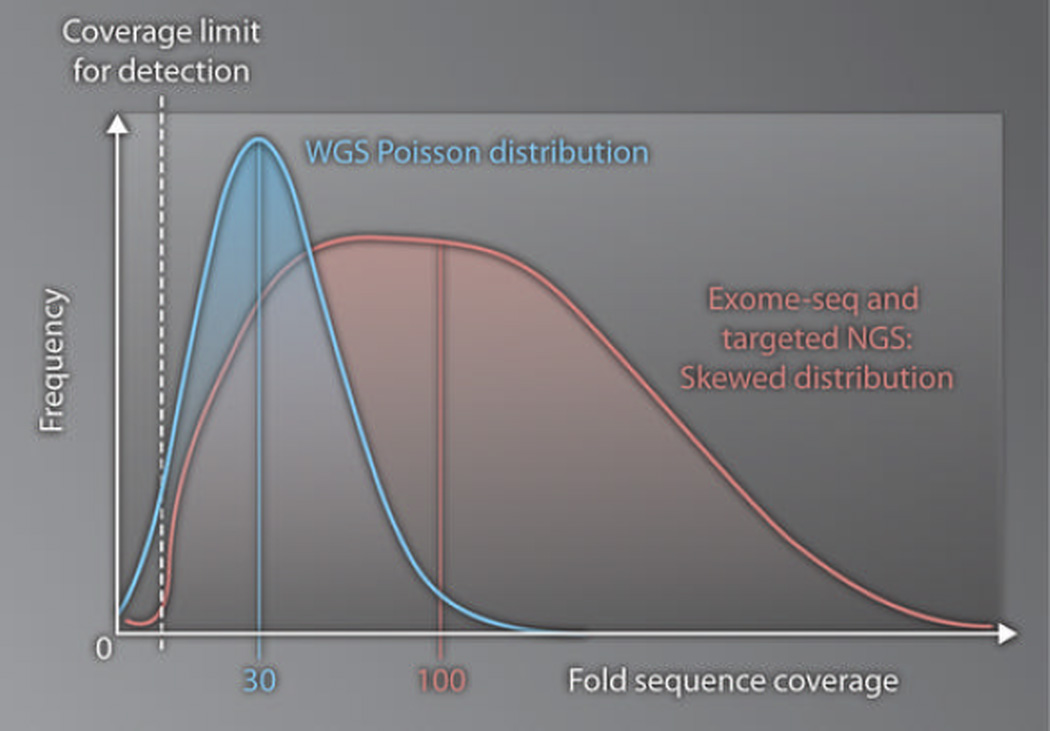
Confirmation of research-grade WGS findings with additional testing and interpretation for diagnostic purposes is important for two reasons. First, research-grade WGS alone is inadequate for clinical interpretation and reporting of variants. This incompatibility arises from the fact that, in order to be economically feasible, research-grade WGS employs genome coverage (or depth of sequencing) and variant filters (bioinformatic standards designed to distinguish true genomic variants from noise) that are of insufficient accuracy for diagnostic use. Because human genomes are diploid, the status of both chromosomes must be delineated at each nucleotide position (genotyping) in order to achieve diagnosis. The coverage distribution of WGS is approximately Poisson in character, which implies that individual genomic regions exhibit a variety of coverages that are symmetrically distributed around a mean (Fig. 1). Research-grade WGS is performed to an average 30-fold coverage (that is, 90 billion bases of sequence for a 3–billion base haploid genome) with a quality score of at least Q20 (1 in 100 chance of a wrong call at each base; raw accuracy of a base call, 99%) (26). As a result, analytical specificity (in this case, the accuracy of genotypes at positions with variants) is sacrificed in favor of analytical sensitivity (the proportion of true variants identified). For example, heterozygous positions are quite often wrongly designated as homozygous. Sixty- or 90-fold coverage would correct this discrepancy and permit genotyping that is likely to be of diagnostic quality, albeit increasing cost substantially (Fig. 1) (26).
Fig. 1. Contrasting technologies.
Frequency of sequences is plotted versus depth of sequence coverage to show, for comparison purposes, the patterns of distribution of sequence coverage afforded by research-grade WGS (blue line) and exome-seq/targeted NGS (red line). The genome coverage achieved by WGS is symmetric about the mean, which at this time is typically 30-fold coverage. Approximately 5% of the genome has insufficient coverage to allow variants to be detected (dotted white line). In contrast, the genome coverage achieved by exome-seq and targeted NGS is right-skewed. Thus, approximately 100-fold average coverage is necessary to achieve a sensitivity of variant detection similar to WGS.
Second, genomic variants identified by research testing should not be reported to patients without confirmatory testing in a Clinical Laboratory Improvement Amendments (CLIA)–certified laboratory (27), which adheres to quality standards appropriate for diagnostic testing (Fig. 2). Our perspective is that WGS and exome-seq are useful for nominating genomic variants, including copy number variants (CNVs), but that CLIA-compliant confirmatory genotyping is required for clinical interpretation of genomic results and reporting of findings to patients. These issues are important because some CLIA-certified commercial laboratories offer WGS results to clients without confirmatory testing. In particular, WGS appears to be a cost-effective and successful strategy for diseases in which the number of gene targets makes individual sequencing too cumbersome and/or expensive (10, 11, 17).
Fig. 2. Fast forward?
Shown are the major similarities and differences in refinements needed for diagnostic use (DX) of WGS, exome-seq, and targeted NGS. The average depth of coverage differs in each approach. Reflex confirmatory testing of all clinically relevant results is necessary for WGS and exome-seq but probably not for targeted NGS; this is because it is possible to obtain a large number of independent observations of each sequence variant. All three methods require pathological interpretation and reporting by a certified laboratory director.
The next question for WGS is whether it is a rational approach for the diagnosis of diseases associated with a modest number of genes that must be sequenced (for example, between 1 and 10). A plethora of such disorders exist, including Mendelian early-onset diabetes mellitus, hereditary cancers of specific tissues, prolonged QT interval, and lysosomal trafficking immunodeficiencies. The current research paper by Bainbridge et al. (11) reports WGS of fraternal twins concordant for dopa-responsive dystonia (DRD), a complex movement disorder caused by three known genes, two of which that had been excluded from the twins’ diagnosis by conventional clinical sequencing. WGS of both twins revealed compound heterozygosity for two previously reported variants in the sepiapterin reductase (SPR) gene, which encodes an aldo-keto reductase enzyme that participates in the biosynthesis of tetrahydrobiopterin, a cofactor for various neurotransmitter biosynthetic enzymes. Sanger sequencing confirmed a molecular diagnosis of SPR-DRD in the twins as well as the carrier status of both parents and two grandparents. These findings illustrate the value of WGS-assisted molecular diagnosis for individualized treatment: In contrast to other forms of DRD, treatment with 5-hydroxytryptamine and serotonin reuptake inhibitors is indicated in patients with SPR defects. Detractors will point out that this diagnosis could have been made by sequencing the third candidate gene alone. However, the Bainbridge et al. report serves as an exemplary model for the diagnostic utility and subsequent therapeutic benefit of combined WGS and reflex confirmatory testing in cases associated with conditions of at least moderate genetic heterogeneity.
Beyond WGS
Exome-seq and targeted NGS differ from WGS in a few key respects. First, these technologies require enrichment of the desired genomic regions before NGS can be performed. Several comparable enrichment methods are currently available, none of which are perfect, given that only a proportion of the product is on-target. That proportion varies with several factors, such as the degree of sample multiplexing— the number of samples that can be given molecular bar codes and pooled during the enrichment and sequencing processes—which, in turn, is a driver of cost-effectiveness. Second, enrichment adds bias, demonstrable by right-skewing of the aforementioned coverage distribution (Fig. 1). This means that for exome-seq to achieve sensitivity comparable to that of WGS, greater depth of coverage must be obtained (Fig. 1), partially offsetting the cost-saving rationale for use of the method in lieu of WGS. However, clinicians and researchers have much more exposure and access to a large body of literature on exome-seq (15–25, 28, 29) followed by confirmatory Sanger sequencing for the diagnosis and discovery of Mendelian mutations, relative to WGS (10, 11, 14). As just one example of such a prospective case study, exome-seq was used to nominate a candidate gene in a child with enteropathy—a disorder with moderate genetic heterogeneity—that failed to be disclosed by extensive conventional molecular testing (21). This report illustrates the value of exome-seq–assisted individualized treatment in that hematopoietic stem cell transplantation ablated the patient’s symptoms. Exome-seq variant detection is limited to mutations located in coding and splice junction regions. However, in none of the cases reported to date did WGS provide diagnostic or therapeutic information beyond that which would have been afforded by whole-exome or targeted-exome sequencing, which is ~fivefold and ~15-fold less expensive than WGS, respectively. Like WGS, exome-seq yields few false-positive identifications of exonic CNVs, but currently has too many false-negative calls. It is not yet possible to offer best-practice guidelines for exome-seq, because the various commercially available kits differ so greatly in the target sizes they accommodate, which range from 20 to 60 Mb. However, exome-seq appears to be an effective adjunct to diagnostic workups for Mendelian diseases.
In our view, targeted NGS is emerging as an ideal interim technology for clinical diagnosis. Enrichment targets vary from the Mendelianome (~0.25% of the genome) to mutation-harboring regions of genes germane to specific clinical presentations. Many clinical laboratories have recently begun to target 10 to 100 genes simultaneously with NGS (30). Unlike WGS, depth of sequencing can be increased economically to achieve diagnostic metrics comparable to conventional capillary sequencing. For example, one of us recently reported results of a retrospective assessment of targeted NGS for 448 recessive childhood diseases in 104 samples, which showed ~95% sensitivity and ~100% specificity for the detection and genotyping of substitution, insertion and deletion, splicing, and gross-deletion mutations at a 160-fold average coverage (13). Multiplexing, enrichment, and sequencing steps were used to achieve a cost of $400 per sample. Custom baits were included to capture known nonexonic mutations and boundaries of known gross deletions, insertions, and rearrangements, which are otherwise technically difficult to detect with NGS. Extensive validation of targeted NGS and interpretation of results using bioinformatic pipelines in conventional clinical testing situations are now needed to identify best practices and inform decisions regarding the necessity of continued confirmatory testing of positive results.
NOT SO FAST
Before a sense of euphoria sinks in, however, we offer perspectives on the four most important hurdles to broad use of NGS-based diagnostic testing in clinical laboratories. First, no clinical-grade general database of disease-associated mutations currently exists. Interpreting the clinical significance of mutations relies on information found in the primary literature and general and locus-specific databases (5, 8); disturbingly, 27% of literature-cited mutations may be incorrect (13). Consortia organized by leading reference laboratories, the National Center for Biotechnology Information, the Human Genome Variation Society, and the Human Variome Project are beginning to develop plans and recommendations to address these issues and create a clinical-grade general database (31).
Second, consensus strategies for standardized, high-throughput interpretation of genetic variants of unknown significance (VUS) must be developed and implemented. The current guidelines for clinical interpretation and reporting of disease variants (5, 8) are largely extensible to NGS but may warrant several considerations for future revisions: (i) Current guidelines for reporting VUS are necessarily conservative, written in the context of testing one or a few candidate genes. There is a high burden to report variants that are unlikely to be causative for fear of under-calling a pathogenic genotype, a fear perhaps less relevant in the context of sequencing all potentially causal genes rather than a single locus. As more public genomic data become available, the knowledge of variant frequencies and improved global insights into their correlation with disease should yield less-ambiguous reports for VUS. (ii) Mutation interpretation guidelines must be extended to include provisions for reporting genotypes and paired haplotypes (genetic constituents of each individual chromosome) with future extensibility to epistasis (gene-gene interactions resulting in modified phenotypes). (iii) Consensus software tools are needed for automated in-process VUS annotation to accommodate increasing test volumes and the numbers of all variants generated by NGS. (iv) In light of ongoing advancements in NGS, the way in which clinical laboratories report incidental findings—genomic variants perceived to be immaterial to the illness for which a diagnosis is sought—requires reassessment in terms of the increasing knowledge of the occurrence of pleiotropy, epistasis, and genetic heterogeneity. For example, broader genetic backgrounds of patients, including variants categorized as incidental, will likely explain why the same disease mutation may result in variable symptoms in distinct individuals. (v) Broader genomic testing creates a much greater need for clinical correlation than was necessary for conventional molecular testing. Adapting to this reality will require both careful collection of phenotypic information and use of a controlled vocabulary when genomic level–sequencing testing is ordered.
Third, genomic training programs must be designed for use in medical school curricula, residency training, and the reeducation of mature physicians. Health care providers will need better interpretive and communication skills regarding genetic information. Clearly, the numbers of clinical geneticists and genetic counselors are, and will continue to be, insufficient to serve as the sole providers of genomic medicine. Thus, a standard for genomic medicine certification for other sub-specialists is urgently needed, as are genomic medicine training tracks for physician assistants and nurse practitioners. In addition, as genomic medicine becomes the standard of care, the role of interpretation and reporting will likely expand to pathologists, who will also require education. Without such initiatives, genomic medicine lacks the infrastructure for broad deployment.
Finally, before clinical practice guidelines can be defined for NGS-based diagnosis, many questions must be answered: What are the analytical gold standards? What are the benefits and harms of using genomic information in health care, and how are these maximized and minimized, respectively? Which sets of disorders benefit from NGS-based diagnostic testing in terms of cost and improved outcomes? What are the implications for preconception carrier testing and neonatal screening? How can improved rates of ascertainment and earlier diagnoses be leveraged to reinvigorate clinical trials of new therapies for orphan disorders? Although NGS has only recently arrived in the clinic and shows great potential as a diagnostic tool, the technology has outpaced the modes of analysis. To remedy this imbalance moving forward will require thoughtful planning by clinical and laboratory geneticists, researchers, bioinformaticians, and ethicists.
Acknowledgments
We thank D. Dinwiddie, N. Miller, and S. Soden for their insights. This work was funded by the Beyond Batten Disease Foundation and Children’s Mercy Hospital. A deo lumen, ab amicis auxilium.
REFERENCES AND NOTES
- 1.McKusick-Nathans Institute of Genetic Medicine. Baltimore, MD: Johns Hopkins University; Online Mendelian Inheritance in Man, OMIM. http://www.ncbi.nlm.nih.gov/omim. [Google Scholar]
- 2.Lialiaris T, Mantadakis E, Kareli D, Mpountoukas P, Tsalkidis A, Chatzimichail A. Frequency of genetic diseases and health coverage of children requiring admission in a general pediatric clinic of northern Greece. Ital. J. Pediatr. 2010;36:9. doi: 10.1186/1824-7288-36-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa T, Scriver CR, Childs B, Opitz JM, Reynolds JF. The effect of Mendelian disease on human health: A measurement. Am. J. Med. Genet. 1985;21:231–242. doi: 10.1002/ajmg.1320210205. [DOI] [PubMed] [Google Scholar]
- 4.McCandless SE, Brunger JW, Cassidy SB. The burden of genetic disease on inpatient care in a children’s hospital. Am. J. Hum. Genet. 2004;74:121–127. doi: 10.1086/381053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maddalena A, Bale S, Das S, Grody W, Richards S. ACMG Laboratory Quality Assurance Committee, Technical standards and guidelines: Molecular genetic testing for ultra-rare disorders. Genet. Med. 2005;7:571–583. doi: 10.1097/01.gim.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- 6.American Society of Human Genetics Board of Directors-American College of Medical Genetics Board of Directors, Points to consider: Ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am. J. Hum. Genet. 1995;57:1233–1241. [PMC free article] [PubMed] [Google Scholar]
- 7.Zoccoli MA, Chan M, Erker JC, Ferreira-Gonzolas A, Lubin IM. NCCLS document MM9-A. Wayne, PA: NCCLS; 2004. Nucleic Acid sequencing methods in diagnostic laboratory medicine; Approved guideline. [ISBN 1-56238-558-5]. [Google Scholar]
- 8.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee, ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 9.Jensen K, Murray F. Intellectual property landscape of the human genome. Science. 2005;310:239–240. doi: 10.1126/science.1120014. [DOI] [PubMed] [Google Scholar]
- 10.Lupski JR, Reid JG, Gonzaga-Jauregui C, RioDeiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, Muzny DM, Hoang LD, Yousaf S, Lupski JR, Gibbs RA. Whole-genome sequencing for optimized patient management. Sci. Transl. Med. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wetterstrand KA. DNA sequencing costs: Data from the NHGRI large-scale genome sequencing program. www.genome.gov/sequencingcosts.
- 13.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, Gumbs CE, Pevsner J, Thomas G, Valle D, Hoover-Fong JE, Goldstein DB. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolze A, Byun M, McDonald D, Morgan NV, Abhyankar A, Premkumar L, Puel A, Bacon CM, Rieux-Laucat F, Pang K, Britland A, Abel L, Cant A, Maher ER, Riedl SJ, Hambleton S, Casanova JL. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Züchner S, Dallman J, Wen R, Beecham G, Naj A, Farooq A, Kohli MA, Whitehead PL, Hulme W, Konidari I, Edwards YJ, Cai G, Peter I, Seo D, Buxbaum JD, Haines JL, Blanton S, Young J, Alfonso E, Vance JM, Lam BL, Peričak-Vance MA. Whole-exome sequencing links a variant in DHDDS to retinitis pigmentosa. Am. J. Hum. Genet. 2011;88:201–206. doi: 10.1016/j.ajhg.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bilgüar K, Oztük AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoğlu D, Tüysüz B, Cağlayan AO, Gökben S, Kaymakçalan H, Barak T, Bakircioğlu M, Yasuno K, Ho W, Sanders S, Zhu Y, Yilmaz S, Dinçer A, Johnson MH, Bronen RA, Koçer N, Per H, Mane S, Pamir MN, Yalçinkaya C, Kumandaş S, Topçu M, Ozmen M, Sestan N, Lifton RP, State MW, Günel M. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, AbuRayyan A, Loulus S, Avraham KB, King MC, Kanaan M. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am. J. Hum. Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, Jiang H, Zhang P, Shen L, Guo JF, Li N, Li YR, Lei LF, Zhou J, Du J, Zhou YF, Pan Q, Wang J, Wang J, Li RQ, Tang BS. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 2010;133:3510–3518. doi: 10.1093/brain/awq323. [DOI] [PubMed] [Google Scholar]
- 20.Simpson MA, Irving MD, Asilmaz E, Gray MJ, Dafou D, Elmslie FV, Mansour S, Holder SE, Brain CE, Burton BK, Kim KH, Pauli RM, Aftimos S, Stewart H, Kim CA, Holder-Espinasse M, Robertson SP, Drake WM, Trembath RC. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 2011;43:303–305. doi: 10.1038/ng.779. [DOI] [PubMed] [Google Scholar]
- 21.Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable infl ammatory bowel disease. Genet. Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 22.Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloğlu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.çalışkan M, Chong JX, Uricchio L, Anderson R, Chen P, Sougnez C, Garimella K, Gabriel SB, dePristo MA, Shakir K, Matern D, Das S, Waggoner D, Nicolae DL, Ober C. Exome sequencing reveals a novel mutation for autosomal recessive non-syndromic mental retardation in the TECR gene on chromosome 19p13. Hum. Mol. Genet. 2011;20:1285–1289. doi: 10.1093/hmg/ddq569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D, Uziel G, Lamantea E, Invernizzi F, Poulton J, Rolinski B, Iuso A, Biskup S, Schmidt T, Mewes HW, Wittig I, Meitinger T, Zeviani M, Prokisch H. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat. Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 26.Kim JI, Ju YS, Park H, Kim S, Lee S, Yi JH, Mudge J, Miller NA, Hong D, Bell CJ, Kim HS, Chung IS, Lee WC, Lee JS, Seo SH, Yun JY, Woo HN, Lee H, Suh D, Lee S, Kim HJ, Yavartanoo M, Kwak M, Zheng Y, Lee MK, Park H, Kim JY, Gokcumen O, Mills RE, Zaranek AW, Thakuria J, Wu X, Kim RW, Huntley JJ, Luo S, Schroth GP, Wu TD, Kim H, Yang KS, Park WY, Kim H, Church GM, Lee C, Kingsmore SF, Seo JS. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Public Law 100–578, the Clinical Laboratory Improvement Amendment of 1988 [CLIA 1988].
- 28.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, Bamshad M, Nickerson DA, Shendure J. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krawitz PM, Schweiger MR, Röelsperger C, Marcelis C, Kölsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, Isau M, Fischer A, Dahl A, Kerick M, Hecht J, Köler S, Jäger M, Grünhagen J, deCondor BJ, Doelken S, Brunner HG, Meinecke P, Passarge E, Thompson MD, Cole DE, Horn D, Roscioli T, Mundlos S, Robinson PN. Identity-by-descent filtering of exome sequence data identifi es PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat. Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 30.GeneTests: Medical Genetics Information Resource (database online) Seattle: University of Washington; 1993–2011. Copyright, http://www.genetests.org. [Google Scholar]
- 31.Kohonen-Corish MR, Al-Aama JY, Auerbach AD, Axton M, Barash CI, Bernstein I, Béroud C, Burn J, Cunningham F, Cutting GR, denDunnen JT, Greenblatt MS, Kaput J, Katz M, Lindblom A, Macrae F, Maglott D, Möslein G, Povey S, Ramesar R, Richards S, Seminara D, Sobrido MJ, Tavtigian S, Taylor G, Vihinen M, Winship I, Cotton RG. Human Variome Project Meeting, How to catch all those mutations—The report of the third Human Variome Project Meeting, UNESCO Paris, May 2010. Hum. Mutat. 2010;31:1374–1381. doi: 10.1002/humu.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]