Highlight text
Transcriptional repression of STP1 is rapidly induced by phosphorylatable sugars through an HXK1-independent signalling pathway involving the participation of sugar-responsive cis elements localized in the promoter.
Key words: Arabidopsis, STP1, sugar repression, sugar signalling, sugar transporter, transcriptional regulation.
Abstract
Sugars regulate the expression of many genes at the transcriptional level. In Arabidopsis thaliana, sugars induce or repress the expression of >1800 genes, including the STP1 (SUGAR TRANSPORTER PROTEIN 1) gene, which encodes an H+/monosaccharide cotransporter. STP1 transcript levels decrease more rapidly after the addition of low concentrations of sugars than the levels of other repressed genes, such as DIN6 (DARK-INDUCED 6). We found that this regulation is exerted at the transcriptional level and is initiated by phosphorylatable sugars. Interestingly, the sugar signal that modulates STP1 expression is transmitted through a HEXOKINASE 1-independent signalling pathway. Finally, analysis of the STP1 5′ regulatory region allowed us to delimit a region of 309bp that contains the cis elements implicated in the glucose regulation of STP1 expression. Putative cis-acting elements involved in this response were identified.
Introduction
As autotrophic organisms, plants produce their carbon skeletons through the photosynthetic process in the form of sugars. These carbon skeletons are essential as structural components and energy sources for plant growth and development. Similarly to many other organisms, plants respond to the carbon fluctuations caused by changes in photosynthetic efficiency or metabolic status and adjust their growth and development accordingly (Baena-Gonzalez and Sheen, 2008; Nunes-Nesi et al., 2010; Eveland and Jackson, 2012). Sugars act as signalling molecules, and plants have evolved mechanisms to efficiently perceive sugar availability and respond by modulating gene expression and protein activity in response to their nutrient status (Gibson, 2005; Rolland et al., 2006; Smeekens et al., 2010). The presence of sugars induces different developmental programmes, including growth, starch biosynthesis, and cell division. In contrast, sugar starvation upregulates photosynthetic activities and carbon remobilization, thus affecting central aspects of development (Koch, 1996; Borisjuk et al., 2003). Thus, understanding the mechanisms involved in sugar perception form an important area of research.
Plants have the capacity to sense different sugars, including sucrose, hexoses, and trehalose, and elicit responses, some that are specific to the type of sugar (Chiou and Bush, 1998; Sheen et al., 1999; Eastmond and Graham, 2003; Price et al., 2004; Wind et al., 2010). However, hexoses appear to be the most common signal detected by plants (Rolland et al., 2006; Smeekens et al., 2010). Diverse lines of evidence have demonstrated that sugar levels are detected by specific receptors and through independent signalling pathways (Rolland et al., 2006; Hanson and Smeekens, 2009). One of those receptors is the HEXOKINASE 1 (HXK1) that, in addition to its enzymatic activity, acts a sugar sensor (Jang et al., 1997; Moore et al., 2003; Cho et al., 2006). Experimental evidence has shown that upon sugar phosphorylation, HXK1 interacts with the VHA-B1 and RPT5B proteins to control the transcription of an important number of target genes (Cho et al., 2006). Evidence exists for additional sugar receptors, including sugar transporters and the negative regulator of trimeric G-protein (RGS1), but their mechanisms of action and downstream components are still poorly understood (Chen and Jones, 2004; Rolland et al., 2006).
Some of these sensors and downstream components of the sugar signalling pathways have initially been identified through genetic approaches with the isolation of sugar-response mutants. The characterization of these mutants has demonstrated the complexity of sugar signalling and the extensive cross-talk with other signalling pathways (León and Sheen, 2003; Gibson, 2005; Rolland et al., 2006; Eveland and Jackson, 2012). Additional components that are required for proper sugar perception were identified from sugar-insensitive mutants, including the enzyme ABA2, the transcription factors ABI4 and ABI5, and the ethylene-insensitive EIN2 protein (Zhou et al., 1998; Arenas-Huertero et al., 2000; Cheng et al., 2002). These factors were originally identified as components of the abscisic acid (ABA) or ethylene biosynthesis and signalling pathways, demonstrating the cross-talk between sugars and these hormones (Finkelstein and Gibson, 2002; León and Sheen, 2003; Yamagishi et al., 2009). Cross-talk has also been reported between sugar signalling and other hormones, such as auxin and gibberellins (Moore et al., 2003; Eveland and Jackson, 2012). Sugar signalling not only interacts with hormones but also with other nutrients, such as nitrogen (Coruzzi and Bush, 2001) and with the energy and stress signalling responses through the participation of the SnRK1 and TOR complexes (Baena-Gonzalez et al., 2007; Baena-Gonzalez, 2010; Xiong et al., 2013).
Based on the genes regulated by different sugars and sugar analogues, several pathways for sugar signalling are recognized and can be grouped into those that depend on HXK1 for signal initiation and those that are independent of this sensor (Rolland et al., 2006). The last group includes several pathways, such as the SnRK1-mediated pathway (Baena-Gonzalez et al., 2007; Jossier et al., 2009), the RGS1 pathway and most likely other undiscovered pathways (Chen and Jones, 2004; Chen, 2008; Sheen, 2010). Due to the complexity of sugar signalling, alternative strategies are required to further understand the molecular basis and additional components of the different sugar signalling pathways.
The regulation of gene expression is one of the most prominent mechanisms by which sugars modulate a variety of responses in plants. Independently of the signalling pathway, sugars positively or negatively affect the transcription of nearly 2000 different genes (Wang et al., 2003; Price et al., 2004; Li et al., 2006; Müller et al., 2007). In spite of the number of genes regulated by sugars, only a few transcriptional factors are known to be involved in this regulation. In fact, the participation of several bZIP and MYB transcription factors was recognized through the use of novel screenings (Rolland et al., 2006; Hanson and Smeekens, 2009; Sheen, 2010). The analysis of target genes has also proven to be a useful approach for identifying the cis-acting elements and trans-acting factors that are involved in sugar regulation. For example, using the promoter region of the α-amylase (α-Amy3) gene from rice allowed the identification of different MYB transcription factors that participate in the sugar regulation mediated by SnRK1A (Lu et al., 2007).
To further elucidate the mechanisms underlying sugar signalling in plants, we characterized the regulation by sugars of the Arabidopsis thaliana STP1 gene (AT1G11260). STP1 encodes a high-affinity sugar transporter that acts as an H+/monosaccharide cotransporter, capable of transporting a wide range of hexoses (Boorer et al., 1994; Büttner and Sauer, 2000). STP1 belongs to a family of 14 members that are highly conserved among plants and mediate hexose transport in cells of different tissues (Stadler et al., 2003; Slewinski, 2011). Several of these transporters are expressed in a tissue-specific manner, or at specific developmental stages (Büttner, 2010). STP1 is the member of the STP family with the highest expression level (Johnson et al., 2006; Johnson and Thomas, 2007). This high expression is detected in photosynthetic tissues, such as leaves and stems, while roots, siliques, and flowers show lower expression levels (Sherson et al., 2003). The expression of this transporter was also detected in guard cells and its accumulation responds to diurnal fluctuation that correlates with the accumulation of sucrose in this cell type. This has led to speculation that this H+/sugar cotransporter might be important for osmoregulation during the day and night periods (Stadler et al., 2003). The expression of several members of the STP family, including STP1, STP4, STP13, and STP14, is strongly repressed by sugars, and STP1 is one of the most repressed genes, as indicated by wide-genome analyses (Price et al., 2004; Büttner, 2010;). However, neither the pathway implicated in this regulation nor the factors involved are known. In this work, we analysed the mechanism that regulates the expression of the STP1 gene in response to sugar levels. This analysis demonstrated that the STP1 transcript is strongly downregulated within minutes after sugars increase. Interestingly, the regulation of this gene by sugars depends on phosphorylated hexoses but is independent of HXK1. We demonstrated that the regulation of this gene occurs at the transcriptional level and that the cis-acting elements responsible for this regulation are within a 309bp fragment of the promoter.
Material and methods
Plant material and growth conditions
For sterile growing conditions, A. thaliana seeds were sterilized following standard protocols (http://www.arabidopsis.org/). To break dormancy, the seeds were incubated for 3 days at 4°C in darkness. Adult plants were grown in Metro-Mix 200 (Grace Sierra, Milpitas, CA, USA). Plants and seedlings were grown under a 16h light/8h dark cycle in 120 µM m2 s–1 light conditions at 22°C. Wild-type Col-0 and the gin2, abi4-1, abi5, kin11, rgs1, and rgs1-2 mutants were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org/). The KIN10 knockout mutant and overexpressing lines were kindly donated by Dr Phillip Rolland (Institute of Botany and Microbiology, Heverlee-Leuven, Belgium). For sugar gene expression analysis, 50 seeds per treatment were grown in liquid 0.1X GM medium containing Murashige and Skoog basal salts (Caisson Laboratories Inc., UT, USA), supplemented with B5 vitamins (Sigma In., MO, USA), 0.05% MES, and 0.5% sucrose, and maintained with agitation at 350rpm. After 10 d, this medium was replaced with carbon (-C) starvation medium (0.1X GM without sucrose) for 2 d. Finally, the treatments were applied in 0.1X GM with or without sugar as indicated in each case using D-glucose monohydrate (Research Organics Cleveland, USA) or mannitol (Sigma-Aldrich, MO, USA) as carbon sources, as indicated.
Transgenic lines were generated through Agrobacterium tumefaciens-mediated transformation by floral dipping (Clough and Bent, 1998) into the Col-0 ecotype. Transgenic lines were selected in 1X GM media with 0.8% Phytagar and supplemented with 50 µg ml–1 kanamycin. At least three independent lines were selected for each construct.
Plasmid constructions
To generate transcriptional fusions of the STP1 upstream region with the GUS reporter gene (Jefferson et al., 1987), the 2.4kb fragment of the intergenic region of STP1 (between the loci AT1G11250 and AT1G11260) was amplified by PCR from DNA using the oligonucleotides STP1-3, 5′-AAG CTT CTC TGA CTG ACG TTA AAT TC-3′, and STP1-5R, 5′-GGA TCC TAA ACA AGA CCC GTA AA-3′. The 1.3kb, 1kb, and 0.5kb deletions were generated from the original 2.4kb fragment through PCR using the following specific forward oligonucleotides: STP1-1327F, 5′-CCA ATG CGG CCG CCC ATG AAA C-3′; STP1-1HF, 5′-GTT GAA GCT TTA GAG CAC TAT G-3′; and STP1-527HF, 5′-GCA AGC TTG TTT CAC ATT TTA AC-3′; and the common reverse STP1-5R oligonucleotide. All the fragments were cloned into the TOPO-TA vector (Invitrogen, Carlsbad, CA, USA) and confirmed by sequencing. Each fragment was subcloned into the pBI101 vector binary vector (Clontech Laboratories, Inc. CA, USA) in the HindIII and BamHI restriction site.
Expression analysis
Total RNA was isolated from frozen tissue using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the protocol provided by the manufacturer. For northern blot analyses, 1–20 µg of total RNA was fractionated in 1.5% agarose denaturing gels with 2% formaldehyde (Mallinckrodt Baker, MEX) and transferred onto a Hybond-N+ nylon membrane (GE, Bucks, UK). Hybridizations and washes were performed in stringent conditions. Probes were 32P-radiolabelled using the Megaprime DNA labelling system, according to the manufacturer’s protocol (GE, Bucks, UK). All probes were obtained by PCR amplification from DNA or cDNA as indicated. The STP1 (At1g11260) probe corresponds to a cDNA fragment of 499bp that was obtained using the oligonucleotides STP1-1, 5′-TGC TAT AGT GGT TGT AAC GTT CAT T-3′, and STP1-2, 5′-GGC TAA TAC ACT TTT TCC TTT ACG ACA-3′. The GUS probe (860bp) was obtained using the oligonucleotides GUS3F, 5′-CGA AAA CTG TGG AAT TGA TCA G-3′, and GUS4R, 5′-ACC ATC AGC ACG TTA TCG AAT C-3′. For DIN6 (At3g47340), a 382bp fragment was obtained using the oligonucleotides DIN6F, 5′-GCC TGA AAG ATC ACG CTG CTC-3′, and DIN6R, 5′-GCC TTT GCA GTC GAA CAA GCC-3′. For β-AMY (At4g15210), a 608bp cDNA fragment was obtained using the oligonucleotides β-Amy-1, 5′-CGG AGA AGG GGA AGT TTT TC-3′, and β-Amy-2, 5′-AAT CTC ATG CCC GTA CTT CG-3′. For SBE2.2 (At5g03650), a 335bp cDNA fragment was obtained using the oligonucleotides SBE2.2A, 5′-GAG TGT CTC TTA CTC CAC GC-3′, and SBE2.2B, 5′-GGG AAC TAT TCT TGG TTT CAC-3′. For APL3 (At4g39210), a 345bp cDNA fragment was obtained using the oligonucleotides Apl3-1, 5′-TTC TTG GGA GAA TGC AGC ATC-3′, and Apl3-2, 5′-TGT TCA TAT CAC AGT ACC GTC-3′. Densitometric analysis was performed using the ImageJ 1.43u program from the National Institutes of Health, USA, http://rsb.info.nih.gov/ij. To evaluate confidence of the data we used ANOVA statistical analysis (http://www.r-project.org/).
GUS histochemical analysis
Twelve-day-old seedlings exposed to different sugar sources for 12h were stained for 2h using the GUS histochemical assay as reported (Jefferson et al., 1987). Plant were clarified using a modified protocol from Malamy and Benfey (1997). Pigments were removed with 70% ethanol and plants were rehydrated by incubations in 50% and 30% ethanol for 15min each, and then transferred to a solution of 0.24 N HCl in 20% methanol and incubated at 62ºC for 1h. This solution was replaced by 7% NaCl in 60% methanol and incubated for 25min at room temperature. Then plants were dehydrated with 40%, 20%, and 10% ethanol for 15min each to finally be mounted in a solution with 50% glycerol and 2% DMSO. Samples were visualized using a stereoscopic (Nikon SMZ1500) and a light microscope (Nikon Eclipse E600).
In silico analysis
The 309bp sequence from the STP1 promoter was analysed using the PLACE (Plant Cis-acting Regulatory DNA Elements) database (Prestridge, 1991; Higo et al., 1999), and released data were analysed to identify the reported motifs involved in sugar regulation. Additional cis elements, which were not included as sugar-responsive elements in this database, were identified by manual comparison with the promoter sequence for DIN6 (At3g47340), a gene that is downregulated in the presence of sugar (Li et al., 2006; Baena-Gonzalez et al., 2007).
Results
STP1 expression is rapidly regulated by glucose supply
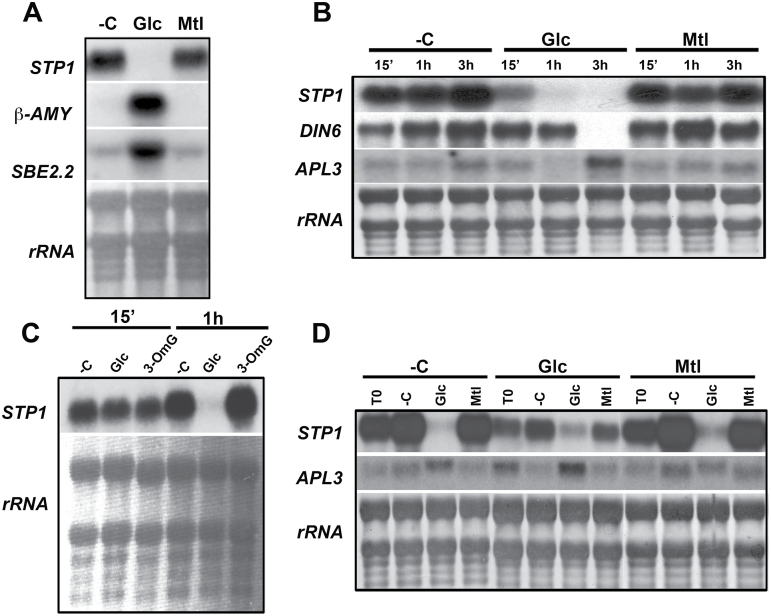
Analysis of the available microarray data indicated that STP1 (AT1G11260) was one of the most prominent downregulated genes in response to sugars in A. thaliana (Price et al., 2004). Thus, we wanted to characterize the regulatory mechanism involved in the STP1 gene response to sugars. To corroborate the effect of sugars on STP1 expression, we analysed the accumulation of its transcript in the presence of glucose (Glc) by northern blotting. Sugar treatments were performed after carbon starvation in liquid media (see Material and Methods). As shown in Fig. 1A and in agreement with published microarray data, the STP1 transcript level was dramatically reduced in the presence of 150mM Glc, relative to the levels in the untreated samples (–C) or in the isosmotic control with mannitol (Mtl). Under these conditions, the expression was induced (Fig. 1A) for the known Glc upregulated genes, such as β-AMY (Beta-amylase) and SBE2.2 (Starch Branching Enzyme 2.2), that were used as controls (Rook et al., 2001). These results confirm that the STP1 transcript is downregulated by the presence of Glc (Price et al., 2004).
Fig. 1.
STP1 expression is repressed by glucose. (A) Northern blot analysis of the total RNA from 12-day-old Col-0 plants that were transferred to media without a carbon source (–C) or with 150mM glucose (Glc) or 150mM manitol (Mtl) for 6h. (B) Time course of the STP1 expression of plants exposed to media without carbon (–C) or with 150mM Glc or 150mM Mtl for 15min, 1h, and 3h. (C) STP1 expression from plants that were treated with 5mM Glc or 3-O-methylglucose (3-OmG) for 15min or 1h. (D) Transcript expression profile from 12-day-old plants grown for 2 days in 0.1X GM without a carbon source (–C) or with 150mM Glc or Mtl and then transferred for 6h to –C medium or medium supplemented with 150mM Glc or Mtl. Each lane in the different blots contains 10 µg of total RNA. The blots were hybridized with radioactive probes for STP1, β-AMY (beta-amylase), SBE2.2 (starch-branching enzyme 2.2), and APL3 (ADP-glucose pyrophosphorylase large subunit), as indicated. The rRNA from the methylene blue-stained membranes is shown as a loading control. The membranes shown are representative of at least two biologically independent experiments.
For more detailed analysis of the regulation of the STP1 gene in response to sugar, the level of its transcript was followed at different times after the addition of Glc. As shown in Fig. 1B, the STP1 transcript level decreased 15min after the addition of Glc. This repression was not observed in the absence of external sugar (–C) or with the addition of Mtl. The STP1 transcript was basically undetectable 1h after the treatment. The rapid response of STP1 contrasts with the slower response for other characterized sugar-repressed genes, such as DIN6/ASN1, which encodes the asparagine synthetase 1 enzyme (Lam et al., 1998; Price et al., 2004). The reduction of the DIN6 transcript was evident only 3h after the addition of Glc (Fig. 1B). A similar situation was observed for the Glc-upregulated APL3 gene (encodes the large subunit of ADP-glucose pyrophosphorylase), whose transcript accumulation in response to Glc was detectable only 3h after the addition of sugar (Fig. 1B). These findings demonstrate that the expression of STP1 is rapidly modulated by the changes in sugar levels.
To establish the sensitivity of STP1 to sugars, the expression of this gene was analysed in the presence of different Glc concentrations (data not shown). As shown in Fig. 1C, the presence of 5mM Glc was sufficient to dramatically decrease STP1 transcript levels, albeit after a longer time (1h) than the 15min that was found with 150mM Glc (Fig. 1B). This reduction was not observed with 3-O-methylglucose (3-OmG), a poorly metabolized Glc analogue (Fig. 1C). These data allowed us to conclusively demonstrate that the accumulation of the STP1 transcript is rapidly downregulated by the presence of low Glc levels and that this regulation is not related to an osmotic response.
STP1 gene expression is dynamically regulated by sugar fluctuation
Experimental evidence has demonstrated that regulation by sugars is complex. For example, sugar regulation of the α-Amy3 gene from rice involves both transcriptional repression and activation in response to the presence or absence of a carbon source. These positive and negative regulations involve the action of different trans-acting factors on the same cis-acting regulatory element (Lu et al., 1998; Lu et al., 2002). To further understand how STP1 responds to changes in the carbon supply, the levels of its transcript were analysed in response to fluctuations in sugar availability. For this purpose, 10-day-old plants grown either in sugar starvation (–C) or in the presence of 150mM Glc or Mtl for 2 days were transferred to media without a carbon source (–C) or with 150mM Glc or 150mM Mtl for 6h, and the levels of the STP1 transcript were analysed. In agreement with our previous results, the initial level of STP1 transcript (T0) was lower in the plants grown in the presence of Glc than in those grown in its absence. Independently of the initial STP1 transcript level, the addition of Glc resulted in a drastic reduction of the STP1 transcript in the plant (Fig. 1D). In contrast, when the plants were transferred to media without sugar (–C), the STP1 transcript accumulated (Fig. 1D). In addition, in accordance with published results, the expression of the APL3 gene increased in the presence of Glc, albeit at different levels depending on the initial media in which the seedlings were grown prior to the treatment (Fig. 1D). Together, these data demonstrated that Glc regulation of STP1 mRNA is rapid and dynamic. This result also indicates that multiple elements may be involved in the Glc regulation of STP1, a regulation that is similar to that of the rice α-Amy3 gene.
Sugar regulation of STP1 responds to the level of phosphorylated hexose
Due to the rapid and sensitive response observed in the accumulation of the STP1 transcript in response to fluctuations in the Glc level, we decided to further investigate the mechanism involved in this regulation. Plants have the capacity to sense different sugars and transmit their signals through different pathways that involve specific components and mechanisms (Xiao et al., 2000; Rolland et al., 2006). The use of Glc analogues has been useful for characterizing the requirements for the regulation of specific genes by sugars (Jang and Sheen, 1994). Thus, we analysed the effect of the Glc analogues mannose (Man) and 3-OmG on STP1 expression. In the presence of 150mM Man, low levels of the STP1 transcript were observed, similar to those found with 150mM Glc (Fig. 2A). Man is a Glc analogue that is transported into cells and phosphorylated by hexokinase (HXK) but is very slowly metabolized (Jang and Sheen, 1994; Xiao et al., 2000). In contrast, in the presence of 3-OmG, no repression of the STP1 transcript was observed, and the expression level remained comparable to the one found in the carbon deprivation (–C) condition (Fig. 2A). 3-OmG is transported into the cell (Jang and Sheen, 1997; Lalonde et al., 1999; Smeekens, 2000) but is not phosphorylated because it is a very poor substrate for HXK (Cortes et al., 2003). Finally, similarly to Glc, the addition of sucrose (Suc) resulted in very low STP1 levels (Fig. 2A). The response observed for STP1 to these different sugar analogues was the same with lower levels (5mM) of these sugars (Fig. 2B). Similar responses to these sugar analogues were found for the DIN6 gene, which is also induced by sugar starvation (Baena-Gonzalez et al., 2007). Under the conditions used in this analysis, only a slight reduction in the expression level of the photosynthetic CAB1 gene was detected (Fig. 2), suggesting that the response of this gene requires a longer treatment time or higher sugar concentrations. Together, these results support the hypothesis that the signal that initiates the regulation of the STP1 transcript requires a phosphorylatable hexose, such as Glc or Man.
Fig. 2.
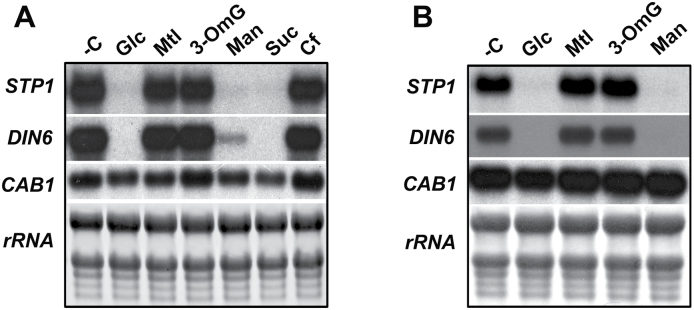
Regulation of STP1 expression by sugars. The expression of STP1, DIN6, and CAB1 was analysed by a northern blot from the RNA extracted from 12-day-old plants grown for 2 days in sugar-depleted (–C) media and then transferred to 150mM (A) or 5mM (B) Glc, Mtl, 3-OmG (3-O-methylglucose), Man (mannose), or Suc (sucrose) for 6h. Each lane was loaded with 10 µg of total RNA. The rRNA from the methylene blue-stained membrane is shown as a loading control. The blots shown are representative of three biologically independent experiments.
An independent HXK1 pathway drives the sugar regulation of STP1
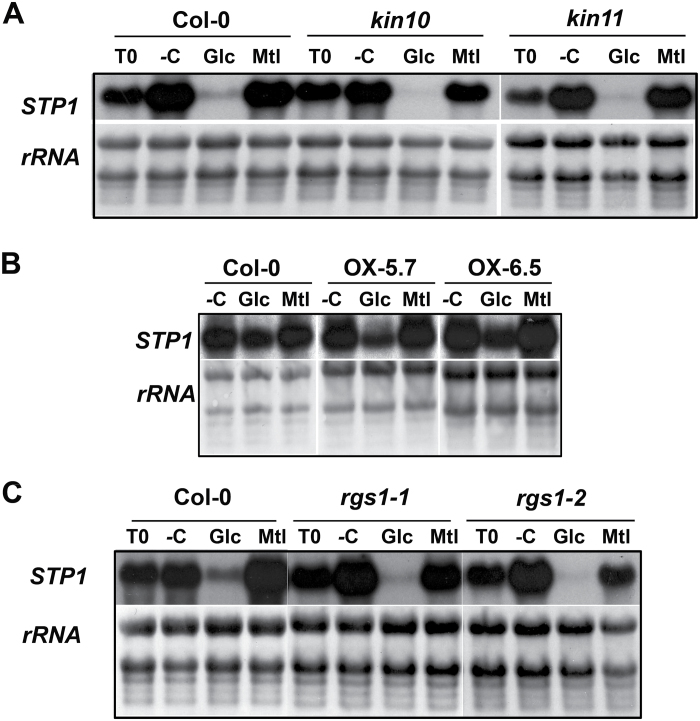
Previous work has demonstrated that HXK1 functions as a primary Glc sensor that initiates a specific sugar signalling pathway; this pathway then induces or represses the expression of many genes in response to phosphorylated sugars (Moore et al., 2003; Price et al., 2004; Li et al., 2006). Additional components of this signalling pathway include the ABI4 and ABI5 transcription factors (Arenas-Huertero et al., 2000). To determine whether any of these components are required for STP1 sugar regulation, we evaluated the STP1 transcript level in response to Glc in the HXK1 (gin2), abi4-1, and abi5 mutants. As shown in Fig. 3, the level of the STP1 transcript in all three mutants decreased upon Glc addition, similarly to wild-type plants. No decrease in the transcript level was observed in the absence of sugar or in the corresponding Mtl isosmotic control (Fig. 3). In this analysis, we observed that the STP1 transcript level in the abi5 mutant prior to sugar treatment (T0) was lower than that of the wild type and the other mutants (Fig. 3). This result suggests that ABI5, independently of its role in sugar regulation, is required to maintain normal levels of the STP1 transcript. However, these results demonstrated that neither HXK1 nor the transcription factors ABI4 or ABI5 participate in the sugar regulation of the STP1 transcript.
Fig. 3.
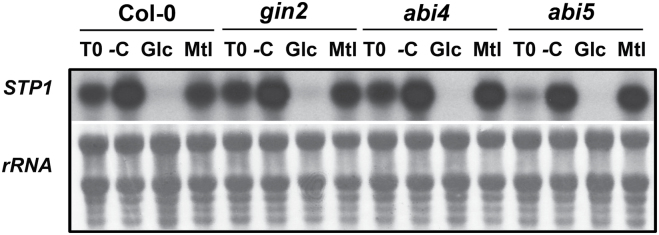
STP1 regulation by Glc is mediated by an HXK1-independent signalling pathway. STP1 expression was analysed in wild-type plants and in the sugar signalling mutants gin2, abi4, and abi5. Samples were obtained from 12-day-old plants grown for 2 days in sugar starvation (–C) conditions and then transferred to –C, 150mM Glc, or Mtl media for 6h. Ten micrograms of total RNA was used for northern analysis and was hybridized with the STP1 probe. An initial control, which was measured before the treatments (T0), is included. The rRNA from the methylene blue-stained membranes is shown as a loading control. The membrane shown is representative of two biologically independent experiments.
In addition to HXK1, other factors have been shown to play important roles in plant sugar responses, including SnRK1 kinase and a heterotrimeric G protein (Baena-Gonzalez and Sheen, 2008; Urano et al., 2013). To analyse the possible role of these factors in the regulation of STP1 by sugars, we measured the expression of STP1 in the corresponding mutants. In the case of SnRK1 kinase, we evaluated knock-out mutants for the two catalytic subunits, kin10 and kin11. Although these catalytic subunits are known to be partially redundant, analysis of a double mutant was not possible due to its lethality (Baena-Gonzalez and Sheen, 2008). In the case of the G protein, we evaluated two independent null mutant alleles of the RGS1 factor (rgs1 and rgs1-2), a protein that modulates G-protein signalling and that has been reported to be an important component for HXK-independent sugar signalling responses (Chen et al., 2003). As shown in Fig. 4A, no difference in the regulation of STP1 by Glc was found in the kin10 or kin11 mutants compared to that in the wild-type Col-0 plants. However, due to the partial redundancy of these subunits, the participation of SnRK1 in the regulation of STP1 in response to Glc cannot be totally excluded. To further address the function of the SnRK1 complex in the Glc regulation of the STP1, two independent lines that overexpress the KIN10 catalytic subunit (KIN10-OX) were analysed (Baena-Gonzalez and Sheen, 2008). We hypothesized that if the SnRK1 kinase has any role in the sugar repression of the STP1 gene, this response should be exacerbated in the KIN10 gain-of-function lines. Because the level of the STP1 transcript in the wild-type plants remain unaltered for the first 15min after the addition of 5mM Glc (Fig. 1C), we analysed the STP1 level in two overexpressing lines, KIN10-OX5.7 and KIN10-OX6.5, under these conditions. However, we did not detect any difference in the STP1 expression level between the overexpressing lines and the wild-type plants 15min after Glc addition (data not shown). We also analysed STP1 levels after exposure to Glc for a longer time (30min). In this case, we detected a slight increase in the repression level in the overexpressing seedlings (Fig. 4B). Densitometric analysis of the STP1 signal from independent biological experiments showed that repression of STP1 expression in the KIN10-OX lines was 31%, compared to 22% in the wild-type plants. Finally, no difference was detected in the STP1 sugar regulation in the two mutant alleles of the RGS1 gene in comparison to that in the wild-type plants (Fig. 4C). These results suggest that none of the factors analysed here play a major role in the regulation of STP1 by sugars.
Fig. 4.
STP1 sugar regulation in SnRK1 and RGS1 mutants. Total RNA was obtained from 12-day-old kin10 and kin11 mutants (A), from two independent overexpressing KIN10 (OX5.7 and OX6.5) lines (B), and from rgs1-1 and rgs1-2 (C). After carbon starvation for 2 days, the plants were transferred to media depleted of sugar (–C) or with 150mM Glc or Mtl for 6h (A and C) or with 5mM Glc or Mtl for 30 min (B). Ten micrograms of total RNA was used from each sample for northern analysis and was hybridized with the STP1 probe. T0 represents the level of STP1 prior to the sugar treatment. The rRNA from the methylene blue-stained membranes is shown as a loading control. Membranes shown are representative of two biologically independent experiments.
The regulation of STP1 by sugars depends on the DNA elements located in the upstream region of this gene
Transcription plays a major role in the sugar regulation of many genes (Price et al., 2004; Bläsing et al., 2005). In various cases, this regulation depends on the presence of one or more cis-acting elements in the promoter region of the sugar-regulated genes (Chen et al., 2006; Li et al., 2006). However, post-transcriptional regulatory mechanisms are also involved in the regulation by sugars (Rolland et al., 2006; Hummel et al., 2009). To characterize the molecular mechanism of the regulation by Glc of the STP1 gene, a 2.4kb fragment upstream from the ATG, which includes the upstream regulatory region and the 5′ UTR, was fused to the β-glucuronidase (GUS) reporter and introduced into A. thaliana plants (Fig. 5). Three independent transgenic lines (L1-2, L3-6, and L4-5) were selected, and the presence of the transgene was confirmed by PCR (data not shown). Homozygous plants from each line were selected.
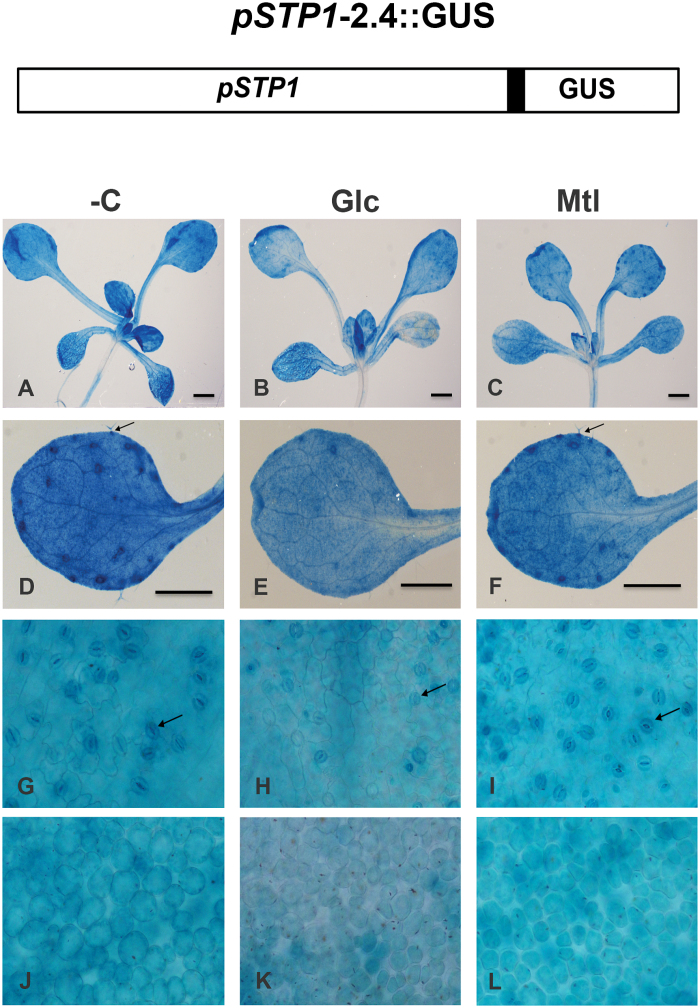
Fig. 5.
Expression pattern of the STP1 gene in seedlings in the presence or absence of sugars. At the top is a diagram of the transcriptional fusion including 2.4kb of the 5’ upstream regulatory region of the STP1 gene (pSTP1-2.4kb) fused to the GUS reporter gene used to generate the transgenic lines. The corresponding 5’ UTR region is shown as a black box. The panels below show GUS staining, including the pSTP1:GUS expression pattern from a representative 12-day-old transgenic line exposed for 12h to media in the absence (–C) or presence of 150mM glucose (Glc) or mannitol (Mtl). Promoter expression pattern in the whole plant (A, B, and C) and in leaves from –C plants (D), with Glc plants (E), or with Mtl plants (F). Arrows mark the trichomes; the base of the mature trichomes strongly stained for GUS activity. (G–L) Epidermal surface of rosette leaves showing stomata (marked by arrows) from plants grown –C (G), with Glc (H), or with Mtl (I); and GUS activity in mesophyll tissue from plants grown –C (J), with Glc (K) and with Mtl (L). Bars correspond to 1mm.
GUS expression patterns of three independent transgenic lines were analysed in 12-day-old seedlings. GUS staining was prominently detected in leaves but it was also present in the vascular system of stems and roots with lower but clearly detectable levels (Fig. 5A). As previously published (Stadler et al., 2003), the site of higher expression at this developmental stage was in leaves (Fig. 5A). A detailed analysis of this organ revealed that STP1 expression was particular strong in trichomes, including the base of the stalk and the cells around them (Fig. 5D), and in stomata (Fig. 5G). The expression in the guard cells was not homogenous with a more intense GUS activity at the membrane near the stomatal pore (Fig. 5G). GUS staining was also present in the mesophyll cells of the leaves at lower levels (Fig. 5J). In contrast to published data (Stadler et al., 2003), we could not detect differences in the GUS expression between the adaxial or abaxial surfaces of the leaves. However, such a difference might exist but be masked by the diffusion of the GUS marker.
Our previous northern analysis demonstrated that the STP1 transcript practically disappears after 1h of Glc addition (Fig. 1B), thus the activity of GUS was followed in the transgenic plants after the addition of 150mM Glc. In contrast to the RNA analyses noticeable differences in GUS accumulation in the Glc-treated plants were not observed prior to 12h of Glc exposure (data not shown). After 12h of Glc treatment a reduction in the GUS activity was observed in all the transgenic lines (Fig. 5B and E). This response is specific for Glc as it is not observed with isosmotic concentrations of Mtl (Fig. 5C and F), which display an undistinguishable GUS level compared to the one observed without the carbon source (Fig. 5A). The decrease in GUS expression in response to the presence of Glc was most prominent in the stomata and trichomes, where the defined patterns observed in these structures were basically lost (Fig. 5E and H). However, even after 12h of Glc treatment, considerable GUS activity was detected in the sugar-treated transgenic plants (Fig. 5) in contrast to the endogenous STP1 RNA response. This apparent discrepancy might be explained because it has been shown that GUS activity persists for long periods beyond its actual promoter activity (Kavita and Burma, 2008). Thus, to unequivocally demonstrate whether the STP1 promoter in these transgenic plants contains the elements responsible for regulation by sugar observed with the endogenous STP1 gene, the expression of GUS and STP1 endogenous transcripts in these lines was analysed by northern blotting. As shown in Fig. 6, high levels of the GUS transcript were detected in the transgenic plants that were transferred to media without sugar (–); this high expression was independent of the initial growing media prior to the transfer (with or without sugar). In contrast, the GUS transcript was barely detectable when these plants were transferred to media containing 150mM Glc (Fig. 6). This regulation was very similar to the one observed for the STP1 endogenous transcript (Figs 1C and 6). From this analysis, we concluded that the cis-acting elements responsible for Glc regulation of the STP1 gene are contained in the 2.4kb fragment, at least under the conditions analysed.
Fig. 6.
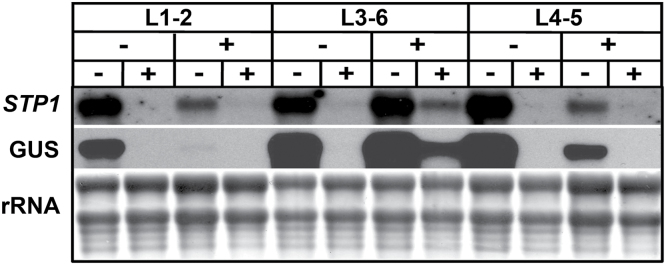
Sugar regulates STP1 expression at a transcriptional level. Total RNA was isolated from 12-day-old plants from independent homozygous transgenic lines containing pSTP1-2.4::GUS. Prior to the treatment, the plants were grown for 2 days in media depleted of sugar (–) or supplemented with 150mM Glc (+) and then transferred to media without (–) or with 150mM Glc (+) for 6h. Each lane contains 10 µg of total RNA, and the blot was hybridized with the STP1 and GUS probes. The rRNA of the methylene blue-stained membrane is shown as a loading control. Membranes shown here are representative of three biologically independent experiments.
Delimiting the cis-acting regions of the STP1 promoter that respond to Glc.
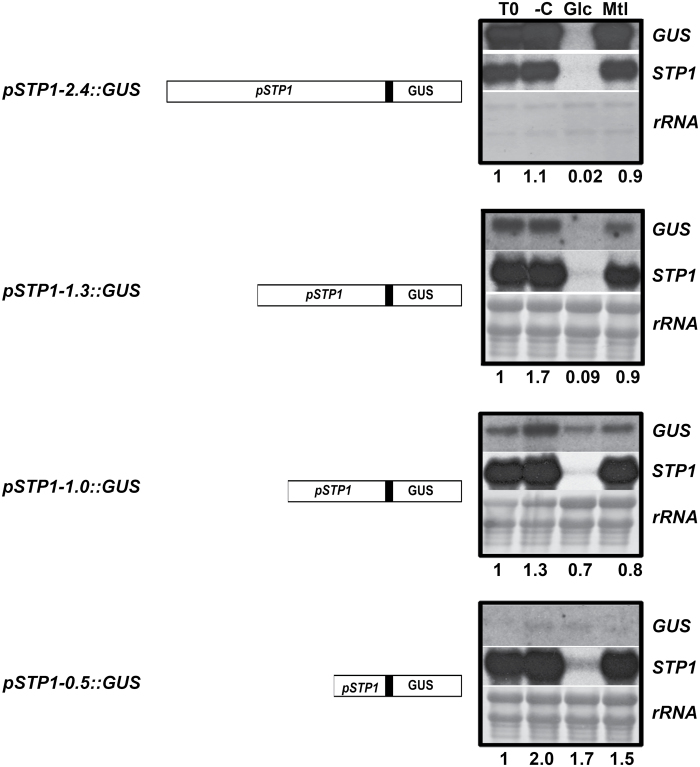
In order to narrow down the specific elements involved in the sugar regulation of STP1, three consecutive deletions of the original 2.4kb upstream fragment were generated and fused to GUS; each containing 1.3kb, 1kb, and 0.5kb from the original fragment (Fig. 7). Independent transgenic lines were selected from each deletion based on their resistance to kanamycin, and the deletion size was corroborated by PCR (data not shown). Homozygous plants from a representative line were selected for each deletion, and the expression levels of STP1 and GUS were analysed after sugar treatments. Treatments were conducted using 10-day-old plants starved of a carbon source for two days, after which the plants were transferred to different media (–C, Glc, or Mtl) for 6h. The expression level of the GUS transgene was compared to the level prior to the transfer (T0) in each deletion. We observed that the basal GUS transcript level (T0) was considerably lower in all deletions than the level observed in the 2.4kb fragment (Fig. 7). This result is particularly evident in the pSTP1-1.3::GUS and pSTP1-0.5::GUS constructs, indicating that important elements required for normal STP1 expression level are localized between the deleted sequences in each case. However, independently of the basal transcript level (T0), the presence of 150mM Glc repressed the GUS transcript level in the pSTP1-1.3::GUS deletion; the extent of repression was similar to the one observed with the initial pSTP1-2.4::GUS construct (Fig. 7). In contrast, minor differences, if any, in the level of GUS transcript were observed in the two additional deletions (pSTP1-1::GUS, and pSTP1-0.5::GUS) in the presence of Glc (Fig. 7). Densitometric analysis from independent lines and independent experiments demonstrated that the presence of Glc results in a 98% reduction in the pSTP1-1.3::GUS lines (P = 0.001) compared with the T0 control plants; only a 30% reduction was detected for the pSTP1-1::GUS lines (P = 0.004) (Fig. 7). The efficiency of the treatments was corroborated by the response of the endogenous STP1 transcript (Fig. 7). Together, these results demonstrate that Glc regulates STP1 expression at the transcriptional level and that repression by sugar depends on cis-acting sequences contained within a 309bp fragment, which is localized between 1.3 and 1kb upstream of the STP1 translational initiation codon (Fig. 7).
Fig. 7.
Deletion analysis of the STP1 promoter region. Total RNA was obtained from representative 12-day-old transgenic homozygous lines containing 2.4, 1.3, 1, and 0.5kb of the upstream sequences of the STP1 gene fused to GUS (pSTP1::GUS), as indicated in each diagram. Plants were deprived of sugars for 2 days prior to being transferred to media without (–C), or with 150mM Glc or Mtl for 6h. T0 corresponds to the RNA from the plants prior to the transfer and was taken as the control. A total of 1 µg of total RNA was used for pSTP1-2.4::GUS, whereas 20 µg was used for the other transgenic lines. Each blot was hybridized against the STP1 and GUS probes as indicated in each deletion. The rRNA staining of the methylene blue membrane is shown as a loading control. The numbers at the bottom of each blot correspond to the level of the GUS transcript relative to the level found in the sample prior to the transfer (T0), which is taken as 1. Densitometric analyses were performed from at least two independent biological experiments.
In silico analysis of putative cis Glc-responsive elements in the 309bp STP1 promoter fragment.
Previous studies have identified cis-regulatory elements for independent genes that participate in repression by sugars (Hwang et al., 1998; Morita et al., 1998; Toyofuku et al., 1998; Tatematsu et al., 2005). Thus, we performed an in silico analysis of the 309bp fragment and searched for motifs that are known to be involved in the repression by sugars. This analysis was performed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/) and by also considering additional cis elements that were found to be overrepresented in the regulatory regions of sugar-repressed genes (Li et al., 2006; Baena-Gonzalez et al., 2007). This analysis revealed 17 potential elements in the 309bp fragment (Fig. 8 and Table 1) that belonged to eight different sugar motifs. One of the most interesting elements found in this region corresponds to the TATCCAOSAMY motif. This motif occurs twice in the 309bp STP1 fragment (at –1134 and –1155 from ATG); the two instances are separated by 15bp (Fig. 7). The TATCCAOSAMY motif was originally found in the 5′ upstream regulatory region of the α-Amy3D gene from rice and has been demonstrated to be essential for the regulation of this gene by sugars (Lu et al., 1998; Lu et al., 2002). Part of the TATCCAOSAMY motif (TATCC) overlaps with two other elements (MYBST1 and I-BOX) in the complementary strand and in reverse orientation (Table 1). The I-BOX is found four times within this sequence (Fig. 8), but only two of these instances overlap with the TATCCAOSAMY motif. An additional element in this region was the CGACGOSAMY3 motif (Hwang et al., 1998), which localized at –1104 in the STP1 promoter (Fig. 8). This motif was also originally described in the promoter of the α-Amy3D gene and is required for the Glc repression of this gene (Hwang et al., 1998). In addition, we identified seven sequences with homology to three elements that are overrepresented in the sugar-repressed genes in the microarray data reported by Li et al. (2006). Four of these sequences share homology with the GATTA motif, two with the EVENINGAT core element and one with the CATCC motif (Fig. 8 and Table 1).
Fig. 8.
Putative sugar regulatory motifs in the 309bp region of the STP1 promoter. The numbers indicate the position of the last base in each motif and refer to the translation initiation site of STP1. The overlapping elements are underlined. The arrowheads indicate elements found in reverse orientation and (–) in the complementary strand.
Table 1.
Known cis-acting elements involved in sugar repression in the 309bp fragment from the STP1 promoter
| Element | Sequence | Reference |
|---|---|---|
| CGACGOSAMY3 | CGACG | Hwang et al., 1998 |
| TATCCAOSAMY | TATCCA | Lu et al., 1998 |
| SREATMSD | TTATCC | Tatematsu et al., 2005 |
| TATCCAYMOTIFOSRAMY3D | TATCCAY | Toyofuku et al., 1998 |
| MYBST1 | GGATA | Baranowskij et al., 1994 |
| I-BOX core | GATAA | Manzara et al., 1991 |
| EVENINGAT core | ATATCT |
Harmer et al., 2000; Li et al., 2006 |
| CATCC | CATCC | Li et al., 2006 |
| GATTA | GATTA | Li et al., 2006 |
| G-box related | ACGTG |
Lu et al., 1998; Baena-González et al., 2007 |
Similar to STP1, the expression of the DIN6 gene is strongly repressed by the presence of sugars and activated during sugar starvation (Baena-Gonzalez et al., 2007). Thus, we decided to compare 309bp of the STP1 fragment with the upstream sequence of DIN6 gene. This analysis revealed only two motifs that were shared between these sequences: one of them is the TATCCAOSAMY motif, and the other is a sequence related to a G-box (ACGTG) (Fig. 8).
Finally, several members of the STP family are also repressed by sugars (Price et al., 2004). Thus, we also searched for common motifs between STP1 and three other known sugar-regulated members. For this analysis, the complete upstream intergenic regions of the STP4 (At3g19930), STP13 (At5g26340), and STP14 (At1g77210) genes were compared against the 309bp fragment of the STP1 promoter. Six out of the eight different motifs previously identified in STP1 were also present at least once in the control regions of the other STP genes (Table 1). Interestingly, the CGACG and the TATCCAOSAMY motifs were found in STP4 and STP13 genes, but not in the STP14 gene.
Discussion
Sugars act as key regulators of gene expression by inducing or repressing the transcription of many genes (Koch, 1996). Many studies have contributed to understanding the mechanisms by which sugars regulate gene expression (Rolland and Sheen, 2005; Eveland and Jackson, 2012). Initial forward genetics studies were valuable for demonstrating the complexity of sugar signalling and provide evidence for the existence of multiple signalling pathways. However, genomics and system biology analyses have been crucial for demonstrating the effect of sugar availability on expression throughout the entire genome (Price et al., 2004; Villadsen and Smith, 2004; Gutierrez et al., 2007; Osuna et al., 2007). The STP family is one of the gene families that has repeatedly been detected in genomic analyses as highly responsive to sugars (Price et al., 2004; Villadsen and Smith, 2004). This family includes genes that encode low- and high-affinity monosaccharide transporters (Stadler et al., 2003; Büttner, 2010; Slewinski, 2011). Compared to other members of the family, STP1 is a high-affinity H+/sugar cotransporter with the highest and broadest expression in A. thaliana (Büttner, 2010). Our data corroborated the findings that the expression of STP1 is rapidly modulated by minor fluctuations in sugars levels (5mM Glc). This response contrasts with the response of other sugar-repressed genes, such as several photosynthetic genes, that require higher sugar levels and a longer time to affect the level of their transcripts (Acevedo-Hernandez et al., 2005). In addition to STP1, other members of the STP family have also been shown to be regulated by sugars; however, whether these involve common mechanisms is unknown.
Previous work demonstrated that STP1 expression is induced by darkness and repressed by light in guard cells. This regulation has been suggested to be important for the import of carbon to these cells, particularly during dark periods (Stadler et al., 2003). Our analysis with transgenic lines containing the pSTP1:GUS fusion corroborated the view that one of the sites with major levels of GUS accumulation corresponds to the stomatal guard cells. Interestingly, this expression is notably decreased with exposure to Glc. Thus it is likely that at least part of regulation previously observed by light is linked to the sugar fluctuations in these cells during dark periods more than a direct downregulation by light. Since guard cells depend on sugar import to maintain their metabolism as they are unable to perform photosynthesis, it is likely that during dark periods the levels of phosphorylatable hexoses become very low and in consequence the expression of the STP1 gene gets induced. Previous work has found an increase in STP1 mRNA at the onset of the dark period. We could speculate that the decrease in sugar import as a result of the lack of photosynthetic activity, together with the start of starch breakdown that will supply carbon skeletons during the next hour, may mean that the actual intracellular phosphorylatable hexose levels are very low. The expression of a high-affinity sugar transporter such as STP1 under these conditions is possibly important for transporting available external hexoses. Although additional experiments will be required to clarify these aspects, the sensitive and rapid response observed here for STP1 expression is very well suited to ensuring proper sugar influx in response to minor fluctuations in sugar availability in guard cells as well as in other plant tissues. The other sites where high GUS expression was detected are the trichomes. However, the physiological reason for the requirement of this transporter in this type of specialized structure is less obvious and will require future analyses.
Considering the mechanisms, we believe that the rapid response of the STP1 transcript to fluctuations in sugar levels suggests that some of the elements involved in the perception of the Glc signal should be present prior to the stimulus. This possibility agrees with the observation that the Glc repression of other STP genes (STP14 and STP4) is normal in the presence of the translational inhibitor cycloheximide (Price et al., 2004). Surprisingly, that study also found that the repression level of the STP1 transcript appears to be less severe in the presence of cycloheximide. Thus, it is possible that the de novo synthesis of some of the trans-acting factors is required either to achieve full repression or to sustain this response (Price et al., 2004). The present analysis also reveals that the half-life of the STP1 transcript is apparently not very long; thus, the repression of the transcription level is reflected in the total transcript level within minutes of Glc addition.
Our analyses of STP1 expression using different Glc analogues demonstrated that the signal that induces the repression of this gene is a phosphorylatable sugar. These data agree with previous reports that found that the non- or poorly phosphorylatable Glc analogues, such as 3-OmG and 6-deoxyglucose, did not change STP1 expression (Cortes et al., 2003; Villadsen and Smith, 2004). Interestingly, our data also demonstrated that the sugar signal that modulates the repression of the STP1 gene is independent of the HXK1 sensor. Therefore, a primary sensor different from HXK1 must perceive the phosphorylated sugars that initiate STP1 regulation. In spite of the important efforts of many groups, still almost nothing is known about alternative receptors for sugar perception with the exception of the regulator of G protein (RGS1). RGS1 has been suggested to bind sugars and attenuate the cell division of the apical root meristem through its interaction with a heterotrimeric G protein independently of HXK1 (Chen et al., 2003; Chen, 2008;). However, in this work, we demonstrated that RGS1 does not appear to play a major role in the sugar regulation of STP1 because the repression of STP1 by sugars is very similar to the repression in wild-type plants in the absence of this regulator.
The genome of most plants encodes various HXK-related genes in addition to HXK1: five in the case of A. thaliana and ten in rice (Granot et al., 2013). Although some of these HXK genes have clear enzymatic activity (type A and B), others apparently lack such activity (HKL) and have been suggested to have regulatory functions (Xiao et al., 2000; Rolland et al., 2006; Karve et al., 2008; Granot et al., 2013). In fact, recent work provided evidence that different HXK genes have signalling roles in different plants. For example, several HXK-type B genes from potato and rice were able to complement the Glc sensitivity of the gin2 mutant (Veramendi et al., 2002; Cho et al., 2009; Karve et al., 2010). In addition, a signalling role was observed for some HKL-type genes in A. thaliana and Physcomitrella (Thelander et al., 2005; Zhang et al., 2010; Karve et al., 2012). Whether any of the additional HXK genes (A, B, or HKL) have a role in the sugar regulation of STP1 remains for future analysis.
Other players that have been shown to participate in sugar signalling are the SnRK1 and TOR kinases (Baena-Gonzalez and Sheen, 2008; Xiong et al., 2013). SnRK1 kinase is highly conserved throughout the evolution of different organisms, including plants, and has been demonstrated to be crucial for energy homeostasis, such as carbon availability (Hardie et al., 1998; Baena-Gonzalez, 2010). Importantly for the present study, alterations in STP1 expression were reported in a microarray analysis from transiently overexpressing KIN10 protoplasts (Baena-Gonzalez et al., 2007). A. thaliana contains two SnRK1 catalytic subunits (KIN10 and KIN11) that are partially redundant (Baena-Gonzalez et al., 2007). However, it was not possible to analyse the double mutant due to its lethality; therefore, in this work, we explored the role of this kinase in the regulation of the STP1 gene using the single kin10 and kin11 mutants (Polge and Thomas, 2007) as well as transgenic lines that overexpress KIN10. KIN10 has been reported to have the most notable activity of the two catalytic subunits (Jossier et al., 2009). In this analysis, we did not observe major differences in the response of STP1 to sugars in any of the various analysed mutants and lines. Thus, although the involvement of this kinase in the regulation of STP1 cannot be completely ruled out, the only difference we observed is a slight reduction in the level of the STP1 transcript in the overexpressing KIN10 lines. Our data indicate that the participation of SnRK1, if any, in the regulation of STP1 is minor.
None of the factors analysed so far play a major role in the regulation of STP1, suggesting the participation of novel factors in the regulation of this gene. Potential additional candidates include factors whose mutants display alterations in STP1 expression. For example, in comparison to the wild-type plants, the sweetie mutant displays an upregulation of the STP1 gene (Veyres et al., 2008). SWEETIE encodes a novel protein of unknown function and is implicated in various processes, including sugar perception, senescence, ethylene biosynthesis, and abiotic stresses (Veyres et al., 2008; Büttner, 2010). Misregulation of the STP1 gene by sugars was also reported in hsr (high sugar-response) mutants. For several genes, these mutants displayed sugar hypersensitivity, and the elements that are affected in these mutants are good candidates for involvement in STP1 sugar regulation. Unfortunately, the identities of the HSR genes are still unknown (Baier et al., 2004).
In this work, we also demonstrated that sugar regulates the STP1 gene at the transcriptional level, and this regulation is similar to that of the sugar-regulated genes DIN6/ASN1 and α-Amy3, whose expression is also induced by sugar starvation and is repressed in its presence (Lam et al., 1998; Lu et al., 2002; Baena-Gonzalez et al., 2007). Similarly to the DIN6 and DIN1 genes, the regulation by Glc of STP1 is independent of the HXK1 pathway (Baena-Gonzalez et al., 2007). These similarities support a common mechanism for the regulation of these genes by sugars. In the present analysis, we were able to delimit the cis-acting elements required for the STP1 sugar repression within 309bp. Our in silico analyses showed the cis-acting elements that are common to the STP1 309bp sequence and the α-Amy3 and DIN6 promoters, including the TATCCA and the G boxes (Lu et al., 2002; Baena-Gonzalez et al., 2007). The TATCCA element (TATCCAOSAMY) was originally identified as the binding site of one MYB-type transcription factor (OsMYBS2) that is essential for the sugar regulation of the α-Amy3 gene in rice (Lu et al., 2002). Moreover, the arrangement of these elements in the STP1 promoter (in tandem and separated by 15bp) is similar to that in the α-Amy3 gene (Lu et al., 1998). Thus, this sequence is an interesting candidate for involvement in the regulation of the STP1 gene by sugars. MYB transcription factors are members of a large gene family in plants with more than a hundred members in A. thaliana (Dubos et al., 2010). Two MYB genes in A. thaliana (At5g47390 and At5g61620) are the closest to the rice MYBS2 factor based just on protein sequence identity. However, the role of this putative orthologue requires further study.
The STP14 gene does contain five TATCCA elements in the 5′ UTR that is shared with both STP1 and DIN6. However, neither the TATCCAOSAMY nor the CGACGOSAMY3 motifs are present in the upstream sequence of the STP14 gene; this gene and STP1 are among the most sugar-repressed genes of the STP family (Price et al., 2004). Thus, the contribution of any of these elements to the control by Glc of the STP genes must be determined in the future.
Finally, a low but reproducible increase in the STP1 transcript at midday was reported and linked to a circadian regulation of this gene (Harmer et al., 2000; Stadler et al., 2003). This is an interesting aspect taking into account that one of the motifs present in the region responsible for sugar regulation includes the EVENINGAT element present in genes regulated by the circadian clock (Harmer and Kay, 2000).This element was also found overrepresented in the sugar-repressed genes in a microarray data reported by Li et al. (2006). It is possible that the expression of the STP1 gene, like many other genes, might be subjected to multiple regulatory mechanisms, in addition to sugars. However, recent evidence supports the view that the levels of sugars directly influence the circadian regulation of many genes (Haydon et al., 2013), supporting possible crosstalk between these regulatory mechanisms. Although that there is still not a direct probe to show that the EVENINGAT element might be directly involved in sugar regulation, this aspect is an interesting possibility that requires further exploration in the future.
Funding
This work was supported by grants 154392 and 127546 from the Consejo Nacional de Ciencia y Tecnología (CONACyT) and IN208211-3 and IB200511 from DGAPA. DLAZ was supported by DGAPA with an undergraduate fellowship from grant IB200511.
References
- Acevedo-Hernández GJ, León P, Herrera-Estrella LR. 2005. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. The Plant Journal 43, 506–519. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. 2000. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes and Development 14, 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Baena-González E. 2010. Energy signaling in the regulation of gene expression during stress. Molecular Plant 3, 300–313. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Sheen J. 2008. Convergent energy and stress signaling. Trends in Plant Science 13, 474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier M, Hemmann G, Holman R, Corke F, Card R, Smith C, Rook F, Bevan MW. 2004. Characterization of mutants in Arabidopsis showing increased sugar-specific gene expression, growth, and developmental responses. Plant Physiology 134, 81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranowskij N, Frohberg C, Prat S, Willmitzer L. 1994. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. The EMBO Journal 13, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. 2005. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. The Plant Cell 17, 3257–3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorer KJ, Loo DD, Wright EM. 1994. Steady-state and presteady-state kinetics of the H+/hexose cotransporter (STP1) from Arabidopsis thaliana expressed in Xenopus oocytes. Journal of Biological Chemistry 269, 20417–20424. [PubMed] [Google Scholar]
- Borisjuk L, Rolletschek H, Wobus U, Weber H. 2003. Differentiation of legume cotyledons as related to metabolic gradients and assimilate transport into seeds. Journal of Experimental Botany 54, 503–512. [DOI] [PubMed] [Google Scholar]
- Büttner M. 2010. The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biology 12, Suppl 1, 35–41. [DOI] [PubMed] [Google Scholar]
- Büttner M, Sauer N. 2000. Monosaccharide transporters in plants: structure, function and physiology. Biochimica et Biophysica Acta 1465, 263–274. [DOI] [PubMed] [Google Scholar]
- Chen JG. 2008. Heterotrimeric G-protein signaling in Arabidopsis: Puzzling G-protein-coupled receptor. Plant Signaling and Behavior 3, 1042–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Jones AM. 2004. AtRGS1 function in Arabidopsis thaliana. Methods in Enzymology 389, 338–350. [DOI] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. 2003. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301, 1728–1731. [DOI] [PubMed] [Google Scholar]
- Chen PW, Chiang CM, Tseng TH, Yu SM. 2006. Interaction between rice MYBGA and the gibberellin response element controls tissue-specific sugar sensitivity of alpha-amylase genes. The Plant Cell 18, 2326–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Endo A, Zhou L, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis abscisic acid biosynthesis and glucose signaling. The Plant Cell 14, 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR. 1998. Sucrose is a signal molecule in assimilate partitioning. Proceedings of the National Academy of Sciences, USA 95, 4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Ryoo N, Hahn TR, Jeon JS. 2009. Evidence for a role of hexokinases as conserved glucose sensors in both monocot and dicot plant species. Plant Signaling and Behavior 4, 908–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Yoo SD, Sheen J. 2006. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cortès S, Gromova M, Evrard A, Roby C, Heyraud A, Rolin DB, Raymond P, Brouquisse RM. 2003. In plants, 3-o-methylglucose is phosphorylated by hexokinase but not perceived as a sugar. Plant Physiology 131, 824–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi G, Bush DR. 2001. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiology 125, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. 2010. MYB transcription factors in Arabidopsis. Trends in Plant Science 15, 573–581. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Graham IA. 2003. Trehalose metabolism: a regulatory role for trehalose-6-phosphate? Current Opinion in Plant Biology, 231–235. [DOI] [PubMed] [Google Scholar]
- Eveland AL, Jackson DP. 2012. Sugars, signalling, and plant development. Journal of Experimental Botany 63, 3367–3377. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI. 2002. ABA and sugar interactions regulating development: cross-talk or voices in the crowd? Current Opinion in Plant Biology 12, 599–609. [DOI] [PubMed] [Google Scholar]
- Gibson SI. 2005. Control of plant development and gene expression by sugar signaling. Current Opinion in Plant Biology 8, 93–102. [DOI] [PubMed] [Google Scholar]
- Granot D, David-Schwartz R, Kelly G. 2013. Hexose kinases and their role in sugar-sensing and plant development. Frontiers in Plant Science 4, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biology 8, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J, Smeekens S. 2009. Sugar perception and signaling--an update. Current Opinion in Plant Biology 12, 562–567. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annual Review of Biochemistry 67, 821–855. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. 2000. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Kay SA. 2000. Microarrays: determining the balance of cellular transcription. The Plant Cell 12, 613–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Hearn TJ, Bell LJ, Hannah MA, Webb AA. 2013. Metabolic regulation of circadian clocks. Seminars in Cell and Developmental Biology 24, 414–421. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Research 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Rahmani F, Smeekens S, Hanson J. 2009. Sucrose-mediated translational control. Annals of Botany 104, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang YS, Karrer EE, Thomas BR, Chen L, Rodriguez RL. 1998. Three cis-elements required for rice alpha-amylase Amy3D expression during sugar starvation. Plant Molecular Biology 36, 331–341. [DOI] [PubMed] [Google Scholar]
- Jang JC, Leon P, Zhou H, Sheen J. 1997. Hexokinase as a sugar sensor in higher plants. The Plant Cell 9, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang JC, Sheen J. 1994. Sugar sensing in higher plants. The Plant Cell 6, 1665–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Sheen J. 1997. Sugar sensing in higher plants. Trends in Plant Science 2, 208–214. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Hill JP, Thomas MA. 2006. The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evolutionary Biology 6, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Thomas MA. 2007. The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Molecular Biology and Evolution 24, 2412–2423. [DOI] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hardie D, Thomas M. 2009. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. The Plant Journal 59, 316–328. [DOI] [PubMed] [Google Scholar]
- Karve A, Rauh BL, Xia X, Kandasamy M, Meagher RB, Sheen J, Moore BD. 2008. Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228, 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve A, Xia X, Moore B. 2012. Arabidopsis Hexokinase-Like1 and Hexokinase1 form a critical node in mediating plant glucose and ethylene responses. Plant Physiology 158, 1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karve R, Lauria M, Virnig A, Xia X, Rauh BL, Moore B. 2010. Evolutionary lineages and functional diversification of plant hexokinases. Molecular Plant 3, 334–346. [DOI] [PubMed] [Google Scholar]
- Kavita P, Burma PK. 2008. A comparative analysis of green fluorescent protein and beta-glucuronidase protein-encoding genes as a reporter system for studying the temporal expression profiles of promoters. Journal of Biosciences 33, 337–343. [DOI] [PubMed] [Google Scholar]
- Koch KE. 1996. Carbohydrate modulated gene expression in plants. Annual Review of Plant Physiology and Plant Molecular Biology 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Boles E, Hellmann H, Barker L, Patrick JW, Frommer WB, Ward JM. 1999. The dual function of sugar carriers: transport and sugar sensing. The Plant Cell 11, 707–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Hsieh MH, Coruzzi G. 1998. Reciprocal regulation of distinct asparagine synthetase genes by light and metabolites in Arabidopsis thaliana . The Plant Journal 16, 345–353. [DOI] [PubMed] [Google Scholar]
- León P, Sheen J. 2003. Sugar and hormone connections. Trends in Plant Science 8, 110–116. [DOI] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. 2006. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Research 16, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Ho TH, Ho SL, Yu SM. 2002. Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of alpha-amylase gene expression. The Plant Cell 14, 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lim EK, Yu SM. 1998. Sugar response sequence in the promoter of a rice alpha-amylase gene serves as a transcriptional enhancer. Journal of Biological Chemistry 273, 10120–10131. [DOI] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. 2007. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. The Plant Cell 19, 2484–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN. 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Manzara T, Carrasco P, Gruissem W. 1991. Developmental and organ-specific changes in promoter DNA-protein interactions in the tomato rbcS gene family. The Plant Cell 3, 1305–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu Y-X, Hwang I, Jones T, Sheen J. 2003. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. [DOI] [PubMed] [Google Scholar]
- Morita A, Umemura T, Kuroyanagi M, Futsuhara Y, Perata P, Yamaguchi J. 1998. Functional dissection of a sugar-repressed alpha-amylase gene (RAmy1A) promoter in rice embryos. FEBS Letters 423, 81–85. [DOI] [PubMed] [Google Scholar]
- Müller R, Morant M, Jarmer H, Nilsson L, Nielsen TH. 2007. Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiology 143, 156–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. 2010. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Molecular Plant 3, 973–996. [DOI] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R, et al. 2007. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. The Plant Journal 49, 463–491. [DOI] [PubMed] [Google Scholar]
- Polge C, Thomas M. 2007. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends in Plant Science 12, 20–28. [DOI] [PubMed] [Google Scholar]
- Prestridge DS. 1991. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Computer Applications in the Biosciences 7, 203–206. [DOI] [PubMed] [Google Scholar]
- Price J, Laxmi A, St.-Martin SK, Jang JC. 2004. Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. The Plant Cell 16, 2128–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. 2006. Sugar sensing and signaling ing plants: Conserved and novel mechanisms. Annual Review of Plant Biology 57, 675–709. [DOI] [PubMed] [Google Scholar]
- Rolland F, Sheen J. 2005. Sugar sensing and signalling networks in plants. Biochemical Society Transactions 33, 269–271. [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW. 2001. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. The Plant Journal 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Sheen J. 2010. Discover and connect cellular signaling. Plant Physiology 154, 562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC. 1999. Sugars as signaling molecules. Current Opinion in Plant Biology 2, 410–418. [DOI] [PubMed] [Google Scholar]
- Sherson SM, Alford HL, Forbes SM, Wallace G, Smith SM. 2003. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. Journal of Experimental Botany 54, 525–531. [DOI] [PubMed] [Google Scholar]
- Slewinski TL. 2011. Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Molecular Plant 4, 641–662. [DOI] [PubMed] [Google Scholar]
- Smeekens S. 2000. Sugar-induced signal transduction in plants. Annual Review of Plant Physiology and Plant Molecular Biology 51, 49–81. [DOI] [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. 2010. Sugar signals and molecular networks controlling plant growth. Current Opinion in Plant Biology 13, 274–279. [DOI] [PubMed] [Google Scholar]
- Stadler R, Büttner M, Ache P, Hedrich R, Ivashikina N, Melzer M, Shearson SM, Smith SM, Sauer N. 2003. Diurnal and light-regulated expression of AtSTP1 in guard cells of Arabidopsis. Plant Physiology 133, 528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E. 2005. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiology 138, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H. 2005. Effect of the energy supply on filamentous growth and development in Physcomitrella patens. Journal of Experimental Botany 56, 653–662. [DOI] [PubMed] [Google Scholar]
- Toyofuku K, Umemura T, Yamaguchi J. 1998. Promoter elements required for sugar-repression of the RAmy3D gene for alpha-amylase in rice. FEBS Letters 428, 275–280. [DOI] [PubMed] [Google Scholar]
- Urano D, Chen JG, Botella JR, Jones AM. 2013. Heterotrimeric G protein signalling in the plant kingdom. Open Biology 3, 120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veramendi J, Fernie AR, Leisse A, Willmitzer L, Trethewey RN. 2002. Potato hexokinase 2 complements transgenic Arabidopsis plants deficient in hexokinase 1 but does not play a key role in tuber carbohydrate metabolism. Plant Molecular Biology 49, 491–501. [DOI] [PubMed] [Google Scholar]
- Veyres N, Danon A, Aono M, et al. 2008. The Arabidopsis sweetie mutant is affected in carbohydrate metabolism and defective in the control of growth, development and senescence. The Plant Journal 55, 665–686. [DOI] [PubMed] [Google Scholar]
- Villadsen D, Smith SM. 2004. Identification of more than 200 glucose-responsive Arabidopsis genes none of which responds to 3-O-methylglucose or 6-deoxyglucose. Plant Molecular Biology 55, 467–477. [DOI] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. 2003. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiology 132, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wind J, Smeekens S, Hanson J. 2010. Sucrose: metabolite and signaling molecule. Phytochemistry 71, 1610–1614. [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC. 2000. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Molecular Biology 44, 451–461. [DOI] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. 2013. Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K, Tatematsu K, Yano R, Preston J, Kitamura S, Takahashi H, McCourt P, Kamiya Y, Nambara E. 2009. CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiology 50, 330–340. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Yuan S, Xu F, Yang H, Zhang NH, Cheng J, Lin HH. 2010. The plastid hexokinase pHXK: a node of convergence for sugar and plastid signals in Arabidopsis. FEBS Letters 584, 3573–3579. [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J. 1998. Glucose and ethylene signal transduction crosstalk revaled by an Arabidopsis glucose-insensitive mutant. Proceedings of the National Academy of Sciences, USA 95, 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]