Abstract
In this study, a continuous flow dielectrophoresis (DEP) microfluidic chip was fabricated and utilized to sort out the microalgae (C. vulgaris) with different lipid contents. The proposed separation scheme is to allow that the microalgae with different lipid contents experience different negative or no DEP force at the separation electrode pair under the pressure-driven flow. The microalgae that experience stronger negative DEP will be directed to the side channel while those experience less negative or no DEP force will pass through the separation electrode pair to remain in the main channel. It was found that the higher the lipid content inside the microalgae, the higher the crossover frequency. Separation of the microalgae with 13% and 21% lipid contents, and 24% and 30%–35% lipid contents was achieved at the operating frequency 7 MHz, and 10 MHz, respectively. Moreover, separation can be further verified by measurement of the fluorescence intensity of the neutral lipid inside the sorted algal cells.
INTRODUCTION
Clean and permanent substitute for petroleum fuel has become a widely discussed subject matter in the territory of energy owing to various concerned issues such as climate change and global warming, potentially increased oil price due to the dwindling reserves of the fossil fuel, need for energy security, etc. As a result, research efforts for development of the alternative energies have been devoted substantially in recent years. There are various alternative energy sources such as sunlight, wind, hydropower, biofuel, geothermal heat, and tidal energy. Among them, biofuel which is extracted from plant and microalgae has been considered as a promising energy source in the future because of the compatibility between now-used vehicle engines and relatively simple acquisition route.1 Compared to the acquisition route of plant, there are several advantages for extraction of lipid inside microalgae: no need to tap into the global food supply chain,2 higher energy density,3 absorbing carbon dioxide to mitigate global warming and producing other valued compounds from the algal cells. To harvest the lipid from microalgae efficiently, several strategies can be exploited such as developing to increase lipid content inside microalgae in short cultivation cycles,4 screening/selecting the lipid-rich microalgae for cultivation or on-site monitoring of the lipid content inside microalgae during the cultivation process in a timely manner, and so on. For the latter, the screening process or determination of the lipid content needs to be completed in a rapid fashion. The conventional techniques for characterization of the lipid fraction inside microalgae such as gas chromatography (GC) analysis and gram measurement5,6 suffer several drawbacks like time consuming, complicated in processes, and requirement of large sample amount. The fluorescence intensity analysis of lipid-conjugated fluorescent dye such as Nile red and BODIPY 505/515 was proposed and can achieve similar lipid fraction results compared to those from conventional GC and gram measurement.7 Based on these results, fluorescence-activated cell sorting (FACS) which is a flow cytometry technique was applied to sort different types of algal cells and analyze the content inside microalgae.5 However, considerable costs for equipment and operation or central facilities are required. Recently, a droplet-based microfluidic system to encapsulate algal cells for subsequent profiling of heterogeneity in the lipid accumulation among individual cells was developed.8 Although the lipid content inside the algal cells can be determined in an online and real-time manner and the cell viability be maintained in the microcapsules for potential follow-up biological analyses, the mechanism of cell sorting is not integrated in this particular microfluidic platform.
As to screening of microalgae based on their lipid contents, there is rare discussion found in the literature. Deng et al. have reported that separation of microalgae with different lipid contents can be achieved under non-uniform AC electric field at proper operating frequencies.9 This is because the microalgae with different lipid contents will experience different dielectrophoresis (DEP). DEP is the motion of the polarized particles as a result of a force exerted on a dielectric particle when it is subjected to a non-uniform electric field and has been widely applied to manipulate microscopic particles in microfluidic system for separation of different sizes of the latex beads, cells, and viruses.10 For a spherical neutral particle, the time-averaged DEP force can be written as follows:
| (1) |
In the equation, a is radius of the particle, εm is the permittivity of the medium, ▽|E|2 is the gradient of the square of the local electric field strength, and Re(fCM) is the real partition of the complex Clausius–Mossotti (CM) factor. The CM factor represents the degree of polarization and can be written as follows:
| (2) |
where ε* = ε − i(σ/2πω) is the complex permittivity, and the subscripts p and m represent particle and medium, respectively. σ is conductivity, ω is the frequency of the applied alternating current, and i=√ − 1. The value of Re(fCM) ranges from −0.5 to 1, The particle experiences positive DEP (pDEP) force and moves toward the high electric field region when Re(fCM) is greater than 0. On the other hand, it experiences negative DEP (nDEP) and moves toward low electric field region when Re(fCM) is less than 0. When Re(fCM) is equal to 0, the particle is at rest and the operating frequency at this condition is called the crossover frequency. The size effect is also included in Eq. (2), where σp = σbulk + 2Ks/a, σbulk is the conductivity of bulk material, and Ks is the surface conductance. For the biomolecules, determination of DEP motion becomes less straightforward owing to their multi-shell structure effect.11–13
DEP in conjunction with pressure-driven flow has been applied to continuously separate the mixture of the particles or biomolecules. A 3-D microelectrode system was constructed with designed electrodes to guide the latex particles or eukaryotic cells over the desired distance for trapping and separation.14 Instead of being guided, the particles can penetrate the electrode pair at/over some threshold velocity, which depends on a number of parameters such as channel height, particle size, dielectric properties, electrode width and local heating, and so on.15 This is due to the relative strength of the DEP force and hydrodynamic force acting on the particles. Furthermore, because of different electric field strengths in the z-direction, Liao et al. have shown that particles with different sizes penetrated at different heights of the microchannel, which caused different lateral positions after penetrating the electrode pair.16 Taking advantage of this phenomenon, separation of particles with different sizes and the yeast cells was demonstrated. The 3-D DEP microfluidic chip was further combined with Raman detection to analyze the bacteria in the human blood.17
In this study, we extended our previous work by developing a continuous flow DEP microfluidic chip to sort the microalgae (Chlorella vulgaris) with different lipid contents. The separation scheme was based on the relative strength of the hydrodynamic force and the nDEP force acting on the algal cells, where the algal cells experience the similar hydrodynamic force but different magnitude of the nDEP force depending on their lipid contents. The frequency sweep was conducted to determine the crossover frequency of the microalgae with different lipid contents that serves as a guideline for the applied operating frequency. Moreover, the neutral lipid inside the microalgae was stained by the Nile red for fluorescence intensity measurement downstream, which was analyzed to verify the separation result.
MATERIAL AND METHODS
Fabrication of continuous flow DEP microfluidic chip
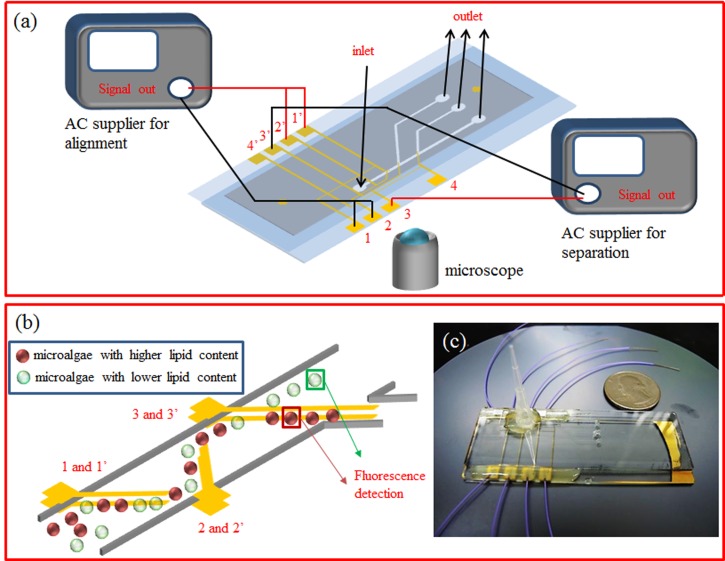
To fabricate the DEP microchip, the glass slides were first cleaned by using basic washing solution (NH4OH:H2O2:H2O = 1:1:5) at 75 °C for 1 h. The 30-nm thick titanium (Ti) adhesion layer was then evaporated onto each cleaned glass slide, followed by deposition of 200-nm thick gold (Au) as the conduction layer using the electron beam evaporator (JEOL, 750 A). The glass slides then went through the etching process to define the patterned electrodes. The microchannel was constructed by spin coating a layer of JSR THB 151N negative type photoresist (JSR Micro, THB 151N) with 15 μm in thickness on the glass slide with bottom electrodes, followed by photolithography to reveal the electrodes at the bottom of the microchannel. The glass slide with top electrodes was carefully aligned with the bottom electrodes under the microscope and clamped together. Since the JSR THB 151N photoresist will become tacky and even melt as the temperature exceeds 130 and 170 °C, respectively,18 the assembly was baked in the oven at 130 °C for 10 min, followed by 150 °C for 5 min to complete bonding of the microfluidic chip. Once bonded, the inlet and outlet of the microchip were connected to Tygon tube and the contacts of microelectrode were soldered with copper lines, which were connected to the AC function generator (AFG3101C, Tektronix) as shown in Figure 1(a). Figure 1(c) shows the photo of the fabricated DEP microfluidic chip.
FIG. 1.
(a) The schematic of the experimental setup. (b) The proposed scheme for separation of microalgae with different lipid contents. (c) The photo of the continuous flow DEP microfluidic chip.
Sample preparation and lipid content characterization
Cultivation of Chlorella vulgaris (C. vulgaris) with different lipid contents can be found elsewhere.19 The average lipid content inside the algal cells was determined through conventional GC method. In this study, the microalgae were sampled at different days during the cultivation process and the lipid contents were determined to be 13%, 21%, 24%, 30%, and 35%.
The conductivity of the microalgae solution was adjusted by adding proper amount of KH2PO4 buffer solution. To distinguish the neutral lipid inside the microalgae, Nile red fluorescence dye was used to label the lipid body inside microalgae. The Nile red working solution was prepared by dissolving the Nile red fluorescence dye in DMSO (Sigma–Aldrich, Inc.), followed by diluting to 5 μM in the solution of 25% DMSO/KH2PO4 medium (v/v). 1 ml of original microalgae solution with 106 cells/ml was concentrated to 107 cells/ml via centrifugation. The centrifuged microalgae were then incubated in 500 μl Nile red working solution for 20 min by using the vortex machine and washed three times with KH2PO4 buffer solution.
DEP separation and fluorescence analysis
Prior to performing continuous sorting of microalgae by DEP, the crossover frequency of the microalgae with different lipid contents needs to be determined. The procedures to carry out the frequency sweep can be found in the literature.9 In brief, the microalgae solution was injected into the sealed microchannel with line patterned electrode array at the bottom of the microchannel. After turning on the AC function generator, the microalgae will experience different DEP force under the non-uniform AC electric field at different applied frequencies. When the microalgae experience pDEP, they attach to the edge of the line electrodes. On the other hand, aggregation of the microalgae near the center of the line electrodes is observed when they experience nDEP. When the operating is around the crossover frequency, the motion of the microalgae becomes less identifiable. Figure 1(b) shows the scheme of continuous sorting of the microalgae. The fabricated DEP microchip was fixed onto the microscope holder and connected to the alternative current power supplier. The solution with Nile red stained microalgae was injected into DEP microchip at the flow rate of 250 μm/s provided by a syringe pump (KDS 100, KD Scientific). The injected algal cells with different lipid contents were all repelled and aligned by the electrode pairs 1–1′ and 2–2′ at the designated operating frequency. Addition of the electrode 2–2′ is to ensure that most of the algal cells can be well aligned into a line pattern for subsequent separation. The aligned algal cells were then separated by the electrode pair 3–3′ downstream according to their lipid content. The fluorescence intensity of the microalgae was measured by the inverted fluorescence microscope (TE-2000S, Nikon), which was connected to digital camera (HQ2, Photometrics) to acquire the sequential images during DEP separation process. The band pass optical filter (535 to 585 nm) was used to minimize the fluorescence intensity of chlorophyll a (640 nm) and, at the same time, allow the acquisition of the fluorescence intensity of the neutral lipid stained by Nile red (570 nm). The detection zone was set to be 30 μm × 30 μm square downstream where the fluorescence intensity of only one single algal cell at a time was measured. The fluorescence intensity of the sorted microalgae was then further analyzed by the image software (image-pro plus, Media Cybernetics).
RESULTS AND DISCUSSION
Continuous sorting of microalgae with different lipid contents
Table I shows the frequency sweep for the microalgae with different lipid contents at the solution conductivity 1.4 mS/cm. It can be seen that the microalgae with 21% lipid content experienced negative DEP at the frequency 1 MHz but positive DEP at 2 MHz, indicating that the crossover frequency was between 1 and 2 MHz. As the lipid content increased to 30% and 35%, the crossover frequencies were around 2 and 3 MHz, respectively. Table II shows the frequency sweep at the solution conductivity 2.93 mS/cm. For the microalgae with 21% lipid content, the crossover frequency increased between 10 and 15 MHz. As to the lipid content around 30%–35%, the microalgae experienced negative DEP in the entire operating frequency range (from 5 to 20 MHz), implying that either there was no crossover frequency or the crossover frequency was beyond 20 MHz. For the microalgae with 13% lipid content, they experienced negative DEP at the frequency 5 MHz but positive DEP at 15 MHz and above. Moreover, their motion is less identifiable at the frequency 10 MHz, indicating that the crossover frequency was around 10 MHz. When the lipid content was between 20% and 30%, the crossover frequency was between 10 and 15 MHz or around 15 MHz. The above showed that (1) the difference between the crossover frequencies of microalgae with different lipid contents is smaller at lower solution conductivity and (2) the higher the lipid content, the larger the crossover frequency. Since the small difference between the crossover frequencies would pose difficulty in applying the proper frequency for microalgae separation, therefore, the solution conductivity was set to be 2.93 mS/cm. As to the latter, it could be reasoned by using the multi-shell model to calculate the change of Re(fCM) of the algal cells.20 The multi-shell model has been used to explain the behavior of the algal cells in electrorotation measurement,21 describe the dielectric collection of algal cells in water quality analysis,22 and calculate the polarizability of the algal cells to account for the chaining behavior23 or the cell properties.24 In this study, the multi-shell model with three concentric spheres was used to describe the algal cells. The inner sphere with radius r1 represents the accumulated lipid content and r1 will change according to the lipid content. The cytoplasm is represented by the shell between the inner sphere and the concentric middle sphere with radius r2. The cell wall is represented by the shell between the concentric middle sphere and the outer sphere with radius r3. The radii r2 and r3 can be assumed to be 2.5 μm and 2.6 μm, respectively (the thickness of the cell wall is usually around 0.1 μm). The effective permittivity for the sphere and shell can be expressed as follows:
| (3) |
| (4) |
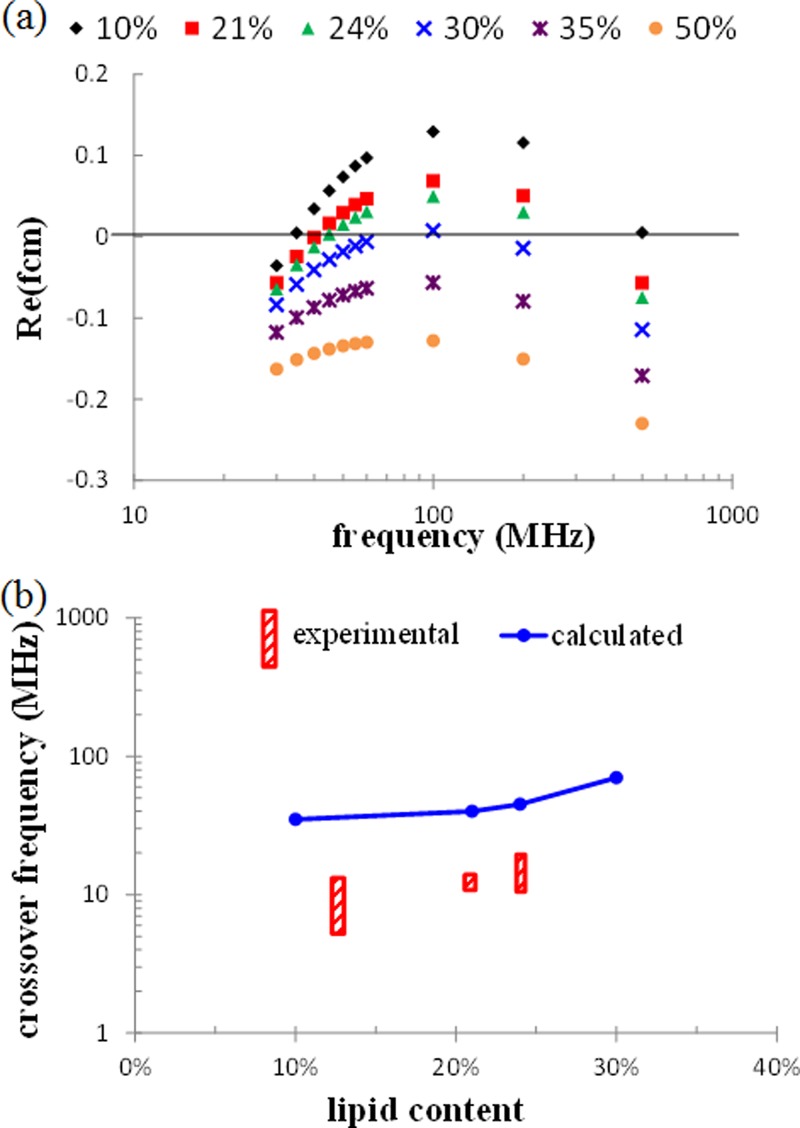
εp* in Eq. (2) was replaced by Eq. (4) to indicate the difference of algal cells with different lipid contents. The permittivity and conductivity for the cell wall (ε3,σ3), cytoplasm (ε2,σ2), and lipid body (ε1,σ1) are assumed to be 5ε0 and 10−8 (S/m), 60ε0 and 0.5 (S/m)13, and 3ε0 and 10−4 (S/m), respectively, where ε0 is vacuum permittivity (8.85 × 10−12 F/m). The permittivity and conductivity of the medium (εm,σm) in Eq. (2) are set at 80ε0 and 0.29 (S/m), respectively. As the lipid content inside algal cells increases from 10% to 50%, the radius of the inner sphere r1 will increase as well and the ratio of decreases from 2.03 to 1.22. Figure 2(a) shows the calculated Re(fcm) vs. the frequencies for the microalgae with different lipid contents. For the lipid content less than or equal to 30%, there are two crossover frequencies, i.e., one is in the lower frequency range and the other in the higher frequency range. For the lipid content larger than 30%, there is no crossover frequencies and the microalgae experience only nDEP force. According to the experimental results where the microalgae experienced from nDEP to pDEP as the operating frequency increased, the first crossover frequency (the one in the lower frequency range) was selected for comparison in this study. The first crossover frequencies for the microalgae with lipid contents 10%, 21%, 24%, and 30% were found to be 35, 40, 45, and 70 MHz, respectively, as shown in Figure 2(b). Despite slightly overestimating the values of crossover frequencies, it does show the dependence of the crossover frequency on the lipid content and the larger the lipid content, the higher the crossover frequency.
TABLE I.
DEP motion of the microalgae with different lipid contents at the operating frequency between 5 and 20 MHz (solution conductivity: under 1.4 mS/cm).
| Frequency | Lipid content (% dry wet) | ||
|---|---|---|---|
| 21% | 30% | 35% | |
| 1 MHz | nDEP | nDEP | nDEP |
| 2 MHz | pDEP | ——a | nDEP |
| 3 MHz | pDEP | pDEP | —— |
“——”means that the motion of the microalgae is less identifiable.
TABLE II.
DEP motion of the microalgae with different lipid contents at the operating frequency between 5 and 20 MHz (solution conductivity: 2.93 mS/cm).
| Frequency | Lipid content (% dry wet) | ||||
|---|---|---|---|---|---|
| 13% | 21% | 24% | 30% | 35% | |
| 5 MHz | nDEP | nDEP | nDEP | nDEP | nDEP |
| 10 MHz | ——a | nDEP | nDEP | nDEP | nDEP |
| 15 MHz | pDEP | pDEP | —— | nDEP | nDEP |
| 20 MHz | pDEP | pDEP | pDEP | nDEP | nDEP |
“——”means that the motion of the microalgae is less identifiable.
FIG. 2.
(a) The calculated Re(fcm) vs. frequency for the microalgae with different lipid contents. (b) The comparison between the experimental and the calculated crossover frequencies for the microalgae with different lipid contents.
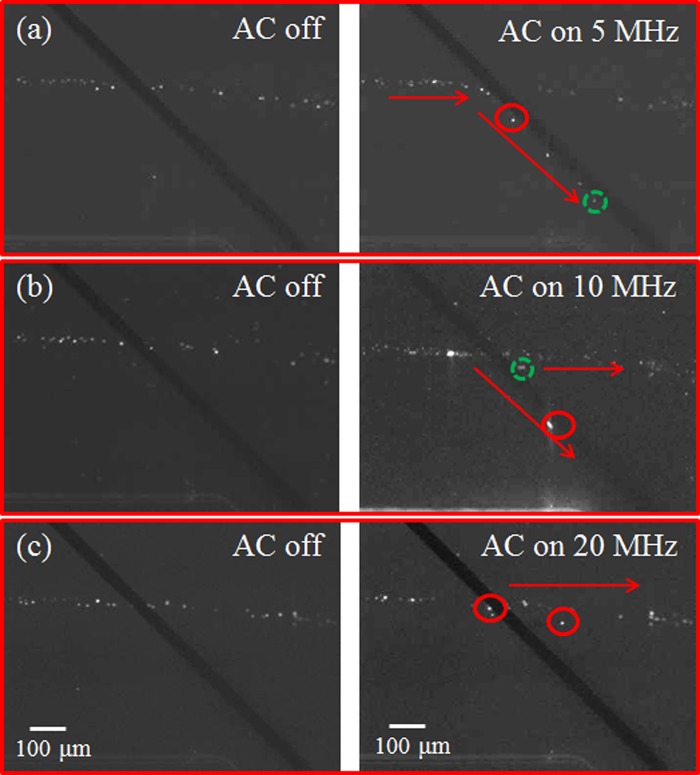
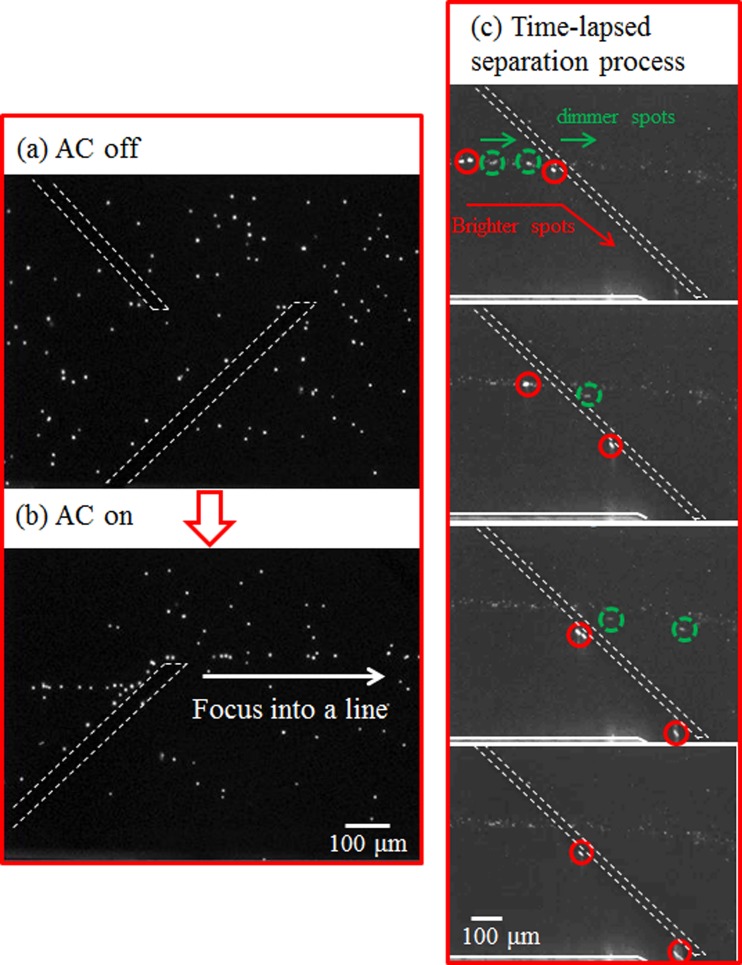
It is worthy of mentioning that, for the lipid content larger than 30%–35%, it becomes difficult to assess the crossover frequency due to the limitation of the equipment. Moreover, the gold electrodes start to dissolve in the solution as the operating frequency exceeds 40 MHz. Nevertheless, from the results, it can be seen that there exist the operating frequencies such that the microalgae with 13% lipid content can be separated from those with lipid content above 21% (5–10 MHz) or the microalgae with 20%–24% lipid content can be separated from those with lipid content above 30% (10–15 MHz). To perform separation of the microalgae with 24% and 35% lipid contents, the flow behavior of the microalgae under different operating frequencies was examined as shown in Figure 3. At 5 MHz, all the microalgae experienced stronger negative DEP force and, hence, were repelled by the electrode pairs and directed to the side channel as shown in Figure 3(a). As the frequency increased to 10 MHz, the microalgae with 24% lipid content passed through the electrode pair and remained in the main channel while those with 35% lipid content were repelled and directed to the side channel as shown in Figure 3(b). This is because the microalgae with 24% lipid content experienced less negative or no DEP force at 10 MHz and the flow rate at 250 μm/s provided enough hydrodynamic force to allow the algal cells to pass through the electrode pair. On the other hand, larger negative DEP force was exerted on the microalgae with 35% lipid content, which overcame the hydrodynamic force and the microalgae were directed to the side channel. As the frequency further increased, the microalgae with less lipid content started to experience positive DEP and might attach to the electrodes. This is detrimental to the performance the electrodes and the effect of DEP, especially when the microalgae are being continuously sorted. Moreover, when the frequency increased to 20 MHz, the microalgae with 35% lipid content also passed through the electrode pair as shown in Figure 3(c). This results in no separation of the microalgae with 24% and 35% lipid contents. Therefore, the working principle for separation of the microalgae using the DEP microchip as fabricated is to make the microalgae with different lipid contents experience different negative DEP force at the proper flow rate where those experiencing less or no negative DEP force remain in the main channel while those experiencing larger negative DEP force are directed to the side channel without fouling the electrodes. For separating the microalgae with 24% and 30%–35% lipid content, the operating frequency at 10 MHz was selected to provide suitable negative DEP force. The same principle was applied to separate the microalgae with 13% lipid content from those with 21% lipid content at the operating frequency 7 MHz. Figure 4 shows the alignment and separation of microalgae with 24% and 35% lipid contents. Before turning on AC power, the microalgae flowed and were randomly distributed inside the microchannel as shown in Figure 4(a). When applying the AC power at the frequency 300 kHz for 5 s, all the microalgae were aligned and guided to downstream as shown in Figure 4(b). From the result of the frequency sweep, it indicates that all the microalgae would experience relatively stronger negative DEP force to overcome the hydrodynamic force and be directed to the side channel. The purpose of alignment prior to separation is to have the microalgae subject to the retardation effect after leaving the paired electrodes,16 which leads to increase of the spacing between two adjacent algal cells. As a result, it is very much likely for the algal cells to pass the detection area one at a time. Figure 4(c) shows the sequential images for separation of the microalgae at 10 MHz. It can be seen that the brighter spots (indicating the microalgae with higher lipid content) slowed down as they approached the electrode pair, being directed to the side channel. For the dimmer spots (indicating the microalgae with lower lipid content), they moved faster than the brighter spots and passed through the electrode pair to remain in the main channel. Note that the trajectory of the dimmer spots was deflected when passing through the electrode pair, which is due to the electric field-induced DEP force gradient in the z-direction.16 Separation of the microalgae with 13% and 21% lipid contents, and 24% and 30% lipid contents, was also achieved (Figures A and B in supplementary materials).25 This demonstrates that the microalgae with different lipid contents can be sorted according to the proposed scheme using the DEP microchip as fabricated.
FIG. 3.
The flow behavior of microalgae with 24% and 35% lipid contents at the operating frequency (a) 5, (b) 10, and (c) 20 MHz. The brighter spots (solid circles) are the microalgae with 35% lipid content and the dimmer spots (dashed circles) are those with 24% lipid content.
FIG. 4.
The alignment of the microalgae (a) before and (b) after turning on the AC function generator. (c) The sequential images for separation of the microalgae with 24% (dashed circles) and 35% (solid circles) lipid contents. The electrodes (dashed lines) and the edge of the microchannel (solid lines) are outlined for clarity.
Continuous fluorescence detection and analysis of the microalgae with different lipid contents
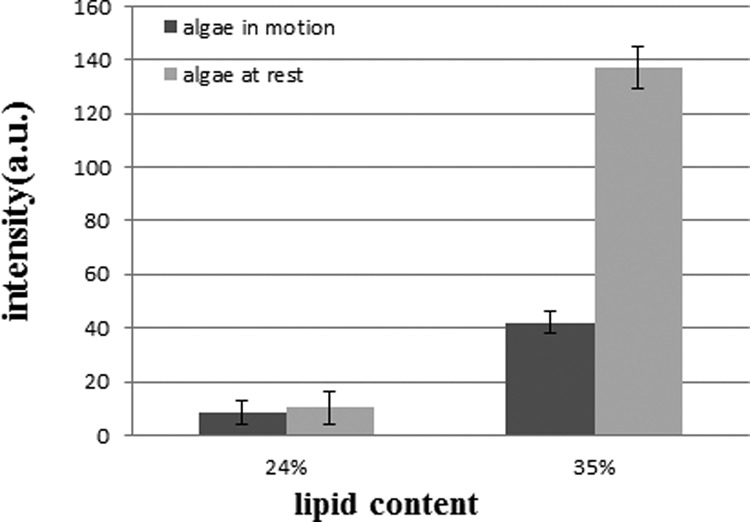
In order to verify separation of the microalgae, the fluorescence intensity measurement of the sorted microalgae with different lipid contents was carried out downstream. Figure 5 shows the fluorescence intensity measurement of the microalgae with different lipid contents. For the microalgae with 13% lipid content, the fluorescence intensity of the neutral lipid was close to the background signal and, thus, not shown in the figure. It can be seen that, when the microalgae were at rest, the measured fluorescence intensity increased as the lipid content increased and there was a discernible difference in the fluorescence intensity among the microalgae with different lipid contents. When the microalgae were in motion, the measured fluorescence intensity decreased compared to that measured at rest, which could be attributed to the sensitivity of the instrument. Still, there exists discernible difference in the fluorescence intensity between microalgae with 24% and 35% lipid contents, separation of these two microalgae samples can be identified and verified based on the measurement of the fluorescence intensity.
FIG. 5.
The measured fluorescence intensity of the microalgae with different lipid contents at rest and in motion.
CONCLUSIONS
In this study, continuous sorting and analysis of microalgae with different lipid contents were demonstrated by using the flow through DEP microfluidic chip. It was found that separation of the microalgae with 13% and 21% lipid content, 24% and 30%–35% lipid content, was achieved at operating frequency 7 MHz and 10 MHz, respectively. Moreover, separation of the microalgae with different lipid contents can be further identified based on the fluorescence measurement. The microfluidic chip as fabricated can be integrated with fluorescence detection system26 for potentially on-site monitoring and optimization of the cultivation process.
ACKNOWLEDGMENTS
The authors are grateful for the financial support from National Science Council in Taiwan (NSC 102-2221-E-006-192-MY2) and NCKU's “Top-University” grant (90080161403049091620).
References
- 1.Chisti Y., Biotechnol. Adv. 25(3), 294 (2007); 10.1016/j.biotechadv.2007.02.001 [DOI] [PubMed] [Google Scholar]; Williams P. J. L., Nature 450(7169), 478 (2007). 10.1038/450478a [DOI] [PubMed] [Google Scholar]
- 2.Ahmad A. L., Mat Yasin N. H., Derek C. J. C., and Lim J. K., Renew. Sustainable Energy Rev. 15(1), 584 (2011). 10.1016/j.rser.2010.09.018 [DOI] [Google Scholar]
- 3.Waltz E., Nat. Biotechnol. 27(1), 15 (2009). 10.1038/nbt0109-15 [DOI] [PubMed] [Google Scholar]
- 4.Chen C. Y., Yeh K. L., Aisyah R., Lee D. J., and Chang J. S., Bioresour. Technol. 102(1), 71 (2011); 10.1016/j.biortech.2010.06.159 [DOI] [PubMed] [Google Scholar]; Chen C. Y., Yeh K. L., Su H. M., Lo Y. C., Chen W. M., and Chang J. S., Biotechnol. Prog. 26(3), 679 (2010); 10.1002/btpr.381 [DOI] [PubMed] [Google Scholar]; Singh A. and Olsen S. I., Appl. Energy 88(10), 3548 (2011); 10.1016/j.apenergy.2010.12.012 [DOI] [Google Scholar]; Satyanarayana K. G., Mariano A. B., and Vargas J. V. C., Int. J. Energy Res. 35(4), 291 (2011). 10.1002/er.1695 [DOI] [Google Scholar]
- 5.Montero M. F., Aristizabal M., and Reina G. G., J. Appl. Phycol. 23(6), 1053 (2011). 10.1007/s10811-010-9623-6 [DOI] [Google Scholar]
- 6.Priscu J. C., Priscu L. R., Palmisano A. C., and Sullivan C. W., Antarct. Sci. 2(2), 149 (1990); [Google Scholar]; Bligh E. G., Curr. Contents 1978(52), 11. [Google Scholar]
- 7.Mutanda T., Ramesh D., Karthikeyan S., Kumari S., Anandraj A., and Bux F., Bioresour. Technol. 102(1), 57 (2011); 10.1016/j.biortech.2010.06.077 [DOI] [PubMed] [Google Scholar]; Cooper M. S., Hardin W. R., Petersen T. W., and Cattolico R. A., J. Biosci. Bioeng. 109(2), 198 (2010); 10.1016/j.jbiosc.2009.08.004 [DOI] [PubMed] [Google Scholar]; Chen W., Zhang C., Song L., Sommerfeld M., and Hu Q., J. Microbiol. Methods 77(1), 41 (2009); 10.1016/j.mimet.2009.01.001 [DOI] [PubMed] [Google Scholar]; Elsey D., Jameson D., Raleigh B., and Cooney M. J., J. Microbiol. Methods 68(3), 639 (2007). 10.1016/j.mimet.2006.11.008 [DOI] [PubMed] [Google Scholar]
- 8.Lee D.-H., Bae C. Y., Han J.-I., and Park J.-K., Anal. Chem. 85(18), 8749 (2013). 10.1021/ac401836j [DOI] [PubMed] [Google Scholar]
- 9.Deng Y.-L., Chang J.-S., and Juang Y.-J., Bioresour. Technol. 135, 137 (2013). 10.1016/j.biortech.2012.11.046 [DOI] [PubMed] [Google Scholar]
- 10.Doh I. and Cho Y. H., Sens. Actuators, A 121(1), 59 (2005); 10.1016/j.sna.2005.01.030 [DOI] [Google Scholar]; Morgan H., Hughes M. P., and Green N. G., Biophys. J. 77(1), 516 (1999); 10.1016/S0006-3495(99)76908-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; Green N. G. and Morgan H., J. Phys. D Appl. Phys. 30(11), L41 (1997); 10.1088/0022-3727/30/11/001 [DOI] [Google Scholar]; Pohl H. A., Pollock K., and Crane J. S., J. Biol. Phys. 6(314), 133 (1978). 10.1007/BF02328936 [DOI] [Google Scholar]
- 11.Minerick A. R., Zhou R. H., Takhistov P., and Chang H. C., Electrophoresis 24(21), 3703 (2003). 10.1002/elps.200305644 [DOI] [PubMed] [Google Scholar]
- 12.Green N. G. and Morgan H., J. Phys. D: Appl. Phys. 30(18), 2626 (1997). 10.1088/0022-3727/30/18/018 [DOI] [Google Scholar]
- 13.Ogata S., Yasukawa T., and Matsue T., Bioelectrochemistry 54(1), 33 (2001). 10.1016/S1567-5394(01)00112-8 [DOI] [PubMed] [Google Scholar]
- 14.Mueller T., Gradl G., Howitz S., Shirley S., Schnelle Th., and Fuhr G., Biosens. Bioelectron. 14(3), 247 (1999). 10.1016/S0956-5663(99)00006-8 [DOI] [Google Scholar]
- 15.Chen D. F., Du H., and Li W. H., J. Micromech. Microeng. 16(7), 1162 (2006). 10.1088/0960-1317/16/7/008 [DOI] [Google Scholar]
- 16.Liao S.-H., Fang Cheng I., and Chang H.-C., Microfluid. Nanofluid. 12(1–4), 201 (2012). 10.1007/s10404-011-0863-9 [DOI] [Google Scholar]
- 17.Cheng I. F., Lin C. C., Lin D. Y., and Chang H., Biomicrofluidics 4(3), 034104 (2010). 10.1063/1.3474638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao V. S., Kripesh V., Yoon S. W., and Tay A. A. O., J. Micromech. Microeng. 16(9), 1841 (2006). 10.1088/0960-1317/16/9/012 [DOI] [Google Scholar]
- 19.Yeh K.-L. and Chang J.-S., Biotechnol. J. 6(11), 1358 (2011); 10.1002/biot.201000433 [DOI] [PubMed] [Google Scholar]; Yeh K.-L., Chang J.-S., and Chen W.-m., Eng. Life Sci. 10(3), 201 (2010). 10.1002/elsc.200900116 [DOI] [Google Scholar]
- 20.Huang Y., Holzel R., Pethig R., and Wang X. B., Phys. Med. Biol. 37(7), 1499 (1992). 10.1088/0031-9155/37/7/003 [DOI] [PubMed] [Google Scholar]
- 21.Wu Y., Huang C., Wang L., Miao X., Xing W., and Cheng J., Colloids Surf. A 262, 57–64 (2005). 10.1016/j.colsurfa.2005.04.008 [DOI] [Google Scholar]
- 22.Hubner Y., Hoettges K. F., and Hughes M. P., J. Environ. Monit. 5, 861–864 (2003). 10.1039/b309131g [DOI] [PubMed] [Google Scholar]
- 23.Suscillon C., Velev O. D., and Slaveykova V. I., Biomicrofluidics 7, 024109 (2013). 10.1063/1.4801870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. M. S. Bono, Jr. , Ahner B. A., and Kirby B. J., Bioresour. Technol. 143, 623–631 (2013). 10.1016/j.biortech.2013.06.040 [DOI] [PubMed] [Google Scholar]
- 25.See supplementary material at http://dx.doi.org/10.1063/1.4903942E-BIOMGB-8-027406 for continuous DEP separation results of 13%–21% and 24%–30% lipid content.
- 26.Chabinyc M. L., Chiu D. T., McDonald J. C., Stroock A. D., Christian J. F., Karger A. M., and Whitesides G. M., Anal. Chem. 73(18), 4491 (2001). 10.1021/ac010423z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4903942E-BIOMGB-8-027406 for continuous DEP separation results of 13%–21% and 24%–30% lipid content.