Abstract
The machinery of mitochondrial DNA (mtDNA) maintenance is only partially characterized and is of wide interest due to its involvement in disease. To identify novel components of this machinery, plus other cellular pathways required for mtDNA viability, we implemented a genome-wide RNAi screen in Drosophila S2 cells, assaying for loss of fluorescence of mtDNA nucleoids stained with the DNA-intercalating agent PicoGreen. In addition to previously characterized components of the mtDNA replication and transcription machineries, positives included many proteins of the cytosolic proteasome and ribosome (but not the mitoribosome), three proteins involved in vesicle transport, some other factors involved in mitochondrial biogenesis or nuclear gene expression, > 30 mainly uncharacterized proteins and most subunits of ATP synthase (but no other OXPHOS complex). ATP synthase knockdown precipitated a burst of mitochondrial ROS production, followed by copy number depletion involving increased mitochondrial turnover, not dependent on the canonical autophagy machinery. Our findings will inform future studies of the apparatus and regulation of mtDNA maintenance, and the role of mitochondrial bioenergetics and signaling in modulating mtDNA copy number.
Keywords: complex V, DNA replication, mitochondrial biogenesis, mitochondrial DNA, mitophagy, nuclease, nucleoid, reactive oxygen species
Introduction
Eukaryotes that use mitochondrial oxidative phosphorylation (OXPHOS) to generate ATP maintain a separate mitochondrial genome (mtDNA), encoding a subset of OXPHOS protein subunits, together with some components of the machinery of intramitochondrial protein synthesis. The maintenance and expression of mtDNA is otherwise dependent on nuclear-coded gene products, constituting a separate apparatus for genome maintenance and gene expression within the cell (McKinney & Oliveira, 2013).
The core machinery of mtDNA replication is broadly conserved among eukaryotes. DNA replication is assumed to depend on the only DNA polymerase consistently found in mitochondria, DNA polymerase γ (PolG; Kaguni, 2004), a member of the family A DNA polymerases (Ito & Braithwaite, 1991). Its closest prokaryotic homologue is the phage T7 DNA polymerase, and it is assumed to function in concert with the mitochondrial helicase Twinkle (Spelbrink et al, 2001), a homologue of phage T7 helicase-primase (gp4). Twinkle is absent from yeast, where other helicases are involved in mtDNA replication. Maintenance of mtDNA requires also mitochondrial transcription factors A (mt-TFA or TFAM), needed for mtDNA compaction and transcription (Larsson et al, 1998; Kang & Hamasaki, 2005; Campbell et al, 2012), and B (mt-TFB2, TFB2M; Matsushima et al, 2004). Three other proteins are essential for mtDNA maintenance, namely mtSSB, the mitochondrial single-stranded DNA-binding protein (Maier et al, 2001), RNase H1 (Cerritelli et al, 2003), and, in some organisms, a second DNA polymerase, PrimPol, that additionally has primase activity (García-Gómez et al, 2013). A number of other proteins are required to maintain normal mtDNA copy number or topology in different organisms (Contamine & Picard, 2000; Copeland, 2012). These include the DNA-binding AAA protein ATAD3A (He et al, 2007), some enzymes of nucleotide metabolism and transport (Saada, 2004), proteins with roles in mitochondrial membrane dynamics (Jones & Fangman, 1992; Wong et al, 2000; Elachouri et al, 2011; Vielhaber et al, 2013), chaperones (Ciesielski et al, 2013), exonucleases (Kornblum et al, 2013), proteases (Herlan et al, 2003; Matsushima et al, 2010; Sesaki et al, 2003), and even cytoskeletal proteins (Reyes et al, 2011). Mitochondria also contain topoisomerases, ligases, and other nucleases, although their specific roles in mtDNA metabolism are unclear. While a crude DNA synthetic machinery can be reconstituted in vitro from a minimal set of these proteins, the full complement of proteins required for faithful mtDNA replication in vivo remains to be determined.
Some components of the mtDNA maintenance machinery are shared with the nuclear compartment, including RNase H1 (Cerritelli et al, 2003) and many proteins implicated in base-excision repair (Alexeyev et al, 2013). Mostly, these are synthesized in two or more isoforms routed to different cellular compartments, for example, via differential splicing, alternative translational start sites (Suzuki et al, 2010), or ambiguous targeting signals (Karniely & Pines, 2005).
mtDNA is packaged together with TFAM and some other proteins into discrete intramitochondrial structures of variable composition, called nucleoids, by analogy with those of bacteria (Spelbrink, 2010; Bogenhagen, 2012). They contain a number of replication proteins whose functional roles are poorly understood, as well as proteins implicated in other cellular processes, including metabolic enzymes and chaperones, and proteins involved in intramitochondrial protein synthesis (Hensen et al, 2014). The apparatus of mitochondrial translation has been functionally implicated in mtDNA maintenance in yeast (Contamine & Picard, 2000), though not in metazoan cells (Storrie & Attardi, 1972).
The importance of mtDNA maintenance for cell physiology and homeostasis is underscored by the finding that its dysfunction leads to diverse types of human disease, including both infantile and late-onset pathologies, showing a bewildering variety of tissue specificities (Shadel, 2008; Rötig & Poulton, 2009; Ylikallio & Suomalainen, 2012). Loss of mitochondrial genome integrity or fidelity is also associated with aging (Oliveira et al, 2010; Bratic & Larsson, 2013). Identifying the full set of gene products involved in faithful mtDNA maintenance is thus of broad interest and importance.
To this end, we implemented a genome-wide (blinded) screen of Drosophila S2 cells, using dsRNA-based RNA interference (RNAi), taking advantage of the fact that S2 cells tolerate loss of mtDNA and continue to grow within the time scale of a typical experiment, despite decreased OXPHOS capacity. Furthermore, mtDNA nucleoids may be identified in these cells on the basis of fluorescence signal from the topology-dependent DNA-intercalating dye PicoGreen (Ashley et al, 2005). Effects on mtDNA copy number were then probed further using quantitative PCR (QPCR), with additional experiments conducted on the cellular phenotypes produced by knockdown of specific genes identified in the screen, notably those encoding subunits of ATP synthase, in order to test aspects of the mechanisms by which they may act.
Results and Discussion
Implementation and outcome of the primary screen
We set out to screen a genome-wide Drosophila dsRNA library in S2 cells, scoring for disappearance of the PicoGreen signal of mtDNA nucleoids as indicative of genes required for mtDNA maintenance. In initial trials, we found it difficult to pick out the nucleoid signal against background cytoplasmic fluorescence. Using a dsRNA against the PolG catalytic subunit (tamas) as a positive control, and a dsRNA directed against GFP as a negative control, we established a protocol whereby it was possible reliably to score (by eye) the disappearance of nucleoid signal (see Fig 1). This involved applying the test dsRNA for 5 days in a 96-well plate format, with addition on day 3 of a dsRNA directed against TFAM. Although prolonged incubation with TFAM dsRNA itself led to mtDNA depletion, the shorter-term treatment conversely enhanced the nucleoid signal in negative control cells, whereas it was decreased to very low levels in cells treated with the positive control dsRNA.
Figure 1. Screening of Drosophila dsRNA library by PicoGreen nucleoid fluorescence in S2 cells.
Micrographs of S2 cells stained with PicoGreen, following 5 days of treatment with the dsRNA indicated. GFP and tamas (Polg, CG8987) were used as negative and positive controls, respectively. In rescreening, CG5924 was also used as a positive control. Both were detected in the blinded screen as positives. Other images show a typical negative (CG31380), a typical positive (CG6413) and a typical case of a target classed as abnormal, in this case CG2028 (CkIα), which showed a decreased number of nucleoid signals per cell. Images are optimized for brightness and contrast but with no other manipulations. Scale bar indicates 50 μm.
The primary screen, conducted blind, was successful in identifying as positives most of the known factors involved in mtDNA metabolism (Table 1, category 1), giving confidence in its validity. In total, 105 dsRNA targets were initially judged as positive (Supplementary Table S1), of which almost half were recorded also as leading to cell death in a fraction of the cells. Consistent with previous studies (Rämet et al, 2002; Boutros et al, 2004), a further 276 targets (Supplementary Tables S1 and S2) gave massive cell death but no specific loss of nucleoid signal and were considered to represent essential genes that could not be studied further. Finally, an additional 132 targets were judged to give an abnormal outcome without complete loss of PicoGreen nucleoid signal (Supplementary Tables S1 and S3), but were not analyzed further, although many fell into similar categories or pathways as those on the positives list. The specificity and knockdown efficiency of this procedure has previously been documented (Clemens et al, 2000; Kleino et al, 2005). Knockdown was here verified at the RNA level by qRT-PCR for 17 specific targets (see SI).
Table 1.
Definitive positives
 |
We compared subjective judgment against a computational method to measure punctate fluorescence intensity (details to be published elsewhere). The latter gave many false positives due to variable background fluorescence, as well as false negatives due to cell debris. Based on rescreening, we judged the manual method to be superior (see SI for details), and we set criteria for defining positives as described below.
Rescreening to identify definitive positives
Positives were considered as confirmed if three positive but no negative findings were obtained. Those where a negative or ambiguous finding was recorded were retained only where three times as many clearly positive findings were obtained upon exhaustive rescreening (double asterisks in column H of Table 1) otherwise they were considered as false positives (double asterisks in column G of Supplementary Table S4).
Five targets that did not give consistently positive findings during rescreening were noted to give rise to multiple splice variants, encoding at least one polypeptide predicted to be mitochondrially localized. For these, we tested dsRNAs targeted specifically on the relevant splice variants, confirming several additional positives (Table 1, green background), whereas dsRNAs targeted against other splice variants or the entire gene were judged negative (Supplementary Table S4, blue background).
Of the original 105 positives, 83 were retained, 20 were reassigned as negative, and one was reassigned as abnormal (pink background in Supplementary Table S3). One was discarded because the dsRNA detected a pseudogene of a gene already in the list, another because a revised gene model combined it with another positive, while another was subsequently re-annotated as two separate genes (but shown as a single entry in Table 1). Positive findings were also obtained for specific splice isoforms of three of the negatives. Thus, the confirmation of 86 out of 106 initial targets indicates a false-positive rate of 18%. One additional positive (CG5794) was unexpectedly identified by a dsRNA nominally targeted against a different gene. The positives fell into seven distinct classes: mitochondrial DNA replication or transcription, cytosolic translation, the proteasome, ATP synthase, mitochondrial dynamics or biogenesis, nuclear gene expression, and a seventh, miscellaneous category.
Although the primary negatives were not rescreened systematically, some that fell into similar functional pathways as definitive positives were re-evaluated using the same criteria. Eleven were promoted to the positives list (yellow background in Table 1), including, for example, most other subunits of ATP synthase. This implies that the initial screen may have missed as many positives as were actually retained, implying a false-negative rate of up to 1%. The overall results of the screen are summarized in Fig 2. The final number of definitive positives was 97, counting only once those with > 1 positive splice variant.
Figure 2. Overall results of the screen.

Schematic diagram illustrating number of dsRNAs analyzed, numbers of primary positives, negatives and other classes, and the results of rescreening.
Copy number of mtDNA
For genes on the definitive positives list, we carried out QPCR to assess changes in mtDNA copy number after 5 days of dsRNA treatment (without concomitant TFAM knockdown), normalized against a single-copy nuclear DNA standard. Based on this assay (see Table 1, column G, raw data in Supplementary Table S5), we classified the positives as showing substantial (++, ≤ 60%) or modest (+) mtDNA depletion, no significant copy number change (0), or an increase in mtDNA (−).
Although these classes seem arbitrary, we found that RNAi knockdown of well-characterized components of the mtDNA replication machinery all gave values in the 20-60% range (++). Surprisingly, we identified only 6 new genes from the screen whose knockdown produced a comparably severe mtDNA depletion. Three of these encode subunits of ATP synthase. The others were CG5794, encoding a de-ubiquitinating enzyme with unknown substrate(s), TweedleY, previously identified as a cuticular protein (Guan et al, 2006), and pointed, a well-studied Ets family transcription factor (Klaes et al, 1994; Morimoto et al, 1996). The fact that knockdown of many genes resulted in loss of PicoGreen signal without major changes in mtDNA copy number suggests that indirect effects may be common, for example, affecting DNA topology, nucleoid architecture, membrane potential or cellular dye uptake.
Positives implicated in mtDNA metabolism
The positives include most of the proteins with known roles in mtDNA replication or transcription, notably the five shown previously to be essential for mtDNA maintenance in Drosophila (Goto et al, 2001; Maier et al, 2001; Iyengar et al, 2002; Matsushima et al, 2004; Humphrey et al, 2012). The list comprises the two subunits of PolG, the catalytic subunit of the mitochondrial RNA polymerase, mtSSB, the Drosophila homologue of the Twinkle helicase, transcription factors TFAM and mtTFB2M, mTERF family members mTTF and mTerf5 (Jõers et al, 2013), plus rnh1 (RNaseH1) and belphegor (homologue of mammalian ATAD3). All gave significant mtDNA depletion except for rnh1 and belphegor. DNA ligase III (lig3, CG17227) and mTERF family members mTerf3 and CG15390 (homologue of mammalian MTERF4) were consistently negative.
DNA ligase III was previously reported as dispensable for nuclear DNA repair but essential for mtDNA maintenance in human cells (Ruhanen et al, 2011) and mouse (Puebla-Osorio et al, 2006; Gao et al, 2011). Our data imply that it is redundant to at least one other mtDNA ligase in Drosophila. Similar arguments may apply to the absence of any topoisomerase, gyrase, recombinase, resolvase or helicase (other than Twinkle). Our study suggests that few dedicated components of the mtDNA replication apparatus remain to be identified, but this does not exclude factors with overlapping roles in other cell compartments.
In mammalian mitochondria, the transcriptional apparatus is considered essential for both leading- and lagging-strand synthesis (Clayton, 1982; Fuste et al, 2010). RNase H1, also required for mtDNA maintenance in mouse (Cerritelli et al, 2003) and human cells (Ruhanen et al, 2011), might be involved in primer removal, but this typically also needs other helicases and nucleases such as Fen1 (CG8648) and Dna2 (CG2990), both implicated in mtDNA replication in mammalian cells (Duxin et al, 2009; Kazak et al, 2013). Their absence from the positives list is unsurprising, however, since both also function in the nucleus. One positive from the miscellaneous category, CG8021, appears to be a Drosophila homologue of SLRP, a mammalian protein involved in mitochondrial mRNA stabilization and processing (Sasarman et al, 2010; Chujo et al, 2012).
The mammalian ATAD3 family has been implicated in nucleoid organization (He et al, 2007), mitochondrial protein synthesis (He et al, 2012), regulation of apoptosis (Huang et al, 2011) and autophagy (Chen et al, 2011), cholesterol trafficking (Rone et al, 2012), mitochondrial dynamics (Gilquin et al, 2010) and stress resistance (Hoffmann et al, 2012). Loss of PicoGreen nucleoid signal with only a minor drop in mtDNA copy number may indicate that belphegor functions also in diverse pathways and that its effects on nucleoids and mtDNA may be indirect.
Nucleases other than RNase H1 have been shown or suggested to have roles in mtDNA metabolism in various organisms, including EXOG (Tann et al, 2011), EndoG (McDermott-Roe et al, 2011) and yeast Exo5 (Burgers et al, 2010). Exo5 has no Drosophila homologue, and the somatically expressed EndoG/EXOG isogene (EndoG, CG8862) was not detected as a positive, although two other nucleases (CG42666 isoforms D and G and Dis3) appear under the miscellaneous category. Both appear to be targeted to multiple locations in the cell, and, like rnh1, their knockdown did not give rise to significant mtDNA depletion at the 5 days time point. CG42666 has a well-characterized 3′ to 5′ (REX1-like) exoribonuclease domain shared with proofreading DNA polymerases, such as E. coli DnaQ (Pol III ε subunit), and with PAN2 deadenylase (Koonin & Deutscher, 1993). Its closest human homologues (REXO1 family) are poorly characterized, thus far implicated only in the suppression of transcriptional pausing (Tamura et al, 2003), but REXO2 has recently been reported to function in mitochondria as an oligoribonuclease required for normal mitochondrial morphology, nucleic acid content and protein synthesis (Bruni et al, 2013).
Dis3 is predicted to have a mitochondrial localization (Mitoprot II score 97%), supported by other studies (Mamolen, 2010; Hou et al, 2012). Its human homologues DIS3 and DIS3L1 are considered to be the main nucleases of the RNA exosome complex (Tomecki et al, 2010), with roles in RNA processing, degradation and surveillance in both nucleus and cytoplasm. Analysis of a putative mitochondrial function for CG42666 and Dis3 will thus be problematic, due to multiple localization and pleiotropy.
Cytosolic translation and the proteasome
These positives include both small and large ribosomal subunit proteins, one translation factor, core and regulatory/subunit components of the proteasome, plus one proteasome assembly chaperone. In total, there are some 91 genes annotated as cytosolic ribosomal proteins in the Drosophila genome, plus 7 others as ‘ribosomal protein-like’. The frequency of annotated ribosomal proteins in the positives list (14/97, 14%) is thus significantly greater than in the genome as a whole (98/13,937, 0.7%, P < 0.01 by χ2 test). Note also that 38 of 132 genes in the abnormal list (29%, Supplementary Table S3) are annotated as encoding ribosomal proteins, in addition to Tor and raptor, which positively regulate cytosolic protein synthesis. For proteasomal genes, the calculations are similar (7/97, 7.2%, versus 60/13,937, 0.4%, P < 0.01 by χ2 test).
Knockdown of proteasomal components gave mtDNA depletion mainly in the moderate (+) range, whereas knockdown of some representative cytosolic ribosomal proteins produced either no change in copy number or a relative increase, which may reflect cell death occurring in a large fraction of the cells by the time they were harvested.
One possible explanation for the link between cytosolic translation and mitochondrial homeostasis is that one or more short-lived and/or poorly translated cytosolic mRNAs encode products essential for maintaining mitochondria. Alternatively, mitochondria might be turned over by enhanced autophagy, which results from a prolonged deficiency of protein synthesis (Neufeld, 2012). Consistent with this, Pitslre, a negative regulator of autophagy (Wilkinson et al, 2011), was positive in our screen.
A more specific impact on mitochondria could involve mitonuclear regulation to minimize proteotoxic stress due to an insufficient supply of nuclear-coded OXPHOS subunits. The apparatus of mitochondrial translation was absent from the list of positives, consistent with antibiotic studies (Storrie & Attardi, 1972), but in contrast with findings in fungi, where translational deficiency leads to loss of mtDNA (Contamine & Picard, 2000). However, a stress signal emanating from the mitochondrial ribosome would most logically produce growth arrest (Richter et al, 2013), rather than mtDNA depletion.
Concomitant knockdown of mitochondrial and cytosolic ribosomal protein genes produced no rescue of cell-death phenotypes, and no obvious rescue of PicoGreen nucleoid signals (Table 1). Mitonuclear imbalance thus does not seem to explain the identification of cytosolic ribosomal protein genes as positives in the study, despite evidence for its involvement in other phenomena, such as aging (Gomes et al, 2013; Houtkooper et al, 2013).
The proteasome represents a crucial intracellular system for turnover of damaged or misfolded proteins, and for post-translational regulation. Proteasomal impairment in yeast has previously been shown to entrain mtDNA instability (Malc et al, 2009). A possible explanation is that proteasomal insufficiency signals the activation of other turnover pathways impacting mitochondria, for example, via the Pink1/parkin system (Narendra et al, 2012). The proteasome may also have more specific targets relevant to mitochondrial homeostasis, as suggested by the identification of a deubiquitinating enzyme (CG5794) as a positive. Two of its yeast homologues, UBP9 and UBP13, regulate mtDNA maintenance via effects on the biosynthesis of ATP synthase (Kanga et al, 2012), although the analogy cannot be exact, since the target is mtDNA-encoded in yeast, but nuclear-coded in animals. UBP9/13 and CG5794 are not orthologous, but the underlying principle may be similar. Known targets of ubiquitylation in mitochondria include proteins involved in mitochondrial dynamics, such as mitofusins (Yonashiro et al, 2006; Cohen et al, 2008). The closest homologues of CG5794 in yeast and human perform diverse tasks related to peroxisomal protein import (Debelyy et al, 2011), cell-cycle regulation (Bozza & Zhuang, 2011), methylmercury resistance (Hwang et al, 2012), nuclear DNA repair (Sy et al, 2013), and signaling (Lui et al, 2011; Poalas et al, 2013).
ATP synthase
The primary screen yielded 5 of the 14 nuclear genes encoding subunits of ATP synthase, once again far above random expectation (P < 0.01, χ2 test). After rescreening, 9 ATP synthase genes were classed as definitive positives (Table 1), including subunits of both the FO and F1 subcomplexes: F1 core subunit β, stalk subunits γ and δ, stator arm subunits b, d, OSCP and Cf6, and FO subunits e and g, implicated in dimerization. F1 core subunit α and stator arm subunit f were negative, while stalk subunit ε and FO subunit c were not consistently positive and thus appear in Supplementary Table S4, although may better be considered as weak positives. A putative assembly factor for ATP synthase, the homologue of yeast Atp12, plus the two isogenes (CG13551 and CG34423) for the inhibitor subunit ATPIF1 were negative.
The requirement for ATP synthase (OXPHOS complex V, cV) to maintain normal mtDNA copy number mirrors observations in yeasts (Giraud & Velours, 1997; Lai-Zhang et al, 1999; Clark-Walker et al, 2000; Contamine & Picard, 2000; Lefebvre-Legendre et al, 2003; Duvezin-Caubet et al, 2006; Wang et al, 2007) and trypanosomes (Schnaufer et al, 2005). This suggests the operation of a universal mechanism, whereby cV knockdown generates a signature of bioenergetic stress, signaling a decrease in mitochondrial biogenesis and/or an increase in mitochondrial turnover. Deficiency of ATP synthase would be expected to disturb mitochondrial membrane potential, impairing the import of nuclear-coded OXPHOS subunits and leading to downregulation of mtDNA as a response to limit proteotoxic stress inside mitochondria. ATP synthase mutations that result in uncoupling are known to lead to mtDNA instability in yeast (Wang et al, 2007), and genes encoding subunits of the mitochondrial import machinery can suppress the petite negativity of yeast mutants lacking ATP synthase (Dunn & Jensen, 2003).
Another predictable effect of ATP synthase knockdown would be an increase in mitochondrial ROS production, resulting from inhibition of electron flow in the respiratory chain and over-reduction of its various electron carriers. To elucidate the mechanism of mtDNA depletion associated with cV knockdown, we therefore measured mitochondrial membrane potential and ROS production by flow cytometry, using the fluorescent probes TMRM and MitoSox, respectively.
To minimize secondary effects, we selected genes whose knockdown produced moderate mtDNA copy number depletion, confirming first that their relative knockdown potency correlated with the amount of mtDNA depletion produced (Fig 3A, Supplementary Fig S2A). Knockdown of either of the two clear positives (CG6105, subunit g, and CG2968, subunit δ), plus one that gave only a weak knockdown and minimal mtDNA depletion (CG9032, subunit ε), produced coherent effects (Fig 3B and C). In all cases, there was a small but significant decrease in TMRM fluorescence per cell and a more variable increase in MitoSox fluorescence, whereas these parameters were unaffected by knockdown of mTTF, which acts directly on mtDNA transcription (Roberti et al, 2006) and replication (Jõers et al, 2013).
Figure 3. Effects of knockdown of cV subunits.
- A–C Relative mtDNA level (A), TMRM fluorescence (B) and MitoSox fluorescence (C) following treatment for 5 days with dsRNA against the genes indicated.
- D MitoSox fluorescence following treatment for 1–3 days with dsRNA against the genes indicated.
- E Whole cell respiration following treatment for 5 days with dsRNA against the genes indicated.
- F, G Relative mtDNA level (F) and MitoSox fluorescence (G) after 5 days of treatment with oligomycin.
Data information: All data normalized to untreated cells. Note that GFP is a negative control, since the cells have no GFP gene, whereas mTTF is a positive control. Means ± SD from at least 4 independent experiments each conducted in triplicate. Asterisks (*) or hashes (#) indicate significant differences from untreated and from GFP values (P < 0.01 and P < 0.05, respectively). Note that correction for the decrease in mitochondrial content mitigates the decrease in TMRM fluorescence, but increases still further the MitoSox fluorescence (Supplementary Fig S4D).
Source data are available online for this figure.
Based on successive measurements over the first 3 days of dsRNA treatment (Fig 3D), the effect on MitoSox fluorescence also reflected the different potencies of knockdown, with CG2968 (subunit δ) producing the most rapid, substantial and sustained ROS increase. Knockdown of ATP synthase also decreased whole cell respiration (Fig 3E). Prolonged treatment with a sublethal dose of oligomycin (50 nM, 5 days) phenocopied the effect of cV knockdown on both mtDNA level (Fig 3F) and MitoSox fluorescence (Fig 3G), implicating the enzymatic activity of cV, rather than any structural role in maintaining cristal architecture (Davies et al, 2012). Oligomycin binds the c ring of cV, most likely at its interface with subunit a (Symersky et al, 2008), so is unlikely to disturb the dimeric structure of the complex, upon which its membrane-deforming action depends.
We next targeted single-copy genes encoding subunits of the respiratory chain, all of which were negative in the initial screen. When genes for subunits of complex I (CG6020, subunit NDUFA9) or complex IV (CG14724, subunit COX5A) were knocked down individually, there was no significant effect on mtDNA copy number (Supplementary Fig S2B), despite a decrease in TMRM fluorescence (Supplementary Fig S2C) and in whole cell respiration (Supplementary Fig S2D), similar to that seen after cV knockdown (compare with Fig 3B and E). Knockdown of cI subunit CG6020 produced a small, though non-significant increase in MitoSox fluorescence (Supplementary Fig S2E), while knockdown of cIV subunit CG14724 resulted in a significant decrease thereof.
Simultaneous knockdown of subunits of cV and either cI (Fig 4A) or cIV (Fig 4B) abolished the copy number decrease seen when cV alone was targeted (compare with Fig 3A) and prevented the concomitant ROS increase (Fig 4C and D). In contrast, mtDNA depletion by mTTF knockdown was not altered by simultaneous knockdown of cI or cIV subunits. Concomitant knockdown of CG2968 (cV) with genes coding for other subunits of cI or cIV also reversed mtDNA depletion and mitigated ROS increase (Supplementary Fig S3A and B). Knockdown of subunits of cI alone did produce a small drop in mtDNA copy number (Supplementary Fig S3C) and an increase in ROS (Supplementary Fig S3D), though these changes were not always statistically significant and were comparable with those produced by knockdown of CG9032 (cV subunit ε), thus probably too small for these genes to have been scored as positives in the primary screen.
Figure 4. Effects of knockdown of cV subunits in combination with other treatments.
- A–D Relative mtDNA level (A, B) and MitoSox fluorescence (C, D) following 5 days of treatment with dsRNA against the genes indicated.
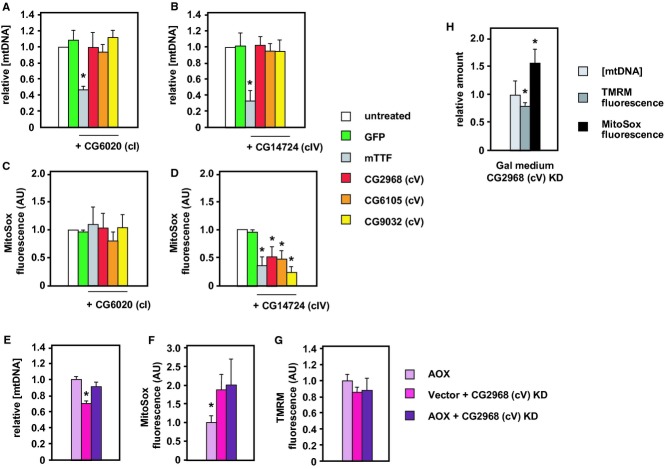
- E–G Relative mtDNA level (E), MitoSox fluorescence (F) and TMRM fluorescence (G) of cells stably expressing AOX or empty vector, as indicated, after 5 days of treatment with dsRNA against CG2968, as shown. Note that these cells were cultured in the presence of hygromycin to maintain the AOX-expressing or control plasmid.
- H Indicated parameters, following 5 days of treatment with dsRNA against CG2968, in cells grown in galactose-containing (Gal) medium.
Data information: All data normalized to untreated cells or (E, F, G) AOX-expressing but otherwise untreated cells grown in parallel. Means ± SD from at least 4 independent experiments, each conducted in triplicate. Asterisks (*) indicate significant differences between experimental and control cell values (P < 0.01).
Source data are available online for this figure.
Overall, these findings indicate increased mitochondrial ROS production as the most consistent marker linking cV knockdown and mtDNA depletion. Accordingly, treatment with the antioxidant TBAP, a chemical mimetic of superoxide dismutase (Faulkner et al, 1994), produced a small but statistically significant mitigation of both effects (Supplementary Fig S3E and F), while TBAP alone produced small effects in the opposite direction. However, increased mitochondrial ROS is not sufficient to induce mtDNA depletion. Stable expression of the Ciona intestinalis alternative oxidase (Fig 4E–G), or growth of cells in medium containing galactose in place of glucose (Fig 3H), enforcing the use of OXPHOS to produce ATP (Robinson et al, 1992), blocked copy number depletion, but preserved the changes in MitoSox (and TMRM) fluorescence produced upon cV knockdown. Copy number depletion must therefore depend on the integration of different metabolic signals.
To investigate the mechanism of mtDNA depletion, we analyzed mitochondrial turnover, using vital dyes for both mitochondria and lysosomes, employing flow cytometry (Fig 5A) and confocal microscopy (Fig 5B and C). Fluorescence per cell of both LysoTracker Red and NAO, a vital dye for the inner mitochondrial membrane, was unaffected by 5 days of dsRNA treatment against mTTF or GFP, but knockdown of cV (CG2968) increased LysoTracker staining by 50%, while halving the NAO signal (Fig 5A). Western blotting indicated a general decrease in mitochondrial content, affecting cI as well as cV (Fig 5D).
Figure 5. Effects of cV knockdown on mitochondrial and lysosomal content.
- LysoTracker Red and NAO fluorescence of cells treated as indicated for 5 days with dsRNA against the indicated genes, normalized against the values for untreated cells. Mean ± SD for four independent experiments; asterisks indicate significant differences from untreated, P < 0.05.
- Representative confocal microscopy images of living cells treated for 5 days with dsRNA against the indicated genes and stained with both MitoTracker Green and LysoTracker Red. The scale bar is 10 μm.
- Mean areas of LysoTracker red staining in cells treated for 5 days with dsRNA against the indicated genes, or with chloroquine. Mean ± SD for three independent experiments, in each of which ≥ 50 cells were analyzed by confocal microscopy in a single plane. Analysis of z-stacks in ≥ 30 cells gave similar results. Note that the increase in lysosome content was intermediate between that of untreated cells and cells treated with chloroquine to block lysosomal turnover of autophagosomes. Asterisks indicate significant differences from untreated, P < 0.01.
- Western blot of total protein extracts from control S2 cells and cells treated with dsRNAs against GFP or CG2968 (cV), probed for NDUFS3 (cI), ATP synthase subunit α (cV) and GAPDH (loading control).
- Relative mtDNA level after 5 days of treatment with dsRNAs against the indicated genes. Mean ± SD for five independent experiments; other data classes were significantly different from untreated, P < 0.05.
Source data are available online for this figure.
Live imaging confirmed the increase in lysosomal signal per cell (Fig 5B and C); MitoTracker Green staining revealed increased mitochondrial fragmentation and a greatly enhanced colocalization of LysoTracker and MitoTracker signal in most cells (Fig 5B). Concomitant knockdown of three core components of the canonical autophagy machinery was unable to block mtDNA depletion (Fig 5E), indicating that other turnover pathways are involved, such as retrotranslocation coupled to proteasomal degradation (Margineantu et al, 2007; Heo et al, 2010), mitochondrial vesicle delivery to lysosomes, via Pink1/parkin signaling (McLelland et al, 2014), or intramitochondrial turnover pathways (see following section).
Co-expression of AOX, which blocked the decrease in mtDNA copy number caused by cV knockdown, attenuated the increase in LysoTracker signal (Supplementary Fig S4A), while chloroquine, which prevents lysosomal acidification, partially rescued mtDNA depletion (Supplementary Fig S4B). These findings are consistent with lysosomal involvement, but do not exclude a contribution from other pathways. Persistent heat stress produced a qualitatively similar effect (Supplementary Fig S4C). Thus, disturbance of the balance between ATP synthase and the respiratory chain complexes may be just one of many processes that produce this outcome. However, given the observations in yeast discussed above, we propose that it represents a conserved homeostatic mechanism for renewal of mitochondria under a variety of metabolic stress conditions.
Mitochondrial biogenesis and homeostasis
Very few mitochondrial proteins appeared in the ‘cell death only’ list (Supplementary Table S2), confirming that their broad absence from the main positives list was not due to their being essential for cell survival. Positives that we did find were opa1-like (orthologue of human OPA1 and yeast MGM1), CG6512 (m-AAA protease complex) and maggie (TOMM22), plus three fly homologues of genes for transcriptional activators or co-activators implicated in regulating mitochondrial biogenesis: erect wing (NRF-1), Ets97D (α subunit of NRF-2), and spargel (PGC-1α). Knockdown of these genes typically produced a modest mtDNA depletion, consistent with the idea that mitochondrial content and quality depends on the balance between mitochondrial turnover and biogenesis, which are independently regulated. The role of mitochondrial quality control in pathological states is already well documented (Martinelli & Rugarli, 2007; Ranieri et al, 2013; Celardo et al, 2014). Clearance of damaged mitochondria is crucial to protection from somatic mtDNA mutations, whereas it may compound defects caused by inherited mutations.
The mitochondrial protein import machinery is required for cell viability because of the essential functions of mitochondria. Therefore, it is surprising that only a single component of this machinery scored positive in the study. Despite extensive functional studies (Shiota et al, 2011), there is little to indicate any special property of TOM22 that may explain this finding, although it has been implicated as a site of regulation of the entire TOM complex by cytosolic protein kinases (Schmidt et al, 2011). It was recently linked to aging (Joseph et al, 2012) and appears to be a specific target of parkin in promoting mitophagy (Bertolin et al, 2013).
Similar questions arise for CG6512. Mammalian OPA1 can be processed by both the m-AAA (Ehses et al, 2009) and i-AAA proteases (Song et al, 2007), and in Drosophila also by the rhomboid protease rho-7 (Rahman & Kylsten, 2011). OPA1 or its isoforms have been previously implicated in mitochondrial genome maintenance (Herlan et al, 2003; Sesaki et al, 2003; Elachouri et al, 2011), although there is no compelling evidence of a direct interaction with mtDNA. The general consensus is that it acts via its documented roles in mitochondrial quality control, affecting cristal morphology and innermembrane fusion (Frezza et al, 2006; Meeusen et al, 2006), as well as metabolism and calcium homeostasis (Kushnareva et al, 2013).
Knockdown of rho-7, CG2658 (paraplegin-like m-AAA component), or CG3499 (i-AAA), all of which were negative in the original screen, produced no significant effects on mtDNA levels (Supplementary Fig S5A). However, we found that these genes, together with opa1-like and CG6512, participate in a mutually interactive network, such that knockdown of any component regulates the others at the RNA level (Supplementary Fig S5B). In particular, knockdown of opa1-like entrained the upregulation of all 4 proteases, while knockdown of CG6512 upregulated CG2658 and CG4399, suggesting a common pathway modulating mitochondrial biogenesis or turnover. Mitochondrial proteases were also upregulated when cV was knocked down.
Nuclear gene expression
Knockdown of most of the previously characterized components of the apparatus of nuclear gene expression that were positive in the screen (category 6 genes in Table 1) produced no significant change in mtDNA copy number. Since most of them are required for cell survival, a mitochondrial defect may therefore be no more than a staging post on the road to cell death. However, downregulation of six genes, RpII215, Art2, pps, omd, Taf12, and Taf1, with documented or hypothesized roles in chromatin modification, Pol II transcription, or RNA processing, led to decreased mtDNA copy number. A possible role in mtDNA metabolism independent from their functions in the nucleus thus needs to be carefully evaluated.
CG10582 (Sin, Sex-lethal interactor) may also be considered a representative of this group. It was originally identified in a yeast 2-hybrid screen as a partner of the splicing factor Sex lethal (Dong & Bell, 1999), but its orthologues are subunits of RNA polymerase III (Hu et al, 2002; Cramer et al, 2008), specifically POL3E, the Pol III homologue of TFIIF (Carter & Drouin, 2010; Kassavetis et al, 2010), required for termination (Landrieux et al, 2006). No other subunits of RNA polymerase III were detected in the screen. One Sin isoform (weakly) predicted to be mitochondrial was also positive in the PicoGreen assay, as was pps (protein partner of snf), also implicated in alternative splicing of Sex lethal (Johnson et al, 2010).
pointed (CG17077) has 4 isoforms created by differential splicing, though none is predicted with high confidence to be mitochondrial. It exhibits functional redundancy in specific contexts (Baltzer et al, 2009) with its homologue Ets97D (NRF-2α), suggesting that the canonical functions of the latter in regulating mitochondrial biogenesis might be shared by the two transcription factors in Drosophila.
Miscellaneous positives: ‘unknowns’ and ‘others’
This heterogeneous group mainly comprises poorly characterized proteins. In addition to several putatively implicated in RNA metabolism, plus a number of zinc finger proteins, they include three that function in vesicle trafficking. Bet1 (CG14084) and Slh (CG3539), conserved from yeast to humans, are members of the syntaxin 5-SNARE complex, involved in ER to Golgi transport (Newman et al, 1990; Hay et al, 1997). Bet1 is a SNARE protein, while Slh, orthologue of mammalian and yeast Sly1 (Dascher et al, 1991; Sogaard et al, 1994; Dascher & Balch, 1996), is a SNARE-interacting SM-family protein. Knockdown of each produced no significant mtDNA depletion, indicating that loss of PicoGreen nucleoid signal is due to something other than decreased copy number. CG10144 is the Drosophila orthologue of yeast (and human) Vps8, involved in vesicle sorting to lysosomes (Chen & Stevens, 1996; Horazdovsky et al, 1996). Our findings suggest a novel role for vesicle trafficking in mitochondrial homeostasis.
Aside from these, no specific pathway or cellular complex is identified more than once in the list. Knockdown mainly produced little or no mtDNA depletion, although three further positives that did so warrant further study, namely IM4, previously implicated in innate immunity, CG14634 and CG32652, all of which encode proteins with no identified homologues outside of insects.
Conclusions
In this study, we use a genome-wide screen to identify novel components of the machinery of mtDNA copy number maintenance and regulation. Most positives belonged to coherent sets, as discussed above (summarized in Fig 6). Unexpectedly, absent from the positives list was any gene involved in mitochondrial protein synthesis or OXPHOS assembly, most known signaling pathways, any subunit of the respiratory chain complexes or of intermediary metabolism, genes involved in handling oxidative stress, or in nucleotide transport and metabolism, which cause mtDNA depletion syndromes in humans (Saada, 2004; Copeland, 2012). The latter is not surprising, since these genes are of pathological importance only in non-proliferating cells, where the standard pathways sustaining the supply of precursors for both nuclear and mtDNA synthesis are unavailable. However, apart from POLG and Twinkle (PEO1), the homologues of most other human mtDNA depletion genes, including MPV17 (Spinazzola et al, 2006), FBXL4 (Bonnen et al, 2013), and SERAC1 (Sarig et al, 2013), were also absent, although some may be false negatives. Others, such as MGME1 (Kornblum et al, 2013), have no Drosophila homologue.
Figure 6. Pathways inferred to be involved in mitochondrial DNA copy number maintenance and homeostasis.
Schematic diagram of the cell, showing categories of identified genes in the positives list (in dark blue), and their possible links to a ROS (or membrane potential, Δψ) surveillance mechanism, orchestrated by ATP synthase.
On the other hand, few novel candidates emerged, for direct involvement in DNA metabolism, despite the burgeoning number of proteins said to be present in nucleoids (Reyes et al, 2011; Bogenhagen, 2012). The involvement of cV, the proteasome, and some key genes for mitochondrial dynamics and quality control suggests that mtDNA copy number is dependent on a balance between mitochondrial turnover and biogenesis, in which specific stresses, notably mitochondrial ROS production and impaired cytosolic protein turnover, may be crucially important.
ATP synthase was inferred to be a key player in homeostatic maintenance of mitochondria, and thus in the amount of mtDNA and its gene products. Mutations in ATP synthase in fungi are already known to play a determining role in whether mtDNA loss can be tolerated (Contamine & Picard, 2000; Lefebvre-Legendre et al, 2003), or even facilitated (Giraud & Velours, 1997; Lai-Zhang et al, 1999; Contamine & Picard, 2000), but with membrane potential implicated as a key parameter (Duvezin-Caubet et al, 2006; Wang et al, 2007). In S2 cells, excess ROS production was better correlated with the strength and kinetics of mtDNA depletion, but this could be an epi-phenomenon. The fact that membrane potential ‘per mitochondrion’ (Supplementary Fig S4D) was restored to its starting value suggests that its disturbance may yet prove to be the primary inducer.
Excess ROS has elsewhere been proposed to lead to mtDNA depletion under pathological conditions (Larosche et al, 2010; Quinzii et al, 2013), and pathological defects in ATP synthase associated with ROS overproduction (Baracca et al, 2007) may downregulate mitochondrial functions (Wojewoda et al, 2010, 2011) and even lead to mtDNA loss (Vergani et al, 1999; Turner et al, 2005). ROS overproduction has been widely suggested both to provoke mtDNA damage, but also to result from it. However, a recent report showed that unrepaired damage leads to mtDNA depletion without increased ROS (Shokolenko et al, 2013), and the role of ROS in producing somatic mtDNA mutations in the PolgA mutator mouse is disputed (Trifunovic et al, 2005; Dai et al, 2010). Thus, the increased mitochondrial ROS seen when cV is knocked down is more logically a cause than a consequence of mtDNA depletion. Furthermore, although ROS may provoke strand breakage, interfering directly with mtDNA replication (Han & Chen, 2013), our data instead suggest that ROS activates mitochondrial turnover before widespread DNA damage would be sustained, as occurs in mammalian cells under TNFα signaling (Nagakawa et al, 2005; Vadrot et al, 2012). However, an opposing pathway has also been suggested, in which ROS over-production promotes mitochondrial biogenesis, not turnover (Moreno-Loshuertos et al, 2006, 2011).
A key aim of future research will be to identify the sensor molecule(s) integrating changes in mitochondrial ROS (or membrane potential) with other metabolic signals, in order to modulate mitochondrial biogenesis and turnover. One possibility consistent with our data is that ATP synthase itself is that sensor.
Materials and Methods
Cell maintenance
Drosophila S2 cells (Invitrogen) were cultured under standard conditions, in Schneider′s medium (Sigma) and diluted 1:6 every 3-4 d. In selected experiments, various drugs were added or glucose was replaced with the same concentration of galactose. S2 cells stably expressing Ciona intestinalis AOX were generated by co-transfection with a plasmid conferring hygromycin resistance.
Screening of Drosophila dsRNA library and fluorescence microscopy of nucleoids
S2 cells were seeded into 96-well plates and treated over 5 days with 0.6–1.2 μg of dsRNA from the library (Open BioSystems), alongside positive and negative controls in each plate, as described previously (Jõers et al, 2013). Nucleoids were visualized by fluorescence microscopy after staining with Quant-iT™ PicoGreen® dsDNA reagent (7.5 μl/ml, Invitrogen). Larger-scale dsRNA treatments for mtDNA copy number evaluation and analysis of cellular parameters were performed essentially as previously (Jõers et al, 2013).
Nucleic acid isolation and QPCR
Total RNA was isolated from S2 cells as previously (Jõers et al, 2013). For DNA isolation, cells from a single well of a 24-well plate were processed by a procedure determined to give consistent results irrespective of cell density, involving SDS lysis, proteinase K digestion, isopropanol precipitation and overnight resuspension at 55°C (see SI). For larger-scale experiments, DNA was prepared from 1.5 × 106 cells cultured in 6-well plates, as previously (Jõers et al, 2013). Mitochondrial DNA copy number was assessed by QPCR using primers against COXII or 16S rRNA (for mtDNA) and RpL32 (nuclear DNA, single-copy, for normalization). Transcript levels were estimated relative to that of RpL32 by a similar procedure, but using cDNA as template.
Measurements of mitochondrial function
Mitochondrial membrane potential, ROS level, and content per cell were determined by flow cytometry (Cannino et al, 2012) of cells stained, respectively, with 200 mM tetramethylrhodamine methyl ester (TMRM), 2.5 μM MitoSox™ (Invitrogen), or either 200 nM 10-nonyl acridine orange (NAO) or 40 nM MitoTracker® Green FM (Life Technologies). Oxygen consumption of living cells (Cannino et al, 2012) was measured using a Clark-type electrode (Hansatech Oxyterm system).
Analyses of lysosomal and mitochondrial content
Lysosome content per cell was measured by flow cytometry of cells stained with 50 nM LysoTracker® Red DND-99 (Life Technologies). MitoTracker® Green FM (Molecular Probes) and LysoTracker Red DND-99 were used for live cell imaging by confocal microscopy of mitochondria and lysosomes, respectively, with spot-area calculation (ImageJ) and image deconvolution (SVI, Huygens software).
Western blotting
Post-nuclear extracts resolved by SDS-PAGE were electroblotted and probed using standard methods (essentially as Fernandez-Ayala et al, 2009; see SI). Primary antibodies used were against NDUFS3 (Abcam, mouse), 1:10,000), ATP5A (Abcam, mouse, 1:1,000), and GAPDH (Everest Biotech, goat, 1:2,000), with appropriate horseradish peroxidase-conjugated secondary antibodies. Visualization used the ECL system (Amersham Biosciences) according to the manufacturer's protocols.
Statistical analyses
Comparisons between populations were performed using unpaired two-tailed Student's t-tests or analyses of variance when more than two samples were compared, with Bonferroni-corrected post hoc t-test.
For further details, see Supplementary Materials and Methods.
Data availability
Original images from the primary screen are deposited at: http://dx.doi.org/10.5061/dryad.v55p5.
Acknowledgments
This work was supported by funding from the Academy of Finland, Tampere University Hospital Medical Research Fund, and the Sigrid Juselius Foundation. AF was supported by FY 2008 Researcher Exchange Program between JSPS and Academy of Finland. We thank Tea Tuomela, Outi Kurronen, Hanna Ojala, and Eveliina Kaulio for technical assistance, and Susanna Valanne, Mika Rämet, Ian Holt, Cory Dunn, Brendan Battersby, Anu Suomalainen, and Laurie Kaguni for useful discussions and advice.
Author contributions
AF initiated the project, conducted the primary screen, and supervised the implementation and interpretation of the secondary screening. GC conducted the functional analysis of ATP synthase knockdown. MG performed the copy number analysis and profiled the expression of mitochondrial proteases and opa1-like at the RNA level. SB and SK assisted technically with copy number analysis. EL and AR devised the computational tool for objectifying the PicoGreen fluorescence signals (details to be published elsewhere). FS generated the AOX-expressing S2 cells. ED provided guidance and input for ATP synthase experiments. HTJ devised the project, analyzed the data, compiled the figures and tables, and wrote the text.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary information for this article is available online: http://msb.embopress.org
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Materials and Methods
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S2
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S5
Review Process File
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
References
- Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity–critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley N, Harris D, Poulton J. Detection of mitochondrial DNA depletion in living human cells using PicoGreen staining. Exp Cell Res. 2005;303:432–446. doi: 10.1016/j.yexcr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Baltzer C, Tiefenböck SK, Marti M, Frei C. Nutrition controls mitochondrial biogenesis in the Drosophila adipose tissue through Delg and cyclin D/Cdk4. PLoS ONE. 2009;4:e6935. doi: 10.1371/journal.pone.0006935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracca A, Sgarbi G, Mattiazzi M, Casalena G, Pagnotta E, Valentino ML, Moggio M, Lenaz G, Carelli V, Solaini G. Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993. Biochim Biophys Acta. 2007;1767:913–919. doi: 10.1016/j.bbabio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Bertolin G, Ferrando-Miguel R, Jacoupy M, Traver S, Grenier K, Greene AW, Dauphin A, Waharte F, Bayot A, Salamero J, Lombès A, Bulteau AL, Fon EA, Brice A, Corti O. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy. 2013;9:1801–1817. doi: 10.4161/auto.25884. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819:914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Bonnen PE, Yarham JW, Besse A, Wu P, Faqeih EA, Al-Asmari AM, Saleh MA, Eyaid W, Hadeel A, He L, Smith F, Yau S, Simcox EM, Miwa S, Donti T, Abu-Amero KK, Wong LJ, Craigen WJ, Graham BH, Scott KL, et al. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93:471–481. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N Heidelberg Fly Array Consortium. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- Bozza WP, Zhuang Z. Biochemical characterization of a multidomain deubiquitinating enzyme Ubp15 and the regulatory role of its terminal domains. Biochemistry. 2011;50:6423–6432. doi: 10.1021/bi200529z. [DOI] [PubMed] [Google Scholar]
- Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123:951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni F, Gramegna P, Oliveira JM, Lightowlers RN, Chrzanowska-Lightowlers ZM. REXO2 is an oligoribonuclease active in human mitochondria. PLoS ONE. 2013;8:e64670. doi: 10.1371/journal.pone.0064670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM, Stith CM, Yoder BL, Sparks JL. Yeast exonuclease 5 is essential for mitochondrial genome maintenance. Mol Cell Biol. 2010;30:1457–1466. doi: 10.1128/MCB.01321-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CT, Kolesar JE, Kaufman BA. Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number. Biochim Biophys Acta. 2012;1819:921–929. doi: 10.1016/j.bbagrm.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Cannino G, El-Khoury R, Pirinen M, Hutz B, Rustin P, Jacobs HT, Dufour E. Glucose modulates respiratory complex I activity in response to acute mitochondrial dysfunction. J Biol Chem. 2012;287:38729–38740. doi: 10.1074/jbc.M112.386060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Drouin G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol. 2010;27:1035–1043. doi: 10.1093/molbev/msp316. [DOI] [PubMed] [Google Scholar]
- Celardo I, Martins LM, Gandhi S. Unravelling mitochondrial pathways to Parkinson's disease. Br J Pharmacol. 2014;171:1943–1957. doi: 10.1111/bph.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM, Frolova EG, Feng C, Grinberg A, Love PE, Crouch RJ. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- Chen TC, Hung YC, Lin TY, Chang HW, Chiang IP, Chen YY, Chow KC. Human papillomavirus infection and expression of ATPase family AAA domain containing 3A, a novel anti-autophagy factor, in uterine cervical cancer. Int J Mol Med. 2011;28:689–696. doi: 10.3892/ijmm.2011.743. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Stevens TH. The VPS8 gene is required for localization and trafficking of the CPY sorting receptor in Saccharomyces cerevisiae. Eur J Cell Biol. 1996;70:289–297. [PubMed] [Google Scholar]
- Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, Suzuki T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–8047. doi: 10.1093/nar/gks506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielski GL, Plotka M, Manicki M, Schilke BA, Dutkiewicz R, Sahi C, Marszalek J, Craig EA. Nucleoid localization of Hsp40 Mdj1 is important for its function in maintenance of mitochondrial DNA. Biochim Biophys Acta. 2013;1833:2233–2243. doi: 10.1016/j.bbamcr.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker GD, Hansbro PM, Gibson F, Chen XJ. Mutant residues suppressing rho(0)-lethality in Kluyveromyces lactis occur at contact sites between subunits of F(1)-ATPase. Biochim Biophys Acta. 2000;1478:125–137. doi: 10.1016/s0167-4838(00)00003-0. [DOI] [PubMed] [Google Scholar]
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. Ubiquitin-Proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol Biol Cell. 2008;19:2457–2464. doi: 10.1091/mbc.E08-02-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contamine V, Picard M. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol Mol Biol Rev. 2000;64:281–315. doi: 10.1128/mmbr.64.2.281-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC. Defects in mitochondrial DNA replication and human disease. Crit Rev Biochem Mol Biol. 2012;47:64–74. doi: 10.3109/10409238.2011.632763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Armache KJ, Baumli S, Benkert F, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, Jawhari A, Jennebach S, Kamenski T, Kettenberger H, Kuhn CD, Lehmann E, Leike K, Sydow JF, Vannini A. Structure of eukaryotic RNA polymerases. Annu Rev Biophys. 2008;37:337–352. doi: 10.1146/annurev.biophys.37.032807.130008. [DOI] [PubMed] [Google Scholar]
- Dai DF, Chen T, Wanagat J, Laflamme M, Marcinek DJ, Emond MJ, Ngo CP, Prolla TA, Rabinovitch PS. Age-dependent cardiomyopathy in mitochondrial mutator mice is attenuated by overexpression of catalase targeted to mitochondria. Aging Cell. 2010;9:536–544. doi: 10.1111/j.1474-9726.2010.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Balch WE. Mammalian Sly1 regulates syntaxin 5 function in endoplasmic reticulum to Golgi transport. J Biol Chem. 1996;271:15866–15869. doi: 10.1074/jbc.271.27.15866. [DOI] [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debelyy MO, Platta HW, Saffian D, Hensel A, Thoms S, Meyer HE, Warscheid B, Girzalsky W, Erdmann R. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem. 2011;286:28223–28234. doi: 10.1074/jbc.M111.238600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z, Bell LR. SIN, a novel Drosophila protein that associates with the RNA binding protein sex-lethal. Gene. 1999;237:421–428. doi: 10.1016/s0378-1119(99)00303-0. [DOI] [PubMed] [Google Scholar]
- Dunn CD, Jensen RE. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 2003;165:35–45. doi: 10.1093/genetics/165.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Rak M, Lefebvre-Legendre L, Tetaud E, Bonnefoy N, di Rago JP. A “petite obligate” mutant of Saccharomyces cerevisiae: functional mtDNA is lethal in cells lacking the delta subunit of mitochondrial F1-ATPase. J Biol Chem. 2006;281:16305–16313. doi: 10.1074/jbc.M513805200. [DOI] [PubMed] [Google Scholar]
- Duxin JP, Dao B, Martinsson P, Rajala N, Guittat L, Campbell JL, Spelbrink JN, Stewart SA. Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol Cell Biol. 2009;29:4274–4282. doi: 10.1128/MCB.01834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, Martinou JC, Westermann B, Rugarli EI, Langer T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elachouri G, Vidoni S, Zanna C, Pattyn A, Boukhaddaoui H, Gaget K, Yu-Wai-Man P, Gasparre G, Sarzi E, Delettre C, Olichon A, Loiseau D, Reynier P, Chinnery PF, Rotig A, Carelli V, Hamel CP, Rugolo M, Lenaers G. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21:12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner KM, Liochev SI, Fridovich I. Stable Mn(III) porphyrins mimic superoxide dismutase in vitro and substitute for it in vivo. J Biol Chem. 1994;269:23471–23476. [PubMed] [Google Scholar]
- Fernandez-Ayala DJ, Sanz A, Vartiainen S, Kemppainen KK, Babusiak M, Mustalahti E, Costa R, Tuomela T, Zeviani M, Chung J, O'Dell KM, Rustin P, Jacobs HT. Expression of the Ciona intestinalis alternative oxidase (AOX) in Drosophila complements defects in mitochondrial oxidative phosphorylation. Cell Metab. 2009;9:449–460. doi: 10.1016/j.cmet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Fuste JM, Wanrooij S, Jemt E, Granycome CE, Cluett TJ, Shi Y, Atanassova N, Holt IJ, Gustafsson CM, Falkenberg M. Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol Cell. 2010;37:67–78. doi: 10.1016/j.molcel.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–244. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Gómez S, Reyes A, Martínez-Jiménez MI, Chocrón ES, Mourón S, Terrados G, Powell C, Salido E, Méndez J, Holt IJ, Blanco L. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52:541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilquin B, Taillebourg E, Cherradi N, Hubstenberger A, Gay O, Merle N, Assard N, Fauvarque MO, Tomohiro S, Kuge O, Baudier J. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol Cell Biol. 2010;30:1984–1996. doi: 10.1128/MCB.00007-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud MF, Velours J. The absence of the mitochondrial ATP synthase delta subunit promotes a slow growth phenotype of rho- yeast cells by a lack of assembly of the catalytic sector F1. Eur J Biochem. 1997;245:813–818. doi: 10.1111/j.1432-1033.1997.00813.x. [DOI] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto A, Matsushima Y, Kadowaki T, Kitagawa Y. Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem J. 2001;354:243–248. doi: 10.1042/0264-6021:3540243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Middlebrooks BW, Alexander S, Wasserman SA. Mutation of TweedleD, a member of an unconventional cuticle protein family, alters body shape in Drosophila. Proc Natl Acad Sci USA. 2006;103:16794–16799. doi: 10.1073/pnas.0607616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Chen JZ. Oxidative stress induces mitochondrial DNA damage and cytotoxicity through independent mechanisms in human cancer cells. Biomed Res Int. 2013;2013:825065. doi: 10.1155/2013/825065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, Walker JE, Holt IJ. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res. 2012;40:6109–6121. doi: 10.1093/nar/gks266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Mao CC, Reyes A, Sembongi H, Di Re M, Granycome C, Clippingdale AB, Fearnley IM, Harbour M, Robinson AJ, Reichelt S, Spelbrink JN, Walker JE, Holt IJ. The AAA+ protein ATAD3 has displacement loop binding properties and is involved in mitochondrial nucleoid organization. J Cell Biol. 2007;176:141–146. doi: 10.1083/jcb.200609158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensen F, Cansiz S, Gerhold JM, Spelbrink JN. To be or not to be a nucleoid protein: a comparison of mass-spectrometry based approaches in the identification of potential mtDNA-nucleoid associated proteins. Biochimie. 2014;100:219–226. doi: 10.1016/j.biochi.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, Ashrafi K, Glickman MH, Rutter J. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Honnen S, Mayatepek E, Wätjen W, Koopman WJ, Bossinger O, Distelmaier F. MICS-1 interacts with mitochondrial ATAD-3 and modulates lifespan in C. elegans. Exp Gerontol. 2012;47:270–275. doi: 10.1016/j.exger.2011.12.011. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, Cowles CR, Mustol P, Holmes M, Emr SD. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J Biol Chem. 1996;271:33607–33615. doi: 10.1074/jbc.271.52.33607. [DOI] [PubMed] [Google Scholar]
- Hou D, Ruiz M, Andrulis ED. The ribonuclease Dis3 is an essential regulator of the developmental transcriptome. BMC Genomics. 2012;13:359. doi: 10.1186/1471-2164-13-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol. 2002;22:8044–8055. doi: 10.1128/MCB.22.22.8044-8055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KH, Chow KC, Chang HW, Lin TY, Lee MC. ATPase family AAA domain containing 3A is an anti-apoptotic factor and a secretion regulator of PSA in prostate cancer. Int J Mol Med. 2011;28:9–15. doi: 10.3892/ijmm.2011.670. [DOI] [PubMed] [Google Scholar]
- Humphrey DM, Parsons RB, Ludlow ZN, Riemensperger T, Esposito G, Verstreken P, Jacobs HT, Birman S, Hirth F. Alternative oxidase rescues mitochondria-mediated dopaminergic cell loss in Drosophila. Hum Mol Genet. 2012;21:2698–2712. doi: 10.1093/hmg/dds096. [DOI] [PubMed] [Google Scholar]
- Hwang GW, Kimura Y, Takahashi T, Lee JY, Naganuma A. Identification of deubiquitinating enzymes involved in methylmercury toxicity in Saccharomyces cerevisiae. J Toxicol Sci. 2012;37:1287–1290. doi: 10.2131/jts.37.1287. [DOI] [PubMed] [Google Scholar]
- Ito JK, Braithwaite DK. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 1991;19:4045–4057. doi: 10.1093/nar/19.15.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B, Luo N, Farr CL, Kaguni LS, Campos AR. The accessory subunit of DNA polymerase gamma is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:4483–4488. doi: 10.1073/pnas.072664899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jõers P, Lewis SC, Fukuoh A, Parhiala M, Ellilä S, Holt IJ, Jacobs HT. Mitochondrial transcription terminator family members mTTF and mTerf5 have opposing roles in coordination of mtDNA synthesis. PLoS Genet. 2013;9:e1003800. doi: 10.1371/journal.pgen.1003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Nagengast AA, Salz HK. PPS, a large multidomain protein, functions with sex-lethal to regulate alternative splicing in Drosophila. PLoS Genet. 2010;6:e1000872. doi: 10.1371/journal.pgen.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BA, Fangman WL. Mitochondrial DNA maintenance in yeast requires a protein containing a region related to the GTP-binding domain of dynamin. Genes Dev. 1992;6:380–389. doi: 10.1101/gad.6.3.380. [DOI] [PubMed] [Google Scholar]
- Joseph AM, Adhihetty PJ, Buford TW, Wohlgemuth SE, Lees HA, Nguyen LM, Aranda JM, Sandesara BD, Pahor M, Manini TM, Marzetti E, Leeuwenburgh C. The impact of aging on mitochondrial function and biogenesis pathways in skeletal muscle of sedentary high- and low-functioning elderly individuals. Aging Cell. 2012;11:801–809. doi: 10.1111/j.1474-9726.2012.00844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni LS. DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- Kang D, Hamasaki N. Mitochondrial transcription factor A in the maintenance of mitochondrial DNA: overview of its multiple roles. Ann NY Acad Sci. 2005;1042:101–108. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- Kanga S, Bernard D, Mager-Heckel AM, Erpapazoglou Z, Mattiroli F, Sixma TK, Léon S, Urban-Grimal D, Tarassov I, Haguenauer-Tsapis R. A deubiquitylating complex required for neosynthesis of a yeast mitochondrial ATP synthase subunit. PLoS ONE. 2012;7:e38071. doi: 10.1371/journal.pone.0038071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniely S, Pines O. Single translation–dual destination: mechanisms of dual protein targeting in eukaryotes. EMBO Rep. 2005;6:420–425. doi: 10.1038/sj.embor.7400394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J Biol Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Reyes A, He J, Wood SR, Brea-Calvo G, Holen TT, Holt IJ. A cryptic targeting signal creates a mitochondrial FEN1 isoform with tailed R-Loop binding properties. PLoS ONE. 2013;8:e62340. doi: 10.1371/journal.pone.0062340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaes A, Menne T, Stollewerk A, Scholz H, Klämbt C. The Ets transcription factors encoded by the Drosophila gene pointed direct glial cell differentiation in the embryonic CNS. Cell. 1994;78:149–160. doi: 10.1016/0092-8674(94)90581-9. [DOI] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymäki H, Enwald H, Stöven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Rämet M. Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J. 2005;24:3423–3434. doi: 10.1038/sj.emboj.7600807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Deutscher MP. RNase T shares conserved sequence motifs with DNA proofreading exonucleases. Nucleic Acids Res. 1993;21:2521–2522. doi: 10.1093/nar/21.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum C, Nicholls TJ, Haack TB, Schöler S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Iuso A, Wieland T, Sciacco M, Ronchi D, Comi GP, Moggio M, Quinzii CM, DiMauro S, Calvo SE, Mootha VK, Klopstock T, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45:214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnareva YE, Gerencser AA, Bossy B, Ju WK, White AD, Waggoner J, Ellisman MH, Perkins G, Bossy-Wetzel E. Loss of OPA1 disturbs cellular calcium homeostasis and sensitizes for excitotoxicity. Cell Death Differ. 2013;20:353–365. doi: 10.1038/cdd.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Zhang J, Xiao Y, Mueller DM. Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J. 1999;18:58–64. doi: 10.1093/emboj/18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larosche I, Lettéron P, Berson A, Fromenty B, Huang TT, Moreau R, Pessayre D, Mansouri A. Hepatic mitochondrial DNA depletion after an alcohol binge in mice: probable role of peroxynitrite and modulation by manganese superoxide dismutase. J Pharmacol Exp Ther. 2010;332:886–897. doi: 10.1124/jpet.109.160879. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L, Balguerie A, Duvezin-Caubet S, Giraud MF, Slonimski PP, Di Rago JP. F1-catalysed ATP hydrolysis is required for mitochondrial biogenesis in Saccharomyces cerevisiae growing under conditions where it cannot respire. Mol Microbiol. 2003;47:1329–1339. doi: 10.1046/j.1365-2958.2003.03371.x. [DOI] [PubMed] [Google Scholar]
- Lui TT, Lacroix C, Ahmed SM, Goldenberg SJ, Leach CA, Daulat AM, Angers S. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/β-catenin signaling. Mol Cell Biol. 2011;31:2053–2065. doi: 10.1128/MCB.01094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D, Farr CL, Poeck B, Alahari A, Vogel M, Fischer S, Kaguni LS, Schneuwly S. Mitochondrial single-stranded DNA-binding protein is required for mitochondrial DNA replication and development in Drosophila melanogaster. Mol Biol Cell. 2001;12:821–830. doi: 10.1091/mbc.12.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malc E, Dzierzbicki P, Kaniak A, Skoneczna A, Ciesla Z. Inactivation of the 20S proteasome maturase, Ump1p, leads to the instability of mtDNA in Saccharomyces cerevisiae. Mutat Res. 2009;669:95–103. doi: 10.1016/j.mrfmmm.2009.05.008. [DOI] [PubMed] [Google Scholar]
- Mamolen MC. 2010. Drosophila melanogaster Dis 3 is dynamic endo- and 3′-5′ exoribonuclease. PhD thesis, Case Western Reserve University; permalink http://rave.ohiolink.edu/etdc/view?acc_num=case1278525341.
- Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS ONE. 2007;2:e1066. doi: 10.1371/journal.pone.0001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli P, Rugarli EI. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim Biophys Acta. 2007;1797:1–10. doi: 10.1016/j.bbabio.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Garesse R, Kaguni LS. Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in Schneider cells. J Biol Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- Matsushima Y, Goto Y, Kaguni LS. Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM) Proc Natl Acad Sci USA. 2010;107:18410–18415. doi: 10.1073/pnas.1008924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott-Roe C, Ye J, Ahmed R, Sun XM, Serafín A, Ware J, Bottolo L, Muckett P, Cañas X, Zhang J, Rowe GC, Buchan R, Lu H, Braithwaite A, Mancini M, Hauton D, Martí R, García-Arumí E, Hubner N, Jacob H, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EA, Oliveira MT. Replicating animal mitochondrial DNA. Genet Mol Biol. 2013;36:308–315. doi: 10.1590/S1415-47572013000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acín-Pérez R, Fernández-Silva P, Movilla N, Pérez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enríquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Ferrín G, Acín-Pérez R, Gallardo ME, Viscomi C, Pérez-Martos A, Zeviani M, Fernández-Silva P, Enríquez JA. Evolution meets disease: penetrance and functional epistasis of mitochondrial tRNA mutations. PLoS Genet. 2011;7:e1001379. doi: 10.1371/journal.pgen.1001379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto AM, Jordan KC, Tietze K, Britton JS, O'Neill EM, Ruohola-Baker H. Pointed, an ETS domain transcription factor, negatively regulates the EGF receptor pathway in Drosophila oogenesis. Development. 1996;122:3745–3754. doi: 10.1242/dev.122.12.3745. [DOI] [PubMed] [Google Scholar]
- Nagakawa Y, Williams GM, Zheng Q, Tsuchida A, Aoki T, Montgomery RA, Klein AS, Sun Z. Oxidative mitochondrial DNA damage and deletion in hepatocytes of rejecting liver allografts in rats: role of TNF-α. Hepatology. 2005;42:208–215. doi: 10.1002/hep.20755. [DOI] [PubMed] [Google Scholar]
- Narendra D, Walker JE, Youle R. Mitochondrial quality control mediated by PINK1 and Parkin: links to parkinsonism. Cold Spring Harb Perspect Biol. 2012;4:pii: a011338. doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld TP. Autophagy and cell growth – the yin and yang of nutrient responses. J Cell Sci. 2012;125:1–10. doi: 10.1242/jcs.103333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira MT, Garesse R, Kaguni LS. Animal models of mitochondrial DNA transactions in disease and ageing. Exp Gerontol. 2010;45:489–502. doi: 10.1016/j.exger.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poalas K, Hatchi EM, Cordeiro N, Dubois SM, Leclair HM, Leveau C, Alexia C, Gavard J, Vazquez A, Bidère N. Negative regulation of NF-κB signaling in T lymphocytes by the ubiquitin-specific protease USP34. Cell Commun Signal. 2013;11:25. doi: 10.1186/1478-811X-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla-Osorio N, Lacey DB, Alt FW, Zhu C. Early embryonic lethality due to targeted inactivation of DNA ligase III. Mol Cell Biol. 2006;26:3935–3941. doi: 10.1128/MCB.26.10.3935-3941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii CM, Garone C, Emmanuele V, Tadesse S, Krishna S, Dorado B, Hirano M. Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 2013;27:612–621. doi: 10.1096/fj.12-209361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Kylsten P. Rhomboid-7 over-expression results in Opa1-like processing and malfunctioning mitochondria. Biochem Biophys Res Commun. 2011;414:315–320. doi: 10.1016/j.bbrc.2011.09.047. [DOI] [PubMed] [Google Scholar]
- Rämet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Ranieri M, Brajkovic S, Riboldi G, Ronchi D, Rizzo F, Bresolin N, Corti S, Comi GP. Mitochondrial fusion proteins and human diseases. Neurol Res Int. 2013;2013:293893. doi: 10.1155/2013/293893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, Harbour ME, Fearnley IM, Crouch RJ, Conti MA, Adelstein RS, Walker JE, Holt IJ. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39:5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myöhänen M, Greco D, Cannino G, Jacobs HT, Lietzén N, Nyman TA, Battersby BJ. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Roberti M, Bruni F, Polosa PL, Gadaleta MN, Cantatore P. The Drosophila termination factor DmTTF regulates in vivo mitochondrial transcription. Nucleic Acids Res. 2006;34:2109–2116. doi: 10.1093/nar/gkl181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BH, Petrova-Benedict R, Buncic JR, Wallace DC. Nonviability of cells with oxidative defects in galactose medium: a screening test for affected patient fibroblasts. Biochem Med Metab Biol. 1992;48:122–126. doi: 10.1016/0885-4505(92)90056-5. [DOI] [PubMed] [Google Scholar]
- Rone MB, Midzak AS, Issop L, Rammouz G, Jagannathan S, Fan J, Ye X, Blonder J, Veenstra T, Papadopoulos V. Identification of a dynamic mitochondrial protein complex driving cholesterol import, trafficking, and metabolism to steroid hormones. Mol Endocrinol. 2012;26:1868–1882. doi: 10.1210/me.2012-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rötig A, Poulton J. Genetic causes of mitochondrial DNA depletion in humans. Biochim Biophys Acta. 2009;1792:1103–1108. doi: 10.1016/j.bbadis.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Ruhanen H, Ushakov K, Yasukawa T. Involvement of DNA ligase III and ribonuclease H1 in mitochondrial DNA replication in cultured human cells. Biochim Biophys Acta. 2011;1813:2000–2007. doi: 10.1016/j.bbamcr.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saada A. Deoxyribonucleotides and disorders of mitochondrial DNA integrity. DNA Cell Biol. 2004;23:797–806. doi: 10.1089/dna.2004.23.797. [DOI] [PubMed] [Google Scholar]
- Sarig O, Goldsher D, Nousbeck J, Fuchs-Telem D, Cohen-Katsenelson K, Iancu TC, Manov I, Saada A, Sprecher E, Mandel H. Infantile mitochondrial hepatopathy is a cardinal feature of MEGDEL syndrome (3-methylglutaconic aciduria type IV with sensorineural deafness, encephalopathy and Leigh-like syndrome) caused by novel mutations in SERAC1. Am J Med Genet A. 2013;161:2204–2215. doi: 10.1002/ajmg.a.36059. [DOI] [PubMed] [Google Scholar]
- Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA LSFC Consortium. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–1323. doi: 10.1091/mbc.E10-01-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Harbauer AB, Rao S, Eyrich B, Zahedi RP, Stojanovski D, Schönfisch B, Guiard B, Sickmann A, Pfanner N, Meisinger C. Regulation of mitochondrial protein import by cytosolic kinases. Cell. 2011;144:227–239. doi: 10.1016/j.cell.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Southard SM, Hobbs AE, Jensen RE. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem Biophys Res Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci USA. 2011;108:15179–15183. doi: 10.1073/pnas.1105921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko IN, Wilson GL, Alexeyev MF. Persistent damage induces mitochondrial DNA degradation. DNA Repair (Amst) 2013;12:488–499. doi: 10.1016/j.dnarep.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard M, Tani K, Ruby YR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–946. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010;62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D'Adamo P, Calvo S, Marsano RM, Donnini C, Weiher H, Strisciuglio P, Parini R, Sarzi E, Chan A, DiMauro S, Rötig A, Gasparini P, Ferrero I, Mootha VK, Tiranti V, Zeviani M. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Storrie B, Attardi G. Expression of the mitochondrial genome in HeLa cells. 13. Effect of selective inhibition of cytoplasmic or mitochondrial protein synthesis on mitochondrial nucleic acid synthesis. J Mol Biol. 1972;71:177–199. doi: 10.1016/0022-2836(72)90345-2. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Holmes JB, Cerritelli SM, Sakhuja K, Minczuk M, Holt IJ, Crouch RJ. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol Cell Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Jiang J, O WS, Deng Y, Huen MS. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013;41:8572–8580. doi: 10.1093/nar/gkt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symersky J, Osowski D, Walters DE, Mueller DM. Oligomycin frames a common drug-binding site in the ATP synthase. Proc Natl Acad Sci USA. 2008;109:13961–13965. doi: 10.1073/pnas.1207912109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Miyata K, Sugahara K, Onishi S, Shuin T, Aso T. Identification of EloA-BP1, a novel elongin A binding protein with an exonuclease homology domain. Biochem Biophys Res Commun. 2003;309:189–195. doi: 10.1016/s0006-291x(03)01556-0. [DOI] [PubMed] [Google Scholar]
- Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J Biol Chem. 2011;286:31975–31983. doi: 10.1074/jbc.M110.215715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Kristiansen MS, Lykke-Abdersen S, Chlebowski A, Larssen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, Dziembowski A, Jensen TH. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Hansson A, Wredenberg A, Rovio AT, Dufour E, Khvorostov I, Spelbrink JN, Wibom R, Jacobs HT, Larsson NG. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc Natl Acad Sci USA. 2005;102:17993–17998. doi: 10.1073/pnas.0508886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CJ, Granycome C, Hurst R, Pohler E, Juhola MK, Juhola MI, Jacobs HT, Sutherland L, Holt IJ. Systematic segregation to mutant mitochondrial DNA and accompanying loss of mitochondrial DNA in human NT2 teratocarcinoma cybrids. Genetics. 2005;170:1879–1885. doi: 10.1534/genetics.105.043653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadrot N, Ghanem S, Braut F, Gavrilescu L, Pilard N, Mansouri A, Moreau R, Reyl-Desmars F. Mitochondrial DNA maintenance is regulated in human hepatoma cells by glycogen synthase kinase 3β and p53 in response to tumor necrosis factor α. PLoS ONE. 2012;7:e40879. doi: 10.1371/journal.pone.0040879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergani L, Rossi R, Brierley CH, Hanna M, Holt IJ. Introduction of heteroplasmic mitochondrial DNA (mtDNA) from a patient with NARP into two human rho degrees cell lines is associated either with selection and maintenance of NARP mutant mtDNA or failure to maintain mtDNA. Hum Mol Genet. 1999;8:1751–1755. doi: 10.1093/hmg/8.9.1751. [DOI] [PubMed] [Google Scholar]
- Vielhaber S, Debska-Vielhaber G, Peeva V, Schoeler S, Kudin AP, Minin I, Schreiber S, Dengler R, Kollewe K, Zuschratter W, Kornblum C, Zsurka G, Kunz WS. Mitofusin 2 mutations affect mitochondrial function by mitochondrial DNA depletion. Acta Neuropathol. 2013;125:245–256. doi: 10.1007/s00401-012-1036-y. [DOI] [PubMed] [Google Scholar]
- Wang Y, Singh U, Mueller DM. Mitochondrial genome integrity mutations uncouple the yeast Saccharomyces cerevisiae ATP synthase. J Biol Chem. 2007;282:8228–8236. doi: 10.1074/jbc.M609635200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S, Croft DR, O'Prey J, Meedendorp A, O'Prey M, Dufès C, Ryan KM. The cyclin-dependent kinase PITSLRE/CDK11 is required for successful autophagy. Autophagy. 2011;7:1295–1301. doi: 10.4161/auto.7.11.16646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojewoda M, Duszyński J, Szczepanowska J. Antioxidant defence systems and generation of reactive oxygen species in osteosarcoma cells with defective mitochondria: effect of selenium. Biochim Biophys Acta. 2010;1797:890–896. doi: 10.1016/j.bbabio.2010.01.035. [DOI] [PubMed] [Google Scholar]
- Wojewoda M, Duszyński J, Szczepanowska J. NARP mutation and mtDNA depletion trigger mitochondrial biogenesis which can be modulated by selenite supplementation. Int J Biochem Cell Biol. 2011;43:1178–1186. doi: 10.1016/j.biocel.2011.04.011. [DOI] [PubMed] [Google Scholar]
- Wong ED, Wagner JA, Gorsich SW, McCaffery JM, Shaw JM, Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylikallio E, Suomalainen A. Mechanisms of mitochondrial diseases. Ann Med. 2012;44:41–59. doi: 10.3109/07853890.2011.598547. [DOI] [PubMed] [Google Scholar]
- Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Table S7
Supplementary Materials and Methods
Source Data for Supplementary Figure S1
Source Data for Supplementary Figure S2
Source Data for Supplementary Figure S3
Source Data for Supplementary Figure S4
Source Data for Supplementary Figure S5
Review Process File
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Data Availability Statement
Original images from the primary screen are deposited at: http://dx.doi.org/10.5061/dryad.v55p5.