Abstract
The pleckstrin homology (PH) domain of the general receptor of phosphoinositides 1 (GRP1) protein selectively binds to a rare signaling phospholipid, phosphatidylinositol (3,4,5)-trisphosphate (PIP3), in the membrane. The specific PIP3 lipid docking of GRP1 PH domain is essential to protein cellular function and is believed to occur in a stepwise process, electrostatic-driven membrane association followed by the specific PIP3 binding. By a combination of all-atom molecular dynamics (MD) simulations, coarse-grained analysis, electron paramagnetic resonance (EPR) membrane docking geometry, and fluorescence resonance energy transfer (FRET) kinetic studies, we have investigated the search and bind process in the GRP1 PH domain at the molecular scale. We simulated the two membrane binding states of the GRP1 PH domain in the PIP3 search process, before and after the GRP1 PH domain docks with the PIP3 lipid. Our results suggest that the background anionic phosphatidylserine lipids, which constitute around one-fifth of the membrane by composition, play a critical role in the initial stages of recruiting protein to the membrane surface through non-specific electrostatic interactions. Our data also reveal a previously unseen transient membrane association mechanism that is proposed to enable a two-dimensional “hopping” search of the membrane surface for the rare PIP3 target lipid. We further modeled the PIP3-bound membrane–protein system using the EPR membrane docking structure for the MD simulations, quantitatively validating the EPR membrane docking structure and augmenting our understanding of the binding interface with atomic-level detail. Several observations and hypotheses reached from our MD simulations are also supported by experimental kinetic studies.
Keywords: pleckstrin homology domain; phosphatidylinositol-3,4,5-trisphosphate or PIP3; anionic lipids; peripheral membrane protein; membrane targeting
Introduction
The cytoplasmic surface of the plasma membrane serves as a platform for many critical cellular signaling processes. In the case of membrane targeting processes, a wide array of signaling events are involved, both at the membrane level and at the protein level, such that signaling proteins are able to respond to second messenger signals in a spatially and temporally precise manner. Pleckstrin homology (PH) domains are regulated membrane targeting motifs found in a wide array of signaling proteins.1–3 They are recruited from the cytoplasm to the plasma membrane during the transient appearance of the signaling lipid phosphatidylinositol (3,4,5)-trisphosphate (PIP3). Many cellular processes such as cell growth, DNA synthesis, cytoskeletal rearrangements, vesicle trafficking, and apoptosis are regulated via PIP3-stimulated recruitment of PH-domain-containing signaling proteins.4–15 Moreover, it has been demonstrated that loss of PIP3 specificity due to PH domain mutations that trigger constitutive membrane binding is linked to the development of disease, including human cancers.16,17 In the present study, we focus on the membrane interactions and PIP3 docking process of the PH domain of general receptor of phosphoinositides 1 (GRP1).
The GRP1 PH domain is used as a representative model system for the current study. The GRP1 PH domain is made up of approximately 120 amino acid residues with a C-terminal α-helix and seven β-sheets. Two β-strands are created from the seven β-sheets and form a β-sandwich as a core structure. There are flexible loops between some of the β-sheets that actively participate in the PIP3-binding process. Structurally conserved PH domains are widely found as membrane targeting motifs that translocate the host protein to the membrane surface and perform various biological functions. More than 560 human proteins containing PH domains have been found, and many of them are believed to selectively bind to PIP lipids.18
The PIP3 lipids belong to an important class of second messenger lipids that are generated from the constitutive plasma membrane lipid, phosphatidylinositol (4,5)-bisphosphate (PIP2), by the phosphatidylinositol 3-kinase family of lipid kinases.19 Upon activation of phosphatidylinositol 3-kinase, transient increase of the PIP3 level is capable of recruiting the PH-domain-containing signaling proteins that selectively bind to the PIP3 lipids. PIP3 is a rare lipid even during a peak signaling event, providing a challenge for rapid PH domain recruitment. Previous studies have shown that the rate of PIP3 targeted by the GRP1 PH domain is increased up to 12-fold by an electrostatic search process that requires native levels of anionic lipids, primarily phosphatidylserine (PS) lipids, in the target bilayer.13,20 Single-molecule studies of GRP1 PH domain bound to PIP3 on supported bilayers show that the protein–PIP3 complex possesses a long bound state lifetime, enabling extended biological activity on the membrane surface, and exhibits the same two-dimensional diffusion rate as a single bulk lipid enabling rapid collisions with membrane-bound substrates. It follows that the friction of the complex against the viscous bilayer is defined mainly by the bound lipid molecule.
The molecular basis of the electrostatic search mechanism that speeds capture of the rare PIP3 lipid on the target membrane surface and the molecular details of the stable PIP3-bound state both remain elusive. Although high-resolution structural data of the GRP1 PH domain [Protein Data Bank (PDB) ID: 1FGY] have provided significant insight into the molecular basis of PIP3 head group binding,21 a detailed molecular-level understanding of the membrane interaction events prior to PIP3 binding is still in its formative stages. Moreover, in the PIP3-bound state, the constraints imposed by the surrounding bilayer on the PH domain–PIP3 complex are different from the constraints imposed by the aqueous crystal on the PH domain–IP4 complex. Hence, a different approach is needed to shed light on the electrostatic search mechanism and provide a deeper molecular understanding of the PH domain bound to its target PIP3 lipid on the bilayer surface.
Atomistic molecular dynamics (MD) simulation is a powerful tool that can provide quantitative information at atomic-level resolution and can be used to capture the details of membrane–protein binding interfaces. By virtue of the viscous nature of membrane environments, it generally takes longer simulation times to equilibrate lipid–protein interactions. It is thus important to carefully select the initial configuration for atomistic MD simulation of membrane–protein systems. An EPR membrane docking geometry provides an ideal starting point for such simulations. EPR membrane docking geometries have been solved for several important peripheral signaling proteins.22–26 The docking geometry obtained from this approach is generally quite accurate and is obtained with minimal perturbation of the native membrane complex.27 Very recently, a GRP1 PH domain EPR membrane docking geometry was determined for GRP1 PH domain bound to its target PIP3 lipid on a PC/PS/PIP3 bilayer.28 Two important features of the PH domain are highlighted by this docking geometry. First, GPR1 PH domain docks in a more a shallow position on the bilayer than the C2 domains previously studied,22–26 such that the PH domain exhibits minimal penetration into the lipid phase.28 Second, the EPR docking structure suggests that the orientation of the PIP3 head group is tilted away from its equilibrium orientation with respect to the membrane surface.28 Despite the rich information available from fluorescence resonance energy transfer (FRET), single-molecule diffusion, and EPR experiments, further studies of the electrostatic search process and the molecular nature of the PH domain–PIP3 complex are needed.
In this article, we investigate two membrane-interaction states to mimic the scenarios before and after PH domain docking to the PIP3 lipids. First, we carried out atomistic MD simulations to reveal the molecular events that occur during the electrostatic search process employed by the PH domain during its hunt for the rare PIP3 target, prior to PIP3 acquisition. In this system, PIP3 is not included in membrane. Our MD data show that the PS lipids direct the GPR1 PH domain to the membrane surface via long-ranged electrostatic rotational and translational steering, as suggested by previous experimental and computational studies.20,29 Interestingly, we observe that besides this longer-ranged electrostatic steering, specific positively charged protein side chains contact the PS lipid at close range. These molecular-level details are critical in explaining the transient association that takes place between the protein and non-PIP3 lipids before the PIP3 docking and are difficult to obtain experimentally because the lifetimes of these events are short.
In order to provide atomistic details of the membrane binding states of the GRP1 PH domain after docking to the PIP3 lipid, we also performed MD simulations with a single GRP1 PH domain bound to a PC/PS/PIP3 target lipid bilayer in a fully solvated system. The EPR membrane docking geometry of the PH domain bound to PIP3 on a PC/PS/PIP3 bilayer was used as the starting point for MD simulations.28 Our MD model quantitatively validates the EPR membrane docking geometry and the simulations reveal several important features of the membrane–protein binding interface. The dynamic behavior of the PIP3 lipid in the binding pocket, which is difficult to observe experimentally, is also presented.
Finally, we employ dipole moment analysis and use a reduced representation to describe the electrostatic character of the protein structure and membrane-interacting loops based on atomistic MD trajectories. This coarse-grained (CG) dipole analysis provides insights to the stepwise membrane associations of the GRP1 PH domain. The implication of the CG analysis for future CG model development is also briefly discussed.
Overall, combining the strengths of EPR and atomistic MD simulations as well as CG dipole moment analysis provides an unprecedented molecular-level picture of the association of GRP1 PH domain with the target membranes. Through the examination of the GRP1 PH domain in different bilayers and with starting configurations at different distances away from the membrane, we dissect the key molecular events that occur during the approach and binding of the PH domain to its rare PIP3 target lipid on the bilayer surface. Our findings provide valuable insights into the regulated membrane targeting of GRP1 PH and other PH domains. Importantly, several observations and hypotheses on the basis of MD simulations are further supported by experimental kinetic studies of site-directed mutants.
Results and Discussion
Electrostatic search of a target membrane surface for rare PIP3
We carried out all-atom MD simulations of a GRP1 PH domain with a PC/PS bilayer to gain atomic-level insights into the early stages of PH domain– membrane interactions prior to PIP3 acquisition. The PH domain is attracted to the membrane surface predominantly by non-specific electrostatic interactions with the background anionic lipids, primarily PS. Once near the anionic membrane, the basic lipid binding face of the PH domain is believed to undergo rapid, transient collisions with the membrane surface before docking to the target PIP3 lipid in a stereospecific manner.20 During these rapid collisions, any binding is fleeting since no equilibrium membrane binding is detectable in the absence of target lipid PIP3, and such binding would defeat the regulated recruitment function of the PIP3 signal. In this section, we discuss the results from our simulations that examine the role of the PS lipids in the GRP1 PH domain electrostatic search process. Experiments have shown that there is a 10-fold increase in the binding affinity between PIP3 and the GRP1 PH domain in the presence of PS lipids.13 It is also seen that this enhancement is mainly due to the increase of the on-rate kinetics of PIP3 binding. This provides evidence that PS lipids are important to the PIP3 electrostatic search process. However, the molecular events that occur during this search remain unclear. By choosing to explore the membrane-affinity behavior of distant PH domain on a PC/PS bilayer, we are able to delineate the key events prior to PIP3 acquisition.
MD simulations of GRP1 PH domain binding with a PC/PS bilayer
Membrane binding mechanism of the GRP1 PH domain in a PC/PS bilayer
Using all-atom MD simulations of a fully solvated PH domain with PC/ PS bilayer systems, we generated four independent 250-ns MD simulations. Figure 1 shows that the PIP3 head group binding pocket of the PH domain is a highly positively charged region. This membrane-binding surface of the GRP1 PH domain is set to face the membrane surface at the beginning of all four MD simulation systems. As shown in Fig. 2a, two of these systems (F1, F2) start off with a separation of at least 20 Å between the closest point on the protein and the membrane phosphate plane. The other two systems (C1, C2) begin with a separation of at least 10 Å from the membrane phosphate plane. Figure 2b shows the position of the V278 Cα atom with respect to the membrane phosphate plane over the course of four trajectories. It shows that V278 binds to the membrane surface and stabilizes at the membrane phosphate plane depth in all four trajectories.
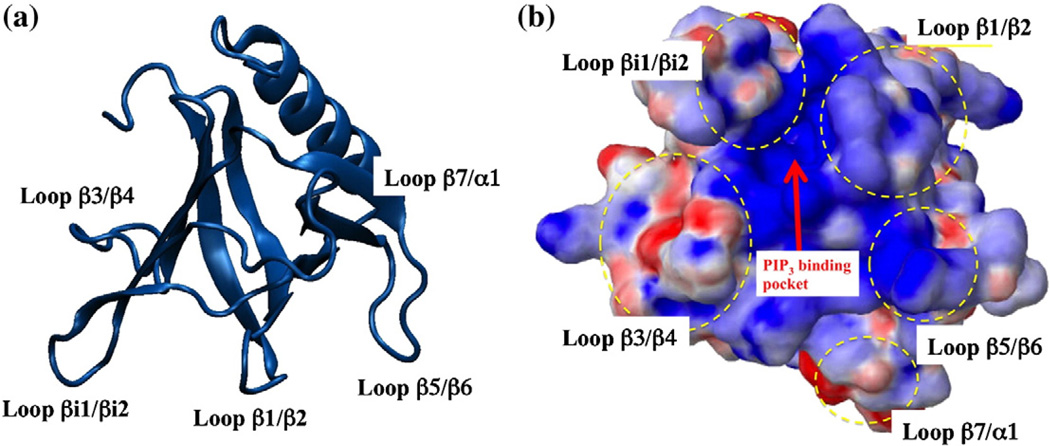
Fig. 1.
Crystal structure and electrostatic potential map of the GRP1 PH domain: (a) The structure of the PH domain with the four membrane-interacting loops and the conserved β-strands along with an α-helix. (b) Molecular surface of the GRP1 PH domain and electrostatic potential map of the membrane-binding surface. The surface is colored according to the electrostatic potential calculated from the APBS software.30 Blue denotes positive potential, and red indicates the negative potential.
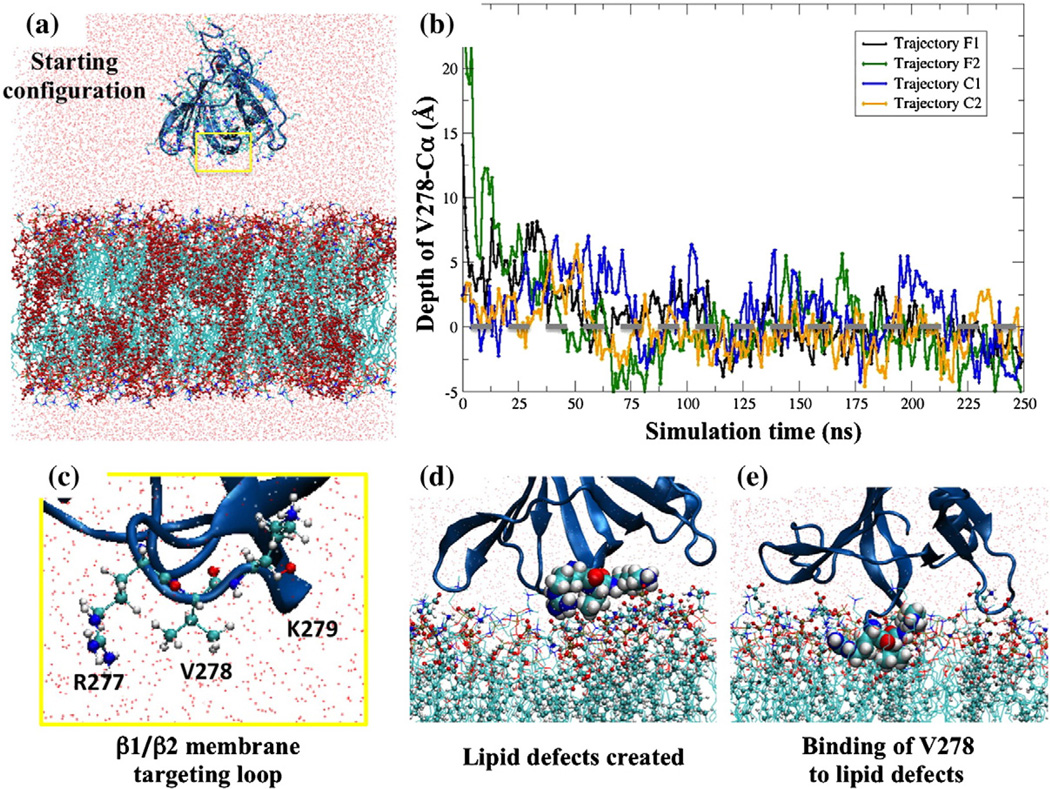
Fig. 2.
Membrane binding of the GRP1 PH domain to a PC/PS membrane: (a) Initial configuration of the MD simulation for the F1 system. (b) Cα positions of V278 with respect to membrane phosphate plane (gray broken line) over the course of the simulations. The starting distances of closest points of the PH domain for F1 and F2 systems are more than 20 Å from the lipid bilayer phosphate plane. The starting distances for the C1 and C2 systems were kept within 10 Å from the lipid bilayer phosphate plane. (c) Close-up view of loop β1/β2 in (a). (d) and (e) are snapshots of the open-and-bind molecular membrane targeting mechanism.
Interestingly, we observe from these trajectories that the residues R277, V278, and K279 of loop β1/β2 play an essential role in initiating the binding of V278 to the membrane surface (Fig. 2c). We observe that there are two coordinated interactions taking place, one electrostatic and the other hydro-phobic in nature, which cause the PH domain to bind to the membrane in a shallow manner and stay on the surface for an extended period. The electrostatic interactions are between the positively charged R277 and K279 residues on the β1/β2 loop in the protein and negatively charged phosphate groups or PS lipid head groups in the bilayer. In addition, the hydrophobic interactions take place between V278 on the β1/β2 loop in the protein and the lipid defects that are created due to exposure of the hydrophobic tails in the bilayer coming from of the aforementioned electrostatic interactions. When the GRP1 PH domain approaches the membrane surface, the positively charged regions of R277 and K279 residues have an affinity for the negatively charged PS lipid head groups or phosphate groups of lipids. This perturbs the membrane locally, and transient lipid defects are created as shown in Fig. 2d. The binding of hydrophobic residue V278 to these short-lived lipid defects stabilizes the GRP1 PH domain at the membrane surface (Fig. 2e). The resulting “open-and-bind” mechanism that requires coordinated interactions between protein side chains and lipids at the atomistic scale is thus hypothesized to generate a transient membrane binding event that rapidly dissociates, enabling a “hopping”-type search of the PS-containing membrane surface for the rare PIP3 target lipid.
Effect of PS lipids on the electrostatic steering of GRP1 PH domain towards the membrane
Our simulations reveal that PS lipids in membrane significantly affect the membrane binding of the GRP1 PH domain. In Fig. 2b, we show that in the presence of PS lipids, the most positively charged side of protein surface faces the membrane surface throughout the duration of all four simulations and eventually attaches to the membrane in that orientation. We further examine the relative protein dipole moment orientation with respect to the membrane normal. Figure 3a shows that in the F1, F2, C1, and C2 systems, the overall protein dipole moment points towards the membrane surface (angle of ~150° with membrane normal), and the protein dipole moment orientations are concentrated in a narrow zone, suggesting the favorable binding orientation of protein in the membrane. It should be qualified that some additional fluctuation in angles might be observed with a longer timescale simulation. This is especially true for the twist angle (the protein's ability to rotate in a cone shape around an orientation) rather than the tilt angle, which is influenced by the dipole interactions. The timescale of the simulation (150 ns) may be attributed to the lack of higher fluctuations in twist angles for Fig. 3a.
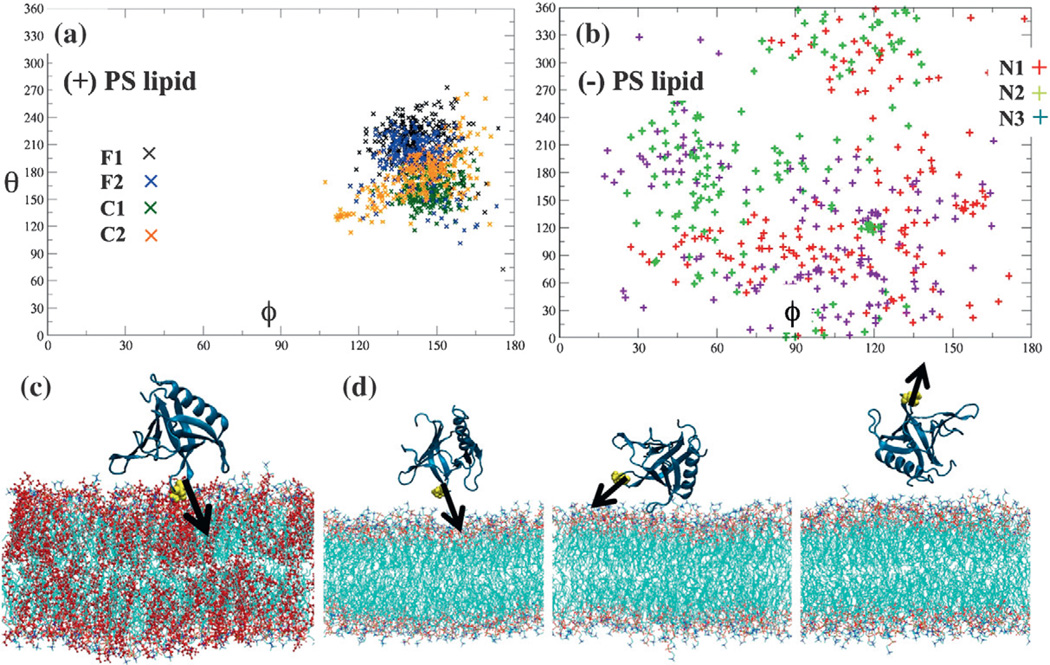
Fig. 3.
Electrostatic steering of the GRP1 PH domain. (a) The orientation of the dipole moment of the protein structure is computed and converted into spherical coordinates with respect to the membrane normal. Data from four different sets of simulations for a PH domain approaching and binding a PC/PS membrane are shown. (b) Same analyses are also applied to three control simulations of a PH domain approaching a pure PC membrane. (c) A snapshot of the initial membrane binding of the PH domain in a PC/PS membrane. PS lipids are shown in red. The V278 residue is shown as yellow spheres. Black arrow indicates protein dipole moment orientation. (d) In the absence of PS lipids in the membrane, these three snapshots show that the GRP1 PH domain is unable to quickly bind to the membrane via an electrostatic steering process (left to right) but instead bounces off the bilayer. The three snapshots (from left to right) are taken from the N1 trajectory at intervals of 0 ns (initial configuration), 10 ns, and 20 ns, respectively.
As a control, we carried out three independent simulations for more than 150 ns in the target bilayer with no PS lipids. The protein dipole moment orientations are also examined for these three control simulations. Strikingly, in the absence of PS lipids, the protein dipole moment orientations sample much larger configuration space with respect to the membrane normal (Fig. 3b). This is greatly different from the systems of PH domain bound to a PC/PS membrane where the protein dipole moment constantly points towards membrane surface in a stable manner (Fig. 3c). In the absence of PS lipids in the membrane, the GRP1 PH domain failed to bind to the membrane in a stable manner in all the three trajectories. The protein was unable to properly associate with the membrane surface for even a few tens of nanoseconds. Instead, we observe that the GRP1 PH domain bounces off the membrane surface (Fig. 3d). It should be noted that the PH domain in two out of three control simulations completely dissociates from the membrane surface within a 150-ns simulation time. Similarly, no stable membrane-binding conformation of protein was found under these conditions.
There are two likely explanations for the failure of the GRP1 PH domain to bind to the membrane in the absence of PS lipids. First, the positive R277 and K279 residues of the β1/β2 loop have lower probability to form stable interactions with the membrane surface without the negatively charged PS lipids. Consequently, the open-and-bind mechanism that lets the hydrophobic V278 residue bind to the lipid defects has a lower chance to take place. Second, the translational and rotational electrostatic steering effect on the PH domain disappears in the absence of PS lipids. It has been previously suggested, both experimentally and computationally, that the abundant background anionic lipids are sufficient to attract the GRP1 PH domain near the membrane and direct the PIP3 binding face of the GRP1 PH domain towards the membrane surface.20,31 The removal of PS lipids in the membrane causes the loss of translational and rotational electrostatic steering and significantly reduces the probability of the GPR1 PH domain from making contacts with the membrane surface by PIP3 binding face of protein surface. It should be noted that the observations from control simulations are not used to claim that the GRP1 PH domain is completely incapable of undergoing the open-and-bind mechanism without PS lipids. Instead, our MD simulations suggest that both the on-rate and residence time of the GRP1 PH domain protein on the membrane increase in the presence of charged PS lipids, prior to binding PIP3.
Ensemble FRET and single-molecule TIRF studies of the GRP1 PH domain detect no stable PH domain binding to PC/PS membranes, indicating that the bound state lifetime is much shorter than 20 ms.13 The stable PH domain binding to a PC/PS membrane during our simulations is fully compatible with this experimental result since the simulations are sub-microsecond. If the simulations could be extended for much longer timescales, the experimental findings suggest that the typical PH domain would dissociate long before the simulation reaches the millisecond regime, since membrane binding in the absence of PIP3 must be transient. However, this transient bilayer-associated state could contribute to the electrostatic search process in two ways: (1) by allowing transient two-dimensional searches of the membrane surface, thereby increasing the probability that a given membrane binding event will yield PIP3 encounter and binding, and (2) by increasing the local concentration of PH domain near the membrane surface due to buffered diffusion. MD simulations of the PH domain approaching PC/PS membranes thus provide atomic-scale insight into the interactions that are experimentally inaccessible and reveal previously undetected transient interactions hypothesized to play a central role in the electrostatic search process needed to rapidly bind the rare target PIP3.
Experimental kinetic studies testing the predictions of the PC/PS simulation
Previous experiments have reported that the presence of PS in the target bilayers significantly increases the on-rate for PH domain binding to PIP3 binding.13 It is important to investigate the role of PS lipids in the membrane sensing mechanism of the PH domain and eventual specific binding of the protein to the PIP3 lipids in the membrane. Using stopped-flow kinetic studies to quantitate membrane on- and off-rates via protein-to-membrane FRET, we investigated how certain positively charged residues (Lys or Arg) affect the ability of GRP1 PH to carry out the PS-enhanced electrostatic search for rare PIP3.
Table 1 summarizes stopped-flow kinetic studies using protein-to-membrane FRET to monitor the efficiency of the electrostatic search process for the WT GRP1 PH domain and the three mutants. The electrostatic search enabled by physiological levels of anionic PS lipids has been previously observed to significantly increase the PIP3 on-rate. This on-rate enhancement can be used to test for protein side chains that are important to the electrostatic search process on the PC/PS regions. Three mutations were selected for the study: K279A of loop β1/β2, K302A of loop β3/β4, and R349A of loop βi1/βi2. One of these mutations, K279A of loop β1/β2, removes the K279 positive charge proposed to be important for interaction with the PC/PS regions during the electrostatic search. The other two mutations, K302A of loop β3/β4 and R349A of loop βi1/βi2, are negative controls in this experiment since their native side chains are not predicted to play a key role in initiating the membrane binding of the PH domain. Table 1 shows that, for the WT PH domain, replacement of PS with PC in the simple PC/PS/PIP3 mixture yields a 3.4-fold decrease in the on-rate for stable binding to rare PIP3, reaffirming the previously observed importance of PS to the electrostatic search process and its on-rate enhancement. (This observed 3.4-fold decrease is within error of that previously observed for the same simple lipid mixtures. A larger decrease, up to 12-fold, is observed when PS is replaced in a more physiological lipid mixture, but here the simple lipid mixtures were the appropriate choice to match the mixtures employed in MD simulations.) Notably, on the PC/PS/PIP3 target membrane, the K279A mutation generates a 1.9-fold slowing of the PIP3 on-rate, while the K302A and R349A mutations yield no such slowing. These experimental findings support the importance of K279 in the electrostatic search process as predicted by the MD simulations of the PH domain interaction with the PC/PS bilayer.
Table 1.
Kinetic parameters and dissociation constants for GRP1 PH domain binding to membranes containing PIP3 lipid
| GRP1 PH domain | Membrane composition | kon (s−1 µM−1) | koff (s−1) | Kd = koff/kon (nM) |
|---|---|---|---|---|
| WT | PC/PS/dansyl-PE/PIP3 | 2.04832 | 0.14892 | 72.7 |
| PC/dansyl-PE/PIP3 | 0.5874 | 0.28296 | 481.7 | |
| K279A | PC/PS/dansyl-PE/PIP3 | 1.05984 | 0.48702 | 459.5 |
| K302A | PC/PS/dansyl-PE/PIP3 | 2.33356 | 0.19492 | 83.5 |
| R349A | PC/PS/dansyl-PE/PIP3 | 2.29164 | 0.56013 | 244.4 |
Errors in these on- and off-rates is ±10%.
Stable docking of the GRP1 PH domain to PIP3 on the target PC/PS/PIP3 bilayer
PH domain membrane docking geometry
Once the PH domain encounters a rare PIP3 lipid on the target membrane surface, it can bind to the PIP3 with high affinity and remain in this membrane-docked state for multiple seconds while it diffuses in the plane of the membrane.20 The docking geometry of the GRP1 PH domain on the bilayer on the basis of the EPR-guided MD simulation is characterized both by the angle and by the z-distance of the protein relative to the membrane phosphate plane. Both measures are discussed in the next two subsections. Moreover, atomistic details of the binding between the PIP3 head group and the protein pocket are also discussed.
Orientation configuration of the GRP1 PH domain
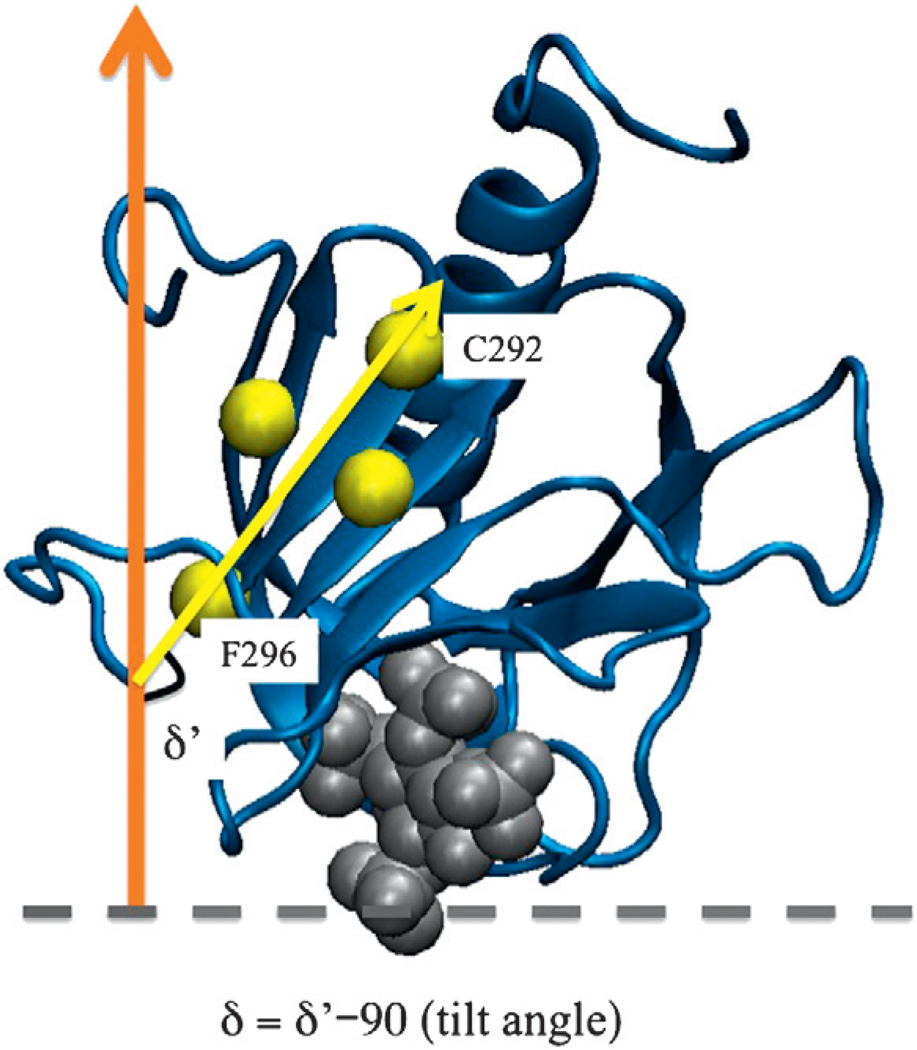
A vector associated with the protein structure was used to quantify the membrane docking geometry of the GRP1 PH domain. The EPR membrane docking geometry of the GRP1 PH domain was quantified by a vector between the Cα of C292 and F296 that represents the long axis of the core β-sandwich (Fig. 4) and the protein tilt angle is defined as the angle between this vector and the plane of the membrane. In the optimized EPR docking geometry, this vector is tilted away from the membrane surface at an angle of 46 ± 7°.28 Based on the calculations carried out from the last 250 ns of the 300-ns MD trajectory, the GRP1 PH domain membrane docking geometry exhibits a tilt angle of 45.7 ± 2.9°. The tilt angle computed from EPR-guided MD simulation is therefore consistent with the EPR membrane docking geometry (46 ± 7°), although large fluctuations are observed during the simulation. The large fluctuation in protein tilt angle may be attributed to the GRP1 PH domain being a shallow membrane-penetrating protein where local interactions between protein side chains and lipid head groups affect the membrane binding structure of the entire domain, especially when the target bilayer is heterogeneous in nature. As a control, we also performed an MD simulation with a different initial membrane docking geometry of the GRP1 PH domain bound to the membrane. Details of simulation setup can be found in Materials and Methods. After about 100 to 150 ns, we found that the GRP1 PH domain re-orientated back to the equilibrium membrane binding geometry.
Fig. 4.
The definition of the tilt angle (δ) for membrane docking geometry of the GRP1 PH domain. The gray broken line indicates the average location of the membrane phosphate plane. z-axis is indicated in an orange arrow.
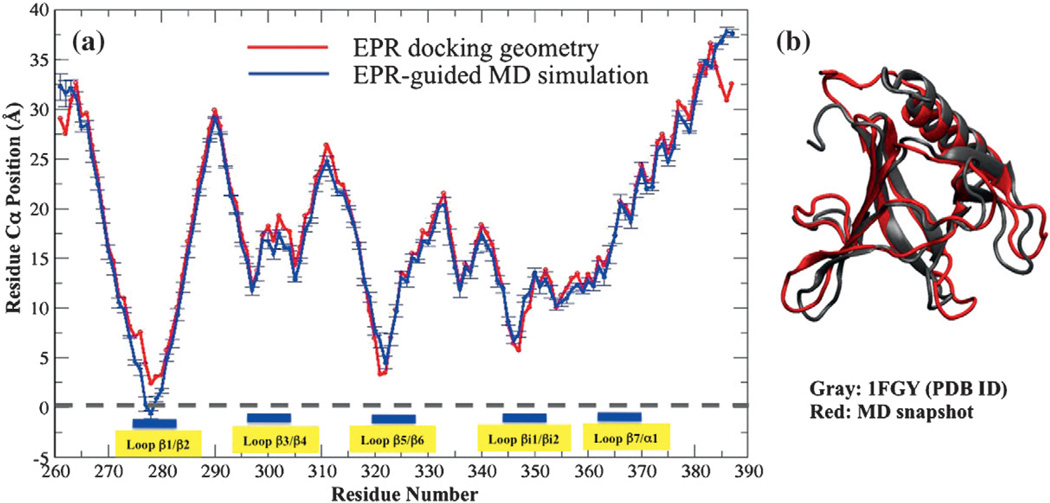
Residue-by-residue Cα positions of the GRP1 PH domain in EPR structure and MD-averaged structure
In the EPR-guided MD simulation, the residue-by-residue averaged heights of Cα positions with respect to the membrane phosphate plane are computed. The relative heights of backbone Cα atoms along the protein sequence are quite similar in the EPR-guided MD simulation and the EPR-deter-mined docking structure (Fig. 5a). Importantly, this observation shows that combined EPR membrane docking geometry and MD simulations is a powerful tool to investigate the details of the protein– membrane binding interface. The MD simulation also agrees with the EPR docking structure showing that the GRP1 PH domain does not penetrate deeply into the hydrocarbon region of the lipid bilayer. In the EPR membrane docking structure, residue V278 is the closest point of the GRP1 PH domain to the membrane surface, and its Cα is located at 2.4 ± 2.6 Å with respect to the lipid bilayer phosphate plane. The MD simulations also confirm that the location of Cα of V278 is the lowest site of the binding structure from simulation with the averaged depth −0.7 ± 0.4 Å. It is evident from Fig. 5a that there are some deviations between the MD structure and the EPR structure, especially in the loop regions. The MD structure is very dynamic and the loops make adjustments upon membrane docking, which can explain the deviation between the EPR membrane docking structure and the membrane-bound MD structure. We confirm this observation by showing an overlay structure for an MD snapshot and 1FGY crystal structure in Fig. 5b. The representative MD snapshot clearly shows deviations from the crystal structure in loop regions.
Fig. 5.
(a) Residue-by-residue Cα positions of the GRP1 PH domain. The red line is computed from the EPR docking geometry of the GRP1 PH domain bound to the POPC/POPS/PIP3 target bilayer, and the lowest point of the black line is shifted to V278 (2.4 Å). The blue line was computed by using the last 250 ns of EPR-guided MD simulation of a GRP1 PH domain bound to the POPC/POPS/PIP3 target bilayer. The gray broken line represents a membrane phosphate plane computed from EPR-guided MD simulations. (b) A comparison of the MD simulation structure (red) with the x-ray structure (gray) of the GRP1 PH domain.
Number of contacts between membrane-interacting loops and the bilayer
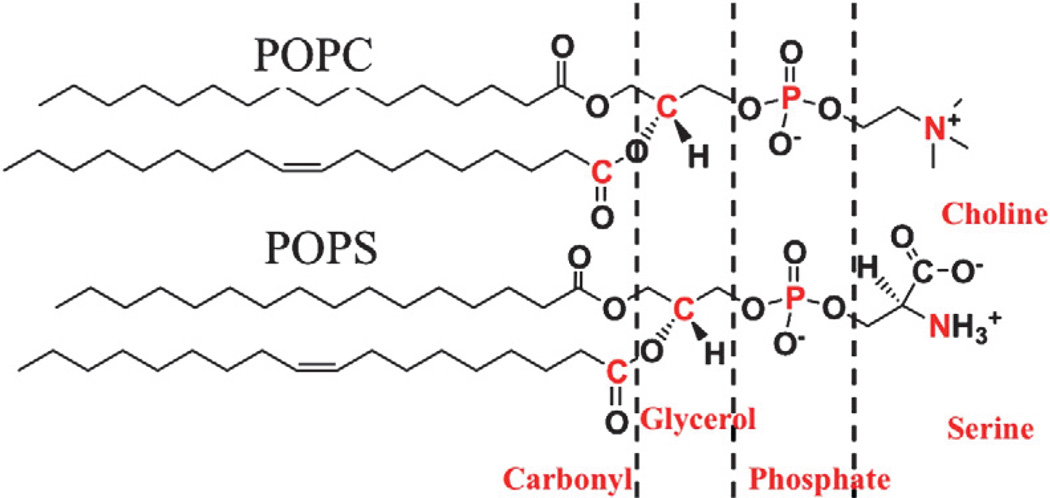
Interactions between extended loops of the GRP1 PH domain and the membrane surface are important to membrane associations of the GRP1 PH domain. To examine the binding interface, we computed the number of contacts between four membrane-interacting loops and the lipid head group parts. The number of contacts between four membrane-interacting loops on the protein and the four different lipid parts (choline or serine, phosphate, glycerol, and carbonyl groups; Fig. 6) is computed and shown in Table 2. Four selected lipid parts are located at the interfacial region of the bulk solvent and the lipid tail groups. The choline (or serine) and the phosphate groups are solvent exposed, while the glycerol and the carbonyl groups are relatively closer to the hydrophobic core of the membrane. We show that the four loops in the protein make limited or no contacts with the glycerol and the carbonyl groups. This agrees with the EPR membrane binding structure in which the GRP1 PH domain binds to the target bilayer surface with limited penetration.
Fig. 6.
The atoms from four groups of the lipid (choline/serine, phosphate, glycerol and carbonyl) that are used to measure contacts between the membrane-interacting loops on the PH domain and the bilayer.
Table 2.
Time-averaged number of contacts between four membrane-interacting loops of GRP1 PH domain and the lipid head group parts within 5 Å
| Bilayer | Loop β1/β2 | Loop β3/β4 | Loop β5/β6 | Loop βi1/βi2 | |
|---|---|---|---|---|---|
| PC/PS/PIP3 | Choline or serine | 3.7 (0.6) | 0.6 (0.2) | 2.5 (0.3) | 3.2 (0.2) |
| Phosphate | 5.1 (0.2) | 0.2 (0.1) | 2.1 (0.2) | 1.7 (0.3) | |
| Glycerol | 3.8 (0.4) | 0 | 0.8 (0.2) | 0 | |
| Carbonyl | 2.3 (0.3) | 0 | 0.3 (0.1) | 0 |
Atoms selected to represent lipid head group parts were colored red, as shown in Fig. 6. The last 250 ns of the MD trajectory was used to compute the averaged number of contacts, and the errors estimated from MD trajectory are shown in parentheses. The following are the residues selected to represent four membrane-interacting loops: extended loop β1/β2 region: residues 275 to 283; loop β3/β4: residues 297 to 304; loop β5/β6: residues 319 to 324; loop βi1/βi2 loop: residues 344 to 349.
Electrostatic interactions between loop side chains and lipid head groups may be important to the direct contacts and the binding structure of the GRP1 PH domain at the interface. Figure 1 clearly shows that the loop β1/β2, loop β5/β6, and loop βi1/βi2 regions are more positively charged than loop β3/β4. These positively charged regions should have favorable interaction with the anionic bilayer and lead to a larger number of direct contacts. Table 2 supports this hypothesis and shows that loop β1/β2, loop β5/ β6, and loop βi1/βi2 have the highest contacts with the lipid head groups, especially making contacts with the solvent-exposed choline (or serine) and phosphate regions.
Specific binding and orientation of the PIP3 head group in the GRP1 PH binding pocket
PIP3 binding of the GRP1 PH domain regulates many cellular processes and is executed with a high level of spatial and temporal precision in cells. It is suggested that the binding between the GRP1 PH domain and PIP3 is highly specific, exclusive, and strong. Directly accessing the details of the tightly bound PH domain–PIP3 can advance our understanding of the PIP3-triggered membrane docking process to a large extent. However, it is experimentally difficult to examine the dynamics of the PIP3 head group in the binding pocket of the PH domain. EPR-guided MD simulations can provide details of the protein–PIP3 binding interactions and the protein–lipid configuration in the binding pocket.
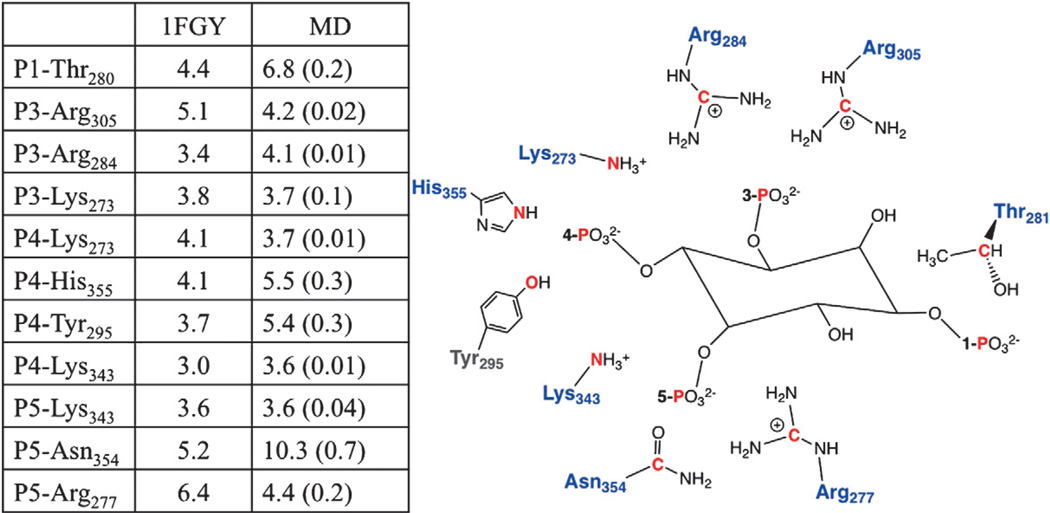
MD simulations were used to reveal atomistic details of the how PIP3 head group binds to the side chains in the protein pocket. We compare the selected side chain–phosphate group distances between the crystal structure (PDB ID: 1FGY) and MD averages shown in Fig. 7. The MD data show that the head group of the PIP3 lipid is stable in the binding pocket, as indicated by the specific interactions between positively charged residues and the 3-, 4-, and 5-phosphate groups of the PIP3 head group. The specific binding between side chains and phosphate groups of the PIP3 head group resembles the contacts observed in the crystal structure. Figure 7 clearly shows that close contacts between the 3- and 4-phosphate group and the corresponding side chains are preserved, with very little fluctuation. It is also seen that the distance between the 5-phosphate group and N354 equilibrates at a longer distance than in the crystal structure, indicating that no stable contacts are formed during the course of the simulations.
Fig. 7.
Comparison of selected distances for the crystal structure (PDB ID: 1FGY) and MD averages between selected side chains in the binding pocket and phosphate groups of the PIP3 head group. Atoms selected to represent phosphate groups of PIP3 head group and selected side chains are colored red. The last 250 ns of MD trajectory was used to compute averaged distances, and errors estimated from the MD trajectory are shown in parentheses.
It has been suggested based on the EPR membrane docking structure that the average orientation of the PIP3 head group with respect to membrane surface changes upon binding with the GRP1 PH domain.28 Here, we use MD simulations to gain insight into the atomic-level details of this binding. To that end, we compute the orientation of the PIP3 head group in the binding pocket with respect to the membrane surface. In our simulations, the PIP3 orientation is defined in the same way as in Li et al.32 The PIP3 orientation is quantified by calculating polar and azimuthal angles of the lipid head group with respect to the membrane surface. The lipid tilt angle is complementary to the polar angle and the twist angle is the same as the azimuthal angle. It is to be noted that the though lipid tilt angle and protein tilt angle may appear to be defined differently, essentially it is the complementary angle to the polar angle of the spherical coordinate system, defined with reference to membrane normal as the z-axis and membrane plane as the xy plane. Our MD simulations show that the PIP3 head group has a tilt angle of 58 ± 4°, which reorients the head group towards the bilayer normal by around 20° (polar angle decreases by 20°) from its protein-free orientation (39 ± 5°).32 This 20° movement towards the bilayer normal is close to that observed experimentally in the EPR docking geometry (27 ± 4°).28 Thus, our MD simulations support the EPR model that the average orientation of the PIP3 head group changes upon binding to the GRP1 PH domain.
Additionally, we explore the orientational dynamics of the PIP3 head group in the binding pocket as well as the structural flexibility of the binding pocket. Our simulations show that the PIP3 head group has a limited orientational flexibility in the binding pocket. This is because of a highly specific binding between the three negatively charged phosphate groups on the PIP3 head group and several positively charged residues in the binding pocket of the PH domain. As a result, any major orientational motions of the PIP3 head group in the binding pocket, such as flipping and drifting of the PIP3 head group, are restricted. We computed the distances between the Cα of residue 296 and four carbon atoms of the PIP3 head group (C1, C3, C4, and C5). The observed fluctuations of these distances are small, which indicates low local dynamics on the head group in the binding pocket.
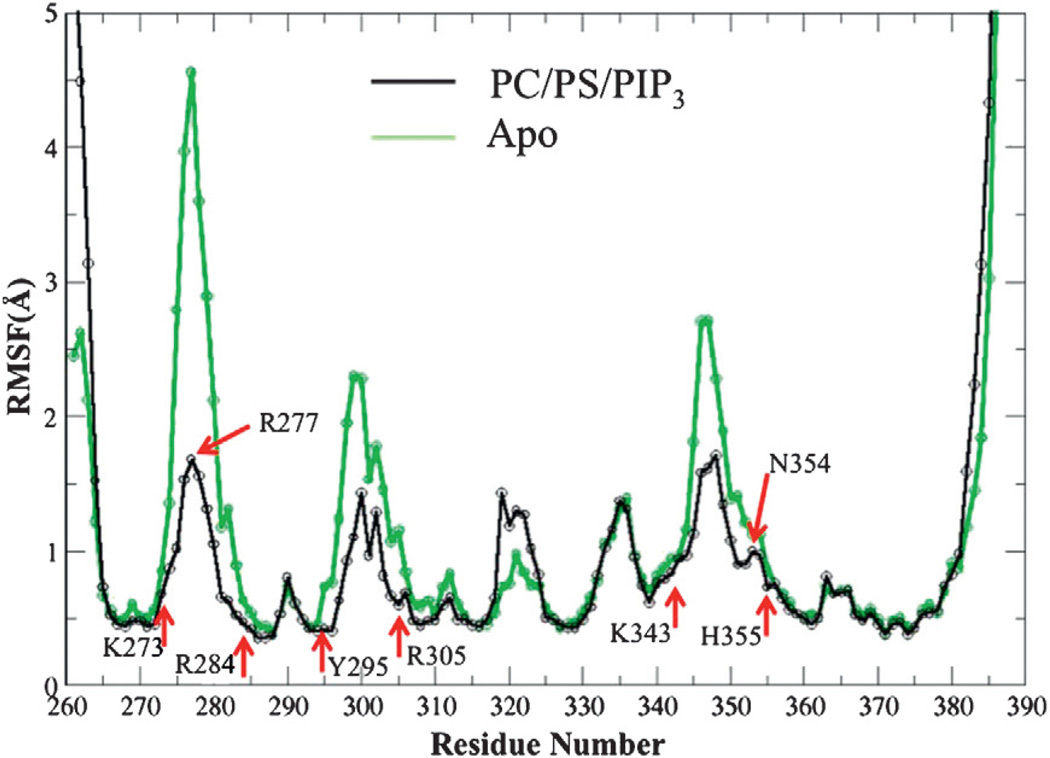
We further utilized the residue-wise root-mean-square fluctuation (RMSF) analysis to investigate the structural flexibility of the protein residues in the binding pocket. We compared the residue-wise RMSF of PIP3-bound PH domain in the PC/PS/ PIP3 bilayer against the apo system. In Fig. 8, the black line shows that the coordination sites (red arrows) of the PIP3 head group mostly exhibit lower fluctuations than other regions of the protein, indicating that the local dynamics of the PIP3 head group-occupied binding pocket are low. The R277 residue of loop β1/β2 is an exception with larger flexibility in structure. This is because R277 is located at the loop β1/β2 where it extensively interacts with the membrane surface, as shown by the number of lipid contact analysis in section B2. RMSF analysis is also applied to the GRP1 PH apo system. We clearly see that loop β1/β2, loop β3/β4, and loop βi1/βi2 are more flexible in the absence of the membrane interactions. Interestingly, PIP3 binding sites exhibit relatively lower RMSF values even in the apo state. This indicates that the PIP3 binding pocket (red arrows) is intrinsically less dynamic with respect to other regions of the protein. It is likely that the binding pocket is “pre-formed” and thus enhances the PIP3 binding efficiency upon the GRP1 PH domain membrane docking.
Fig. 8.
RMSF analysis for membrane-bound PH domain and apo systems. RMSF values are computed from the last 150 ns of MD trajectories for both systems.
Experimental kinetics studies testing the predictions of the EPR-guided MD docking model
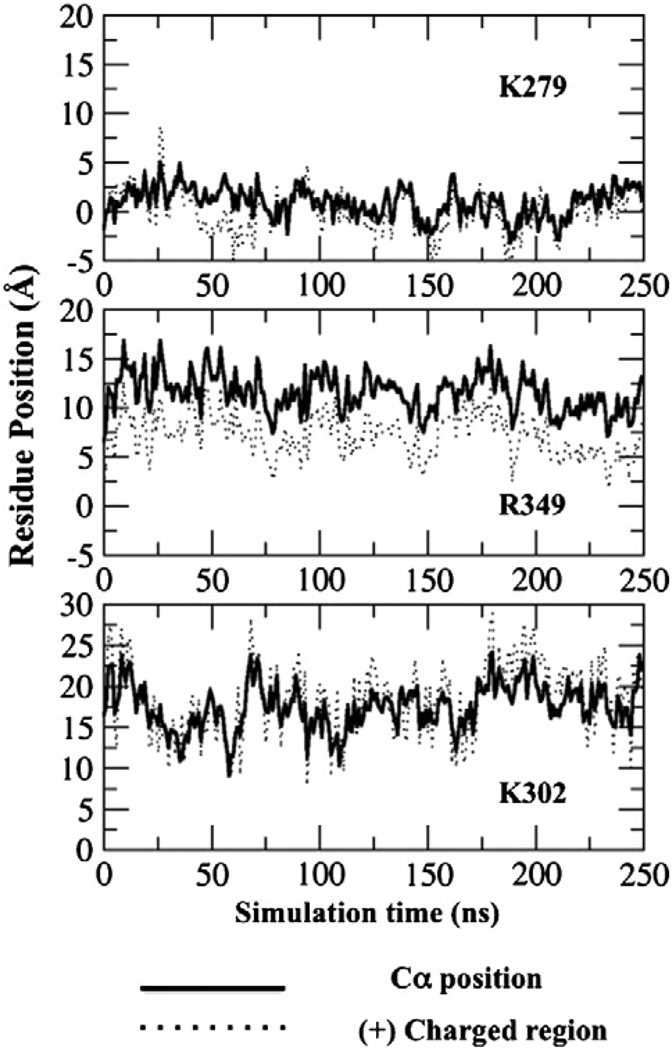
The same three mutants (K279A of loop β1/β2, K302A of loop β3/β4, and R349A of loop βi1/βi2) created to test the predicted PH domain interaction with the PC/PS membrane surface during electrostatic searching also help in testing the model for the PH domain docked to its target PIP3 lipid. In the PIP3-docked state, the EPR-guided MD simulation predicts that K279 and R349 will contact the anionic bilayer surface, while K302 will contact the bilayer less frequently (see Fig. 9). We measure the relative positions of Cα atoms and positively charged regions of K279, R349, and K302 from the membrane phosphate plane. Figure 9 shows that both the Cα position and the charged region of K279 are close to the membrane phosphate plane throughout the course of the simulation, allowing K279 to contact the bilayer frequently. As for R349, although the Cα is about 12 Å from the membrane phosphate plane, we found that the charged region of R349 is relatively closer to the membrane surface. This interesting observation indicates that R349 is still able to contact the bilayer surface. By contrast, both the Cα position and the charged region of K302 are farther from the membrane phosphate plane, implying that K302 plays a minor role in membrane association of the PIP3-docked state. Table 1 shows that the K279A and R349A mutations increase the off-rate for PH domain dissociation from the target membrane by 3.5-fold and 3.7-fold, respectively. On the other hand, the K302 mutation increases the off-rate by only 1.3-fold. These findings support the proposed docking geometry. To sum up, the side-chain interactions with the anionic bilayer have a significant impact on the target membrane affinity. Table 1 shows that the K279A mutation causes a 6.3-fold loss of target affinity due to the combined effects of its slower on-rate during electrostatic searching and its faster off-rate from the PIP3-bound state. The R349A mutation yields a 3.3-fold loss of target affinity due to its faster off-rate, while the K302A mutation has little or no effect on the on-rate, off-rate, or affinity due to its lack of bilayer contact.
Fig. 9.
Plots of Cα positions and positively charged region of selected residues with respect to membrane phosphate plane. The amine nitrogen and the central, charged carbon represent the positively charged regions for lysine and arginine side chains, respectively. The data were collected during the last 250 ns of MD trajectory of the GRP1 PH domain bound to the PC/PS/PIP3 target bilayer.
CG dipole analysis reveals insights into electrostatic-driven membrane associations of the GRP1 PH domain
In the previous two sections, we demonstrated that electrostatic interaction between the GRP1 PH domain and the lipid bilayer surface is the key driving force for membrane association. In this section, we use reduced (CG) representations of protein structure to analyze the electrostatic membrane interactions of the PH domain. We construct CG models of the PH domain at different resolutions to examine the scenarios that occur at different length scales. Our CG analysis of atomistic MD trajectories revealed an interesting phenomenon of these systems and can be used to determine the optimal representation required to capture essential information about them.
In order to complete the search process for the target PIP3 lipid in the membrane, the GRP1 PH domain is known to undergo a stepwise electrostatic search process. Before the PH domain makes contact with the membrane surface, it can sense the negative electrostatic field created by the PS lipids from a distance and orientate itself towards the surface using the positively charged residues on the protein surface. During the membrane associations, direct interactions between side chains of membrane-interacting loops and negatively charged PS lipids and phosphate groups are important in many aspects for the protein membrane binding. This includes initiation of membrane binding and stabilization of the equilibrium membrane binding structure. Experimental kinetic studies in Table 1 further confirm that mutations (K279A and R349) on membrane-interacting loops reduce the PH domain binding affinity to target PIP3 lipids. These observations imply that electrostatic-driven interactions of PH domain membrane binding play an important role, both for the long-ranged and for the short-ranged distances. It is thus important to analyze the electrostatic character of the PH domain structure globally and locally to gain insights into these electrostatic-driven membrane associations.
First, we looked at the global electrostatic character of the GRP1 PH domain by measuring the protein dipole moment for both the apo system and the PH domain bound to a PC/PS/PIP3 target bilayer. Because the positively charged side chains (Arg and Lys) of the PH domain concentrate around the PIP3 head group binding pocket, it is not surprising that the vector of the protein dipole moment points towards the PIP3 head group binding site; specifically, the protein dipole moment points towards the loop β1/ β2 region as shown in Fig. 10a. This is in agreement with our prediction that loop β1/β2 is a key structural motif for the PH domain membrane targeting. We also monitor the flexibility of protein dipole moment orientation by plotting a two-dimensional spherical coordinate φ–θ plot. Figure 10b shows that the protein dipole moment is rigid for both the apo and the membrane-bound state of the PH domain. It is possible that this intrinsic electrostatic character causes the PH domain to effectively respond to the negative electrostatic field of the membrane surface via an electrostatic steering process.
Fig. 10.
Dipole moment orientation analysis for the GPR1 PH domain structure. (a) The dipole moment for protein structure is shown in a blue arrow. (b) φ–θ angle plot of protein structure dipole moment orientation. Protein structure (residue 266 to residue 380) dipole moment analyses are obtained based on the last 200 ns of MD trajectories for both systems. All frames of MD trajectories are firstly aligned with crystal structure (PDB ID: 1FGY). Dipole moment of selected regions are computed via the “measure dipole” function of the VMD software. The dipole moment orientations (vectors) are converted to spherical coordinate φ–θ angles.
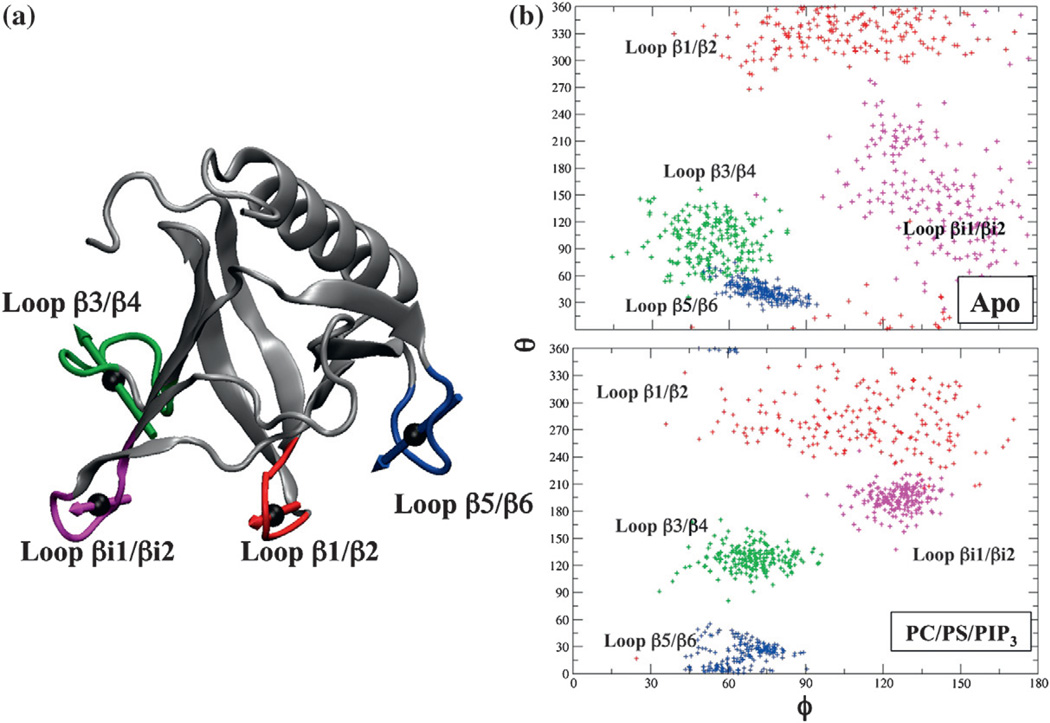
As shown in Fig. 11a, CG dipole moment analysis is applied at the local level of protein structure by focusing on the four membrane-interacting loops. Unlike Fig. 10b, orientations of the local dipole moments on loop regions shown in Fig. 11b exhibit greater flexibility not only in the apo system but also in the membrane-bound state. Figure 11b also shows that the dipole moment orientation of loop β3/β4 and loop βi1/βi2 samples more concentrated φ–θ space in the membrane-bound state. Interestingly, the loop with the most membrane contacts, loop β1/β2, has a similar degree of flexibility in the dipole moment orientation for both states. This intrinsically flexible electrostatic character in the membrane-interacting loop might allow the protein to adapt to the dynamic nature of membrane environment and successfully execute the electrostatic search process. For example, the flexibility of loops can allow positively charged side chains to respond to negatively charged sources and adapt to new orientations within its energetically accessible ranges.
Fig. 11.
(a) Dipole moment orientation analyses of four membrane-interacting loops of the GRP1 PH domain. The calculations for dipole moment orientation are as described in the Fig. 8 caption. Selected residues within four membrane-interacting loops are as follows: loop β1/β2 region: residues 275 to 279; loop β3/β4: residues 297 to 304; loop β5/β6: residues 319 to 324; loop βi1/βi2 loop: residues 344 to 349. (b) Dipole moment orientation analyses for the systems of apo (top) and PIP3-bound GRP1 PH domain in a PC/PS/PIP3 target membrane (bottom).
Overall, CG dipole moment orientation analysis offers insights about membrane associations by the PH domain structure. These observations provide guidance for the future development of a CG protein model. To be able to fully simulate the electrostatic search process of current membrane protein complexes, it is desirable to construct such reduced or simpler CG models for protein and membrane structures to access larger length scales and longer timescales. Our CG dipole moment analysis suggests that the dynamic character of membrane-interacting loops and the membrane itself should be included in order to achieve the desired goal while capturing the essential physics of the system.
Summary and Conclusions
In this study, we investigated the docking reaction of the GRP1 PH domainon the membrane surface using a combination of MD simulations and CG analysis, coupled with experimental studies. Two important GRP1 PH domain membrane-docking states were examined, before and after the PH domain docks to its target PIP3 lipid. Electrostatic-driven membrane associations were found to play key roles in various stages of GRP1 PH domain membrane binding. The electrostatic interactions between the PH domain side chains and individual lipids are also crucial to the stabilization of the equilibrium membrane binding structure of the PH domain.
Our findings are that the GRP1 PH domain likely undergoes a two-dimensional hopping search of the membrane surface before docking to PIP3 lipids. Although the lifetime of the membrane-interacting state during hopping must be transient, it is likely that the early stages of non-PIP3 membrane interactions are also critical for the formation of the PIP3–PH domain complex. In this study, we first carried out four independent atomistic MD simulations of the PH domain with a PC/PS membrane. From the simulations, it was shown that the interactions between PS lipids and positively charged protein side chains play a central role in forming the short-lived membrane-bound state. We observed that the presence of PS lipids is capable of inducing electrostatic rotational steering of the PH domain. Consequently, the PH domain with its highly positively charged surface facing the membrane is guided towards the membrane. In all of the four trajectories, we observed that loop β1/β2 plays a key role to initiate and stabilize membrane binding of the PH domain. In the loop β1/ β2, V278 is flanked by a positively charged arginine (R277) and lysine (K279). Analysis of the MD trajectories reveals that the R277 and K279 have close interaction with the negatively charged PS lipids or nearby lipid phosphate groups on the membrane surface, resulting in membrane perturbation and the creation of transient lipid defects. The hydrophobic residue V278 is then able to penetrate deeply into the bilayer head group region, enabling the protein to bind in a weak but stable manner.
This weakly membrane-bound state with V278 penetrating into the head group region could facilitate a brief period of lateral diffusion on the membrane surface, thereby helping to generate the 10-fold increase in PIP3 on-rate observed when the target membranes contain native levels of PS.13 Notably, this state might exhibit faster lateral diffusion than the PIP3-bound state and might diffuse even faster than free lipids in the bilayer, since the V278 penetration in the head group region would likely yield less friction against the bilayer than a lipid that penetrates deeply into the hydrocarbon region,34 including the PIP3 lipid that defines the single lateral diffusion rate of the PIP3-bound state.20 However, the equilibrium affinity and bound state lifetime of this state must be small and transient, respectively, to ensure that membrane recruitment in the absence of the PIP3 signal stays minimal since biological activation is provided purely by membrane recruitment. Consistent with this biological imperative, no stable recruitment is detected in the absence of PIP3 by in vitro FRET.13
These atomic-scale insights indicate that PS lipids reduce the dimensionality of the search process and increase the residence time of the PH domain on the membrane surface. As a result, the PH domain has an improved probability of binding to the rare PIP3 lipids. This idea is further supported by experimental kinetic studies. Mutation on loop β1/β2 (K279A) generates a 1.9-fold decrease in the PIP3 on-rate than in the WT system. Two other mutations, K302A of loop β3/β4 and R349A of loop βi1/βi2 that serve as negative controls in this experiment, have little or no effect to the on-rate of PIP3 binding. These observations again reinforce our hypothesis that electrostatic-driven membrane association is critical to the early stage of PIP3 search by the GRP1 PH domain.
A recent paper from Lucas and Cho proposed a PS-binding site for the PDK1 PH domain and highlights the important role of PS binding in the biological function of the PH domain.35 In vivo experiments further demonstrate that specific positively charged side chains of the PDK1 PH domain modulate the plasma membrane localization and signaling function. In the work of Lucas and Cho, it is hypothesized that such specific PS binding might be conserved in other PH domains. Interestingly, they specifically pinpoint R277 and K279 of the GRP1 PH domain, showing that these side chains are the candidate residues for PS binding via partial sequence alignment of PH domains. Their data strengthen our argument that R277 and K279 of loop β1/β2 are important to the initiation of stable membrane binding of the GRP1 PH domain.
Another important membrane binding state, that the GRP1 PH domain binds to a PIP3 in a PC/PS/PIP3 lipid bilayer, was also investigated. The EPR membrane binding structure was used as the starting configuration for atomistic MD simulations. The EPR-determined membrane binding structure provides a reasonable starting configuration for the all-atom MD simulation of the complete system. Our EPR-guided MD simulations quantitatively confirm PH domain membrane docking geometry determined by EPR spectroscopy. The MD simulations also reveal subtle deviations of loop regions of the PH domain upon membrane binding. By examining the residue-by-residue membrane docking structure and computing the number of contacts between loop regions and lipid head group parts, we show from our MD model that the GRP1 PH domain is a shallow membrane docker. The shallow penetration of the PH domain into the membrane might reduce the friction and permit rapid diffusion of the PH domain on membrane surface.
Given the essential role of strong binding between the PIP3 head group and the GRP1 PH domain, MD simulations offer high-resolution details of the PIP3 binding pocket. Single-molecule studies show that this is a very stable membrane binding state and the lifetime of the membrane-bound PH domain can reach up to few seconds.20 This is mainly due to the strong affinity between the PH domain and the PIP3 head group. We have shown that the coordination between protein side chains and phosphate groups of the PIP3 head group are stereospecific in nature, which can explain the high binding affinity. We further examined the integrity and flexibility of the PIP3 head group binding pocket in the presence or absence of PIP3. Our data suggest that the core region of the binding pocket (3-, 4-phosphate group binding site) has low flexibility, even in the solution state. This indicates that the binding pocket is a stable structural motif and no additional structural rearrangement for the formation of the PIP3 binding pocket of the PH domain is required upon docking to the PIP3 head group. Such intrinsic characteristics of the pre-formed PIP3 binding pocket might be important to achieve high PIP3 binding efficiency. This is unlike another PIP-binding protein, the Epsin N-terminal homology (ENTH) domain, which is known to bind specifically to PIP2 lipids. In the ENTH domain, the 4-, and 5-phosphate groups of PIP2 lipids also form stereospecific coordination with positively charged side chains in the PIP2 binding pocket. However, unlike the PH domain, the complete binding pocket in the ENTH protein is not formed until the formation of the N-terminal amphipathic helix (H0) upon ENTH membrane binding.
Overall, we demonstrate that combining EPR-determined docking structures and MD simulations is a powerful approach to gain insight into the molecular mechanism of membrane docking reactions of peripheral membrane protein systems. This combined approach has provided important insight into the molecular mechanism regarding how the GRP1 PH domain can locate a PIP3 lipid in the membrane via an electrostatic process and PIP3 binding specificity. Future work will focus on investigating the equilibrium membrane structure of the PH domain under different membrane compositions as well as examining the molecular mechanism of how the GRP1 PH domain binds selectively to the PIP3 while simultaneously excluding relatively abundant PIP2 lipids in the membrane.
Materials and Methods
System setup of GPR1 PH domain lipid bilayer-bound simulations
PC/PS lipid bilayer-bound system
The GRP1 PH domain–lipid bilayer MD simulations utilized a single GRP1 PH domain and a PC/PS lipid bilayer. Four independent trajectories (F1, F2, C1, and C2) were set up and simulated in a PC/PS lipid bilayer system. The F1 and F2 systems started with positions of the GRP1 PH domain far from the lipid bilayer surface. The closest point of the GRP1 PH domain is more than 20 Å from lipid bilayer phosphate plane. The F1 and F2 systems have different membrane targeting orientations. The C1 and C2 systems start with the initial positions of the GRP1 PH domain closer to the lipid bilayer surface. The closest point of the GRP1 PH domain is kept within 10 Å from the lipid bilayer phosphate plane. The C1 and C2 systems also have different membrane targeting orientations. In all four systems, the targeting orientation of the GRP1 PH domain is set up with the PIP3 binding pocket facing towards the lipid bilayer surface.
For the PC/PS lipid bilayer system, subsequent solvation, ionization, minimization, and MD equilibration protocols were carried out as follows. The protein–membrane system was equilibrated using a stepwise relaxation procedure. After placement of the PH domain on the membrane surface, systems were solvated with TIP3P water. In the PC/PS system, the simulation system was neutralized by Na+ counterions. Initially, all PH domain Cα and phosphorus atoms of phosphate groups were harmonically restrained with a force constant of 5 kcal/(mol Å2), and a conjugate gradient minimization of 10,000 steps was applied, followed by heating to 310 K and 10 ns of constant NPT equilibration. After all restraints on Cα atoms of the PH domain and phosphorus atoms were removed, constant NPT ensemble MD simulations were used to generate 250 ns of trajectory for each system.
The system setup and equilibration for the three 150-ns control simulations with a single GRP1 PH domain targeting the pure PC lipid bilayer followed the same procedures as described in the F1 and F2 systems of PC/ PS lipid bilayer. A 0.15 M NaCl concentration was set for three control simulations. Different initial membrane targeting poses of GRP1 PH domain are used for these three control simulations. In all three targeting orientations, the protein surface of the PIP3 head group binding pocket faces towards the lipid bilayer surface.
PC/PS/PIP3 lipid bilayer-bound system
The GRP1 PH domain–lipid bilayer MD simulations utilized a single GRP1 PH domain bound to a lipid bilayer. The coordinates for the GRP1 PH domain and the PIP3 head group (IP4) were the crystal structure from Lietzke et al. (PDB ID: 1FGY).21
The GRP1 PH domain lipid bilayer-bound configuration in both target bilayer systems was built in the following way: the relative orientation of the IP4-bound PH domain structure with respect to the target bilayer surface was guided by the EPR docking geometry.28 Once the IP4–PH domain structure matched the EPR structure, the PH domain–IP4 complex was translated to the DPPIP3 lipid location of the target bilayer where the DPPIP3 head group matches the IP4 orientation in the PH domain-binding pocket. The IP4 coordinates were removed and replaced by PIP3 lipid coordinates, yielding a protein–lipid complex for both systems.
For the PC/PS/PIP3 target bilayer system, subsequent solvation, ionization, minimization, and MD equilibration protocols were carried out as follows. The protein– membrane system was equilibrated using a stepwise relaxation procedure. After placement of the PH domain on the membrane surface, systems were solvated with TIP3P water. In the PC/PS/PIP3 system, the simulated system was neutralized by Na+ counterions. Initially, all PH domain Cα and carbon atoms on the PIP3 were harmonically restrained with a force constant of 5 kcal/(mol Å2), and a conjugate gradient minimization of 10,000 steps was applied, followed by heating to 310 K and 5 ns of constant NPT equilibration. This was followed by another 5 ns of constant NPT equilibration where the restraints on the PIP3 were removed. After all restraints on Cα of the PH domain were removed, we continued simulations for 300 ns. The last 250 ns of trajectories was taken for data analysis.
For the control simulations in the PC/PS/PIP3 target bilayer system, different membrane docking structures of the GRP1 PH domain were used. After the placement of the PH domain on the membrane surface was done as described above, we further adjusted the docking geometry of the PH domain in the following ways. First, the tilt angle of the structure was adjusted to around 80° (from 46° of EPR-guided orientation) by rotating the protein structure. The second adjustment is to further orient the structure by moving the less positively charged region (loop β3/β4) towards the membrane surface, while maintaining the same tilt angle of the docking structure. The subsequent solvation, ionization, minimization, and equilibration procedures are the same as for the EPR-guided MD system. A greater than 500 ns of trajectory was generated for this control simulation.
Protein-only apo system
The GRP1 PH domain of crystal structure 1FGY was solvated with TIP3P water. The simulation system was neutralized by Cl− counterions. Initially, all PH domain Cα were harmonically restrained with a force constant of 5 kcal/(mol Å2), and a conjugate gradient minimization of 5000 steps was applied, followed by heating to 310 K and 5 ns of constant NPT equilibration. After the dimensions of the simulation box were equilibrated, it was followed by another 250 ns of constant NVT equilibration, where all restraints on Cα of the PH domain were removed.
General simulation details
All MD simulations employed the CHARMM22 force field for the proteins with CMAP corrections36 and CHARMM36 for lipids.37 The original parameterization for PIP3 head groups was used as previously described.38 Tail groups of PIP3 lipid from the original parameterization were replaced with DP tail groups in order to be consistent with EPR experiments. Simulations were performed under constant NPT conditions, and periodic boundary conditions are applied. A Langevin thermostat with a damping coefficient of 0.5 ps−1 was used to maintain the system temperature at 310 K. The system pressure was maintained at 1 atm using a Langevin piston barostat.39 Short-range non-bonded interactions were truncated smoothly between 10 Å and 12 Å. The particle mesh Ewald algorithm40 was used to compute long-range electrostatic interactions at every time step. All covalent hydrogen bonds were constrained by the SHAKE algorithm (or the SETTLE algorithm for water),41 permitting an integration time step of 2 fs. System minimization, equilibration, and dynamics were performed using the NAMD 2.7 software package.42 System construction and image generation were performed using the VMD 1.9 software package.33
Preparation and equilibration of lipid bilayers
POPC/POPS lipid bilayer
The CHARMM-GUI43 was employed to generate a 75%/25% POPC/POPS mixed lipid bilayer with a total of 72 lipids, which has 36 lipids on each leaflet. This lipid patch is replicated along the x and y coordinate directions to create a 2 × 2 system that consist of a total of 288 lipids. The 288-lipid POPC/POPS system was then solvated with TIP3P waters and neutralized with Na+ ions. The fully solvated mixed lipid bilayer system was simulated for 80 ns under constant NPT conditions. After 10 ns, the value of the area per lipid fluctuates between 59 Å2 and 63 Å2, and the averaged area per lipid over the last 40 ns is 61.3 ± 0.2 Å2. The force field and software for MD simulations are described in General simulation details.
POPC/POPS/DPPIP3 lipid bilayer
A 2 × 2 POPC/POPS lipid bilayer system was created as described above. Two POPS lipids, each one located in the central region of each leaflet, were replaced with DPPIP3 lipids. This new POPC/POPS/DPPIP3 mixed lipid bilayer was solvated with TIP3P waters and neutralized with Na+ ions. Followed by the same simulation procedures, the averaged area per lipid over the last 40 ns of constant NPT simulations is 61.4 ± 0.1 Å2. The resulting area/lipid is the same as in the POPC/POPS mixed lipid bilayer system.
POPC lipid bilayer
The CHARMM-GUI43 was employed to generate a pure POPC lipid bilayer with a total of 288 lipids, which has 144 lipids on each leaflet. The 288-lipid POPC system was solvated with TIP3P waters. The fully solvated lipid bilayer system was simulated for 100 ns under constant NPT conditions. The force field and software for MD simulations are described in General simulation details. The averaged area per lipid over the last 60 ns is 64.5 ± 0.2 Å2. This value is close to previously reported simulation value for a smaller patch of pure POPC lipid bilayer with a total of 72 lipids (64.7 ± 0.2 Å2/lipid).37
Experimental determination of target membrane on- and off-rates
Reagents
Wild-type GRP1 PH domain and site-directed mutants were constructed, expressed, and purified as described previously.28,44 Sonicated unilamellar vesicles composed of PC/dPE/PIP3 (92/5/3) or PC/PS/dPE/PIP3 (69/23/5/3) were prepared as detailed previously.13,28
Stopped-flow kinetic measurements
Kinetic studies were carried out as previously described13,44 on an Applied Photophysics SX.20 stopped-flow fluorescence instrument. Specifically, protein-to-membrane FRET was employed to monitor the time course of PH domain binding to, or dissociating from, SUVs in physiological buffer containing 10 mM DTT at 25 °C. PH domain (1 µM) was rapidly mixed (dead time < 1 ms) with plasma-membrane-like SUVs (total lipid concentration of 600 µM) containing excess PIP3 target lipid (6 µM accessible) yielding final concentrations of PH domain (0.5 µM) and accessible PIP3 (3 µM). Under these conditions, the forward binding reaction is virtually irreversible because of the high affinity of the PH domain for PIP3 lipid, and best-fit analysis of the binding time course with a single-exponential component for binding to homogenous sites yielded the apparent on-rate constant [kon (app)], calculated as previously described.13,44 Dissociation kinetics were measured by rapidly mixing the pre-formed PH domain–membrane complex (0.5 µM protein and 200 µM total lipid) with excess IP6 (40 mM final). Best fitting of a single-exponential function to the dissociation time course yields the off-rate constant (koff) as previously detailed.13,44
Acknowledgements
This study was supported by grants from National Institutes of Health (R01-GM063796 to G.A.V. and R01-GM063235 to J.J.F.). Computational resources were provided by the National Science Foundation through XSEDE computing resources of the Texas Advanced Computing Center (Ranger), the San Diego Supercomputing Center (Gordon), and the National Institute for Computational Sciences (Kraken). The authors thank Gard Nelson and Dr. Joseph Baker for assistance and helpful discussions, and Dr. Brian Ziemba (Falke Laboratory, University of Colorado, Boulder) for sending the EPR docking geometry coordinates.
Abbreviations used
- PH
pleckstrin homology
- GRP1
general receptor of phosphoinositides 1
- PIP3
phosphatidylinositol (3,4,5)-trisphosphate
- EPR
electron paramagnetic resonance
- MD
molecular dynamics
- CG
coarse-grained
- FRET
fluorescence resonance energy transfer
- PS
phosphatidylserine
- PIP2
phosphatidylinositol (4,5)-bi-sphosphate
- PDB
Protein Data Bank
- RMSF
root-mean-square fluctuation
- ENTH
Epsin N-terminal homology
References
- 1.Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA. Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol. Cell. 2000;6:373–384. doi: 10.1016/s1097-2765(00)00037-x. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell. Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 3.Moravcevic K, Oxley CL, Lemmon MA. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012;20:15–27. doi: 10.1016/j.str.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, et al. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 5.Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- 6.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, et al. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 7.Newton AC. Lipid activation of protein kinases. J. Lipid Res. 2009;50:S266–S271. doi: 10.1194/jlr.R800064-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 9.Czech MP. PIP2 and PIP3: complex roles at the cell surface. Cell. 2000;100:603–606. doi: 10.1016/s0092-8674(00)80696-0. [DOI] [PubMed] [Google Scholar]
- 10.Insall RH, Weiner OD. PIP3, PIP2, and cell movement—similar messages, different meanings? Dev. Cell. 2001;1:743–747. doi: 10.1016/s1534-5807(01)00086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Herzmark P, Weiner OD, Srinivasan S, Servant G, Bourne HR. Lipid products of PI(3)Ks maintain persistent cell polarity and directed motility in neutrophils. Nat. Cell Biol. 2002;4:513–518. doi: 10.1038/ncb810. [DOI] [PubMed] [Google Scholar]
- 12.Czech MP. Dynamics of phosphoinositides in membrane retrieval and insertion. Annu. Rev. Physiol. 2003;65:791–815. doi: 10.1146/annurev.physiol.65.092101.142522. [DOI] [PubMed] [Google Scholar]
- 13.Corbin JA, Dirkx RA, Falke JJ. GRP1 pleckstrin homology domain: activation parameters and novel search mechanism for rare target lipid. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- 15.Hurley JH. Membrane binding domains. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 17.Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 20.Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys. J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG. Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol. Cell. 2000;6:385–394. doi: 10.1016/s1097-2765(00)00038-1. [DOI] [PubMed] [Google Scholar]
- 22.Frazier AA, Wisner MA, Malmberg NJ, Victor KG, Fanucci GE, Nalefski EA, et al. Membrane orientation and position of the C2 domain from cPLA2 by site-directed spin labeling. Biochemistry. 2002;41:6282–6292. doi: 10.1021/bi0160821. [DOI] [PubMed] [Google Scholar]
- 23.Frazier AA, Roller CR, Havelka JJ, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 24.Kohout SC, Corbalan-Garcia S, Gomez-Fernandez JC, Falke JJ. C2 domain of protein kinase calpha: elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malmberg NJ, Van Buskirk DR, Falke JJ. Membrane-docking loops of the cPLA2 C2 domain: detailed structural analysis of the protein–membrane interface via site-directed spin-labeling. Biochemistry. 2003;42:13227–13240. doi: 10.1021/bi035119+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landgraf KE, Malmberg NJ, Falke JJ. Effect of PIP2 binding on the membrane docking geometry of PKC alpha C2 domain: an EPR site-directed spin-labeling and relaxation study. Biochemistry. 2008;47:8301–8316. doi: 10.1021/bi800711t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malmberg NJ, Falke JJ. Use of EPR power saturation to analyze the membrane-docking geometries of peripheral proteins: applications to C2 domains. Annu. Rev. Biophys. Biomol. Struct. 2005;34:71–90. doi: 10.1146/annurev.biophys.34.040204.144534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen HC, Ziemba BP, Landgraf KE, Corbin JA, Falke JJ. Membrane docking geometry of GRP1 PH domain bound to a target lipid bilayer: an EPR site-directed spin-labeling and relaxation study. PLoS One. 2012;7:e33640. doi: 10.1371/journal.pone.0033640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lumb CN, Sansom MSP. Finding a needle in a haystack: the role of electrostatics in target lipid recognition by PH domains. PLoS Comp. Biol. 2012;8:e1002617. doi: 10.1371/journal.pcbi.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosys-tems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumb CN, He J, Xue Y, Stansfeld PJ, Stahelin RV, Kutateladze TG, Sansom MSP. Biophysical and computational studies of membrane penetration by the GRP1 pleckstrin homology domain. Structure. 2011;19:1338–1346. doi: 10.1016/j.str.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Venable RM, Rogers LA, Murray D, Pastor RW. Molecular dynamics simulations of PIP2 and PIP3 in lipid bilayers: determination of ring orientation, and the effects of surface roughness on a Poisson–Boltzmann description. Biophys. J. 2009;97:155–163. doi: 10.1016/j.bpj.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Ziemba BP, Falke JJ. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of 1) bound lipids and 2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids. 2013 doi: 10.1016/j.chemphyslip.2013.04.005. in press. http://dx.doi.org/10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed]
- 35.Lucas N, Cho W. Phosphatidylserine binding is essential for plasma membrane recruitment and signaling function of 3-phosphoinositide-dependent kinase-1. J. Biol. Chem. 2011;286:41265–41272. doi: 10.1074/jbc.M111.300806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 37.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lupyan D, Mezei M, Logothetis DE, Osman R. A molecular dynamics investigation of lipid bilayer perturbation by PIP2 . Biophys. J. 2010;98:240–247. doi: 10.1016/j.bpj.2009.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feller SE, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 40.Darden T, York D, Pedersen L. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 41.Miyamoto S, Kollman PA. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 42.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jo S, Kim T, Iyer VG, Im W. Software news and updates—CHARNIM-GUI: a web-based grraphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 44.Pilling C, Landgraf KE, Falke JJ. The GRP1 PH domain, like the AKT1 PH domain, possesses a sentry glutamate residue essential for specific targeting to plasma membrane PI(3,4,5)P3. Biochemistry. 2011;50:9845–9856. doi: 10.1021/bi2011306. [DOI] [PMC free article] [PubMed] [Google Scholar]