Highlight
This study demonstrates that OsIG1 plays an essential role in the regulation of empty-glume identity, floral organ number control and female gametophyte development in rice.
Key words: embryo sac, empty glume, female gametophyte, OsIG1, ovule.
Abstract
The indeterminate gametophyte1 (ig1) mutation was first characterized to modulate female gametophyte development in maize (Zea mays). However, the function of its rice orthologue, OsIG1, remains unknown. For this, we first analysed OsIG1 localization from differential tissues in rice. Real-time quantitative PCR (qRT-PCR) and histochemical staining results demonstrated that the expression signal of OsIG1 was strongly detected in young inflorescence, moderately in mature flower and weakly in leaf. Furthermore, RNA in situ hybridization analyses exhibited that OsIG1 was strongly expressed in inflorescence meristems, floral meristems, empty-glume- and floret- primordia, especially in the primordia of stamens and immature ovules, and the micropylar side of the mature ovary. In OsIG1-RNAi lines, wrinkled blade formation was accompanied by increased leaf inclination angle. Cross-section further showed that the number of bulliform cells located between the vasculatures was significantly increased, indicating that OsIG1 is involved in division and differentiation of bulliform cell and lateral growth during leaf development. OsIG1-RNAi suppression lines showed pleiotropic phenotypes, including degenerated palea, glume-like features and open hull. In addition, a single OsIG1-RNAi floret is characterized by frequently developing double ovules with abnormal embryo sac development. Additionally, down-regulation of OsIG1 differentially affected the expression of genes associated with the floral organ development including EG1, OsMADS6 and OsMADS1. Taken together, these results demonstrate that OsIG1 plays an essential role in the regulation of empty-glume identity, floral organ number control and female gametophyte development in rice.
Introduction
The LATERAL ORGAN BOUNDARIES DOMAIN/ASYMMETRIC LEAVES2-LIKE (LBD/ASL) proteins (hereafter referred to as LBD) were identified as plant-specific transcription factors harbouring a conserved LATERAL ORGAN BOUNDARIES (LOB) domain (Ori et al., 2000; Semiarti et al., 2001; Chalfun-Junior et al., 2005; Borghi et al., 2007). The LOB domain includes a zinc-finger motif responsible for DNA binding and a leucine-zipper-like motif mediating protein–protein interactions (Husbands et al., 2007). The LBD family comprises 43 members in Arabidopsis (Iwakawa et al., 2002; Shuai et al., 2002), 35 members in rice (Oryza sativa; Yang et al., 2006), and 57 in poplar (Zhu et al., 2007), respectively. Previous studies indicated that LBD proteins play a vital role in determining of leaf adaxial–abaxial polarity in Arabidopsis and maize (Lin et al., 2003; Xu et al., 2003; Evans, 2007) through repressing the expression of KNOTTED-LIKE HOMEOBOX (KNOX) genes in lateral organs (Ori et al., 2000; Semiarti et al., 2001; Chalfun-Junior et al., 2005; Borghi et al., 2007; Evans, 2007). Additionally, several LBD genes, such as RTCS in maize and Crown rootless1 in rice, were implicated in the modulation of crown and lateral root development in an auxin-dependent pathway (Inukai et al., 2005; Liu et al., 2005; Okushima et al., 2007; Lee et al., 2009).
It has also been demonstrated that LBD proteins are involved in reproductive organogenesis as well as vegetative growth (Xu et al., 2003; Evans, 2007). For instance, in maize, the ramosa2 gene containing an LOB domain was revealed to determine inflorescence branch formation (Bortiri et al., 2006). The maize indeterminate gametophyte1 (ig1) gene was found to regulate female gametophyte development (Evans, 2007). The ig1 mutant fails to restrict the proliferative phase in the embryo sac, resulting in an indeterminate number of eggs, polar nuclei and synergids (Evans, 2007). The rice LBD gene degenerated hull1 (DH1) is required for glume formation, as demonstrated by the phenotype of its T-DNA insert mutant with degenerated hull and naked pistils and stamens (Li et al., 2008). Despite substantial investigations, little is known about the links between LBD proteins and floral organ development.
In angiosperms, the ovule is located within the pistil, which consists of one or more carpels. As the female reproductive structures, ovules are eventually developed into seeds to generate new population of offspring. By far, only a few genes which regulate ovule and carpel development have been characterized in plants (Lopez-Dee et al., 1999; Pagnussat et al., 2004; Skinner et al., 2004). Most of the isolated ovule identity genes belong to MADS-box genes. In Arabidopsis thaliana, cell fate in proliferating ovule primordia is specified by the MADS box gene STK (SEEDSTICK; Pinyopich et al., 2003). In rice, a loss-of-function mutant of D-lineage MADS-box gene OsMADS13 had the ovule replaced with carpelloid structures (Dreni et al., 2007; Yamaki et al., 2011). Both the YABBY gene DROOPING LEAF (DL) and MADS-box gene OsMADS3 were shown to genetically interact with OsMADS13 in the regulation of ovule identity and floral meristem determinacy (Li et al., 2011a ). Furthermore, OsMADS6 and OsMADS58 are genetically associated with rice AGAMOUS (AG) subfamily genes to regulate the identity of stamens, carpels and ovules (Dreni et al., 2011; Li et al., 2011b ). However, LBD family proteins potentially involved in the regulation of ovule development have not been identified.
Although rice displays a high degree of conserved synteny with maize, it remains unclear whether LBD genes are also involved in mediating female gametophyte development and other unknown functions in rice. In this study, we characterized a LBD gene OsIG1 from rice (O. sativa), an orthologue of maize IG1, and uncovered its novel roles in regulating floral organ development, in addition to its conserved function in female gametophyte development, using genetic and molecular approaches. We further revealed that OsIG1 might participate in mediating floral development through regulating the expression of some developmentally associated genes including jasmonic acid-synthesizing gene EG1 (EXTRA GLUME1), AGAMOUS-LIKE6-like gene OsMADS6 and E-class gene OsMADS1. Hence, our results allowed us to infer previously undescribed regulatory roles for OsIG1 in rice.
Materials and methods
Plant materials and growth conditions
Rice transgenic lines in a Nipponbare (O. sativa ssp. japonica) background and wild-type (WT) Nipponbare were used throughout this study. The plants were grown in farm fields at Sichuan University, Chengdu, China. Primary transformants (T0) were first grown in the artificial climate incubators (Binder, Tuttlington, Germany) under standard conditions (28 °C day/20 °C night, 12h light/12h dark) and transplanted into the field 5 weeks later.
Plasmid construction and rice transformation
To generate an RNAi construct, a 485 base pair (bp) OsIG1 fragment (Os01g0889400; positions 461–810 from the ATG of the coding region plus 135bp of the 3′ untranslated region) was amplified by RT-PCR. An inverted-repeat fragment was constructed into vector pSK int and transferred into the pHB plasmid. To produce the OsIG1 promoter::GUS fusion construct, the open-reading-frame sequence of β-glucuronidase (GUS) gene was obtained from pBI121 by digesting with BamHI and SacI. Then the obtained fragment was inserted into pHB and the resulting construct was denoted GUS-pHB. About 2.1kb of the OsIG1 gene fragment upstream of the translational start codon was isolated by PCR from Nipponbare genomic DNA (primers are listed in Supplementary Table 1). Products were digested with EcoRI and BamHI and ligated into EcoRI-/BamHI-digested GUS-pHB to generate plasmid pOsIG1-GUS. The constructs were used to transform Nipponbare callus using engineered Agrobacterium tumefaciens (strain EHA101) as described previously (Hiei et al., 1994).
Tissue preparation and microscopic analyses
Fresh tissues from WT and RNAi plants were analysed with a stereoscopic Leica M420 microscope (Leica Microsystems, Bannockburn, IL, USA) equipped with Leica DFC280 digital camera system. For histological analysis, samples were fixed in FAA solution (3.7% formaldehyde/50% ethanol/5% glacial acetic acid), dehydrated with a series of ethanol solutions, embedded in paraffin and cut into sections 8 µm thick. The sections were stained with ferrovanadium hematoxylin. To evaluate the viability of OsIG1-RNAi pollen grains, anthers from transgenic mature flowers were dissected in a drop of 1% I2/KI solution. The sections and pollen grains stained were photographed using a Leica DFC280 camera. For scanning electron microscopy, samples were fixed in 2.5% glutaraldehyde solution. Fixed samples were dehydrated with graded ethanol series, dried by critical-point drying, sputter-coated with platinum and observed using a scanning electron microscope [JSM-5900LV; SEMTech (STS), North Billerica, MA, USA].
RT-PCR and real-time quantitative PCR analysis
Total RNAs were isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) followed by treatment with DNase I to digest genomic DNA (TaKaRa, Dalian, China). About 1μg of total RNA from each sample was used for first-strand cDNA synthesis (Toyobo, Tokyo, Japan). Triplicate quantitative assays were performed using the SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) with an ABI PRISM Step One Plus real-time PCR system (Applied Biosystems) according to the manufacturer’s instructions. The relative expression level was determined based on the 2−∆∆CT method (Livak and Schmittgen, 2001) by using rice actin and ubiquitin as internal reference genes. Primers used for real-time quantitative PCR (qRT-PCR) analysis are listed in Supplementary Table 1.
In situ hybridization and histochemical staining
The hybridization and immunological detection were performed as described by Dai et al (2007). The gene-specific regions of OsIG1 were amplified and cloned into pGEM-T Easy (Promega, Madison, WI, USA) for synthesis of an RNA probe; the primers used are shown in Supplementary Table 1. Digoxigenin-labelled sense and antisense probes were synthesized with T7 or SP6 RNA polymerase (Roche, Indianapolis, IN, USA), respectively. GUS staining was performed according to the method described previously (Sieburth and Meyerowitz, 1997).
Subcellular localization analysis
The coding sequence of the OsIG1 gene was directionally cloned into the expression vector pART27-mcs:GFP (kindly provided by Professor Shuqing Cao, Hefei University of Technology, Hefei, China), containing the green fluorescent protein (GFP) coding sequence under the control of a 35 S promoter. This generated the pART27-OsIG1-GFP construct. Plasmid vectors pART27-OsIG1-GFP and pART27-GFP were introduced into protoplasts of Arabidopsis according to the methods described previously (Yoo et al., 2007). After incubation for 16–20h at 23 °C, protoplasts were observed under a Leica TCS SPII confocal microscope using 488 and 633nm excitation wavelengths and three-channel measurement of emission: 435nm (blue/DAPI), 522nm (green/GFP) and 680nm (red/chlorophyll).
Results
The expression pattern of OsIG1
According to the previous information (Evans, 2007) and database (http://rapdb.dna.affrc.go.jp/), the genomic sequence of OsIG1 (Os01g0889400) spans 3399bp, and contains three introns and four exons (Supplementary Fig. 1A). It has a deduced coding protein of 270 amino acids with a conserved LOB domain containing C block, GAS block and the predicted coiled-coil structure (Supplementary Fig. 1B; Evans, 2007). Phylogenetic analysis showed that OsIG1 is closely related to maize IG1, and together they are clustered in the same clade with maize IAL1, rice IAL1, and Arabidopsis AS2 (Supplementary Fig. 1C; Evans, 2007).
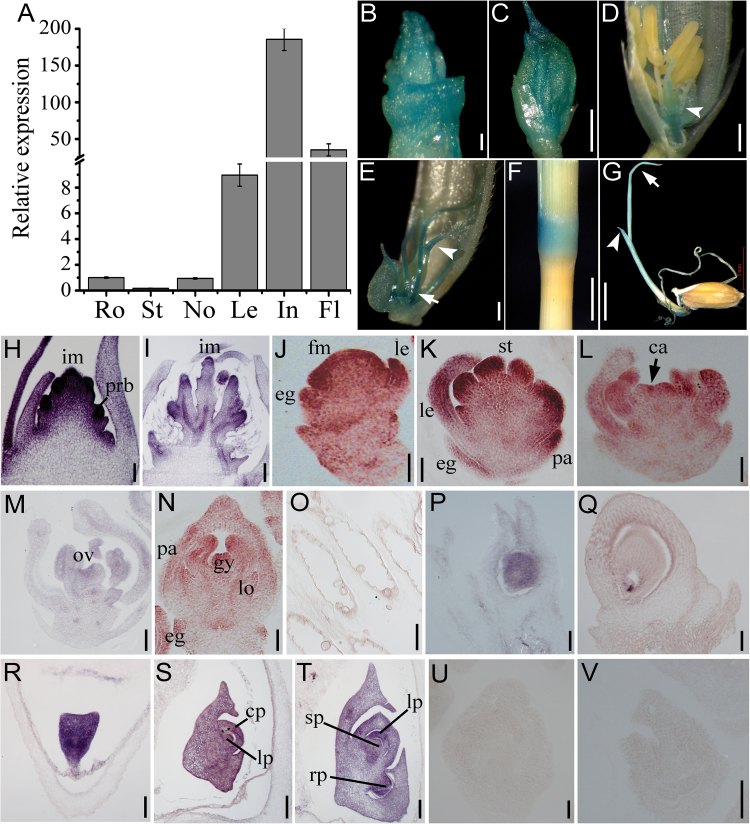
To investigate the function of OsIG1, we analysed the OsIG1 expression pattern using qRT-PCR, promoter-GUS fusions and in situ hybridization. First, qRT-PCR analysis demonstrated that OsIG1 was strongly expressed in the young inflorescence, moderately in mature flower and weakly in leaf (Fig. 1A). OsIG1 expression was lowest in the root and stem relative to other tissues examined (Fig. 1A). To further confirm this result, a 2.1 kbp promoter region of OsIGI was used to direct GUS expression in transgenic rice. As a result, GUS activity was mainly detected in young inflorescence (Fig. 1B), empty glumes and floret organs (Fig. 1C–E), such as in palea, lemma (Fig. 1C) and pistil (Fig. 1D) during early spikelet development stages. Additionally, GUS activity in the lodicules (Fig. 1E), filaments (Fig. 1E) and leaf (Fig. 1G) was observed as well. Furthermore, the elongating internode also showed a strong GUS signal in the transgenic plant driven by the OsIG1 promoter (Fig. 1F), whereas very weak signal was detected in root (Fig. 1G). Overall, the expression patterns of OsIG1 detected by qRT-PCR were consistent with the GUS signal shown in the transgenic lines.
Fig. 1.
Expression patterns of OsIG1 in WT of rice. (A) Real-time quantitative RT-PCR analyses of OsIG1 expressions in various organs of WT rice. RNAs were collected from 3-week-old seedlings: root (Ro), shoot (St), leaf (Le) and inflorescence (In) at stage In5, with the node (No) and flower (Fl) at the mature embryo sac stage. Actin was used as a control. The expression of IG1 in root was set as 1. Error bars represent SD from three independent biological replicates. (B–G) Localization of GUS activity under the control of the OsIG1 promoter. (B) A 5 mm-long inflorescence; scale bar, 1mm. (C) A 3 mm-long spikelet; bar, 1mm. (D) Postmeiosis spikelet: arrowhead indicates ovary; bar, 200 µm. (E) The mature embryo sac stage: arrow and arrowhead indicate filament and style, respectively; bar, 200 µm. (F) Node; bar, 5mm. (G) Young seedling: arrow and arrowhead indicate primary leaf and second leaf, respectively; bar, 5mm. (H–V) In situ hybridization of cross-sections with an OsIG1-specifc antisense probe. (H, I) Longitudinal section of a young inflorescence. (J–N) Longitudinal sections of a developing WT spikelet. (O) Anther in nearly mature flowers. (P) Immature ovary. (Q) Mature ovary. (R–T) Sections of WT embryos at 2 days after pollination (DAP) (R), 5 DAP (S) and 7 DAP (T). (U, V) In situ analysis using a sense probe of OsIG1 in the early spikelet as negative controls. ca, carpel; cp, coleoptilar; eg, empty glume; fm, floral meristem; gy, gynoecium; im, inflorescence meristem; le, lemma; lo, lodicule; lp, leaf primordia; ov, ovule; pa, palea; prb, primary rachis branches; rp, root primordium; sp, shoot primordium; st, stamen. Scale bars, 50 μm in (H, J–O, R), 400 μm (I) and 100 μm in others.
To more precisely determine the spatial and temporal patterns of OsIG1 expression in floral organs, we performed RNA in situ hybridization analysis. Strong signals of OsIG1 were detected in the inflorescence meristems (Fig. 1H, I), floret primordia (Fig. 1J–L) and the empty glume (Fig. 1J–N). During organogenesis of florets, OsIG1 transcripts were also detected in the primordia of stamens and ovules (Fig. 1L, M). Notably, OsIG1 showed constitutive expression in nucellus (Fig. 1P) and later the detected signal was restricted to the micropylar end of the mature ovaries (Fig. 1Q). During embryogenesis, the OsIG1 transcripts were detected throughout various stages of embryo development (Fig. 1R–T). As controls, no signals were detected in floral meristems and floral organ primordia with the sense probe (Fig. 1U, V).
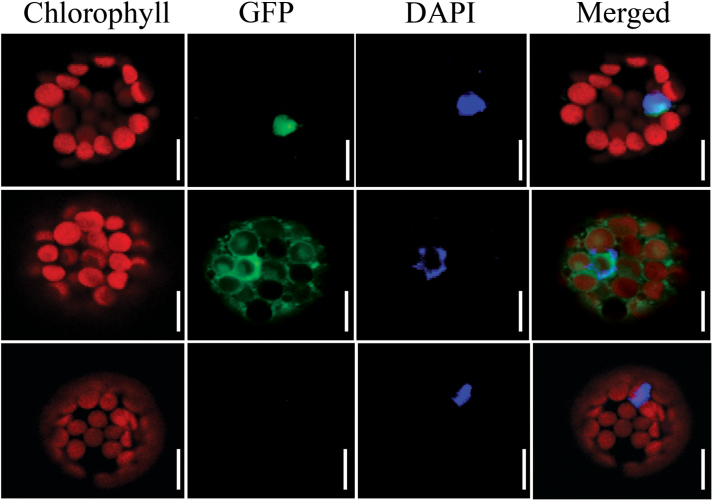
To determine experimentally subcellular localization, OsIG1 was fused in frame with GFP and transiently expressed in Arabidopsis mesophyll protoplasts. As expected, we found that the OsIG1-GFP fusion protein was unambiguously localized in the nucleus (Fig. 2), confirming that OsIGI is a nuclear protein.
Fig. 2.
Localization of OsIG1-GFP fusion protein transiently expressed in Arabidopsis protoplasts. Upper panels, OsIG1-GFP; middle panels, 35S-GFP; bottom panels, an untransformed plant as a negative control. Left to right: red, chlorophyll autofluorescence; green, GFP fluorescence; blue, nucleus stained with DAPI; merged, combined fluorescence from GFP, chlorophyll and DAPI. Scale bars, 20 µm.
Alteration of floral development in OsIG1-RNAi repression plants
Because of the obvious expression of OsIG1 in the floral tissues, we investigated the potential effects of OsIG1 on rice floral development by constructing an inversely repeated RNAi vector (Supplementary Fig. 2A). Over 20 independent transgenic lines were obtained. Three representative transformants with 60–68% reduction of endogenous OsIG1 expression level were selected for detailed phenotypic analysis (Supplementary Fig. 2B). To determine the specificity of OsIG1 silencing, we searched the rice genomic sequence database Rice BLAST (http://RiceBLAST.dna.affrc.go.jp/) using the 485bp RNAi fragment, and found that four genomic loci (three loci located on Os02g0318851 and one locus located on Os05g34450) with continuous 21–24bp short sequence identities to the RNAi fragment. The RNAi fragment showed 27% identity to OsIAL1 (Os05g0417000) and 34% to Os02g0318851. Using qRT-PCR analysis we found that the expression pattern of the OsIAL1 gene, with highest abundance in young inflorescence (Supplementary Fig. 3A), is similar to that of OsIG1 (Fig. 1A). Subsequent measurement of expression levels for Os05g0417000 and Os02g0318851 revealed that their expression was nearly unaffected in the down-regulated OsIG1 lines (Supplementary Fig. 3B, C). These results suggest there was only a low probability of incorrect targeting caused by OsIG1-RNAi and that the observed phenotypic alterations most likely resulted from down-regulation of OsIG1 alone.
Next, we examined the effects of OsIG1 deficiency on plant floral development in detail. Generally, a typical WT rice floret normally consists of a pistil in a central whorl (whorl 4), six stamens in whorl 3, two lodicules adjacent to the lemma in whorl 2 and two interlocking organs, palea and lemma, surrounding the inner floral organs in whorl 1. A floret with two pairs of sterile glumes (rudimentary glumes and empty glumes), which subtend at its base, constitutes a spikelet (Fig. 3A, B). Compared with WT, OsIG1-RNAi plants exhibited a wide variation in the number and position of floral organs as well as glumes (Fig. 3C–K). In the RNAi spikelets, empty glumes (also called sterile lemmas) were elongated and widened, resembling lemma/palea in shape and size (Fig. 3C). In addition to increased numbers of empty glumes (Fig. 3D) and extra glume-like organs (Fig. 3E), it was found that the formation of velum-like organs occurred in RNAi lines (Fig. 3F). In whorl 1, the paleae of the RNAi plants exhibited severe defects, including some smaller paleae (Fig. 3G) and others underdeveloped or distorted (Fig. 3H). In whorl 2, some lodicules were often fused at the base and transformed into carpel-like organs (Fig. 3I), suggesting that OsIG1 may be required for lodicule specification in rice. In whorl 3, the filament-plumed stigma chimeras coupled with reduced stamen numbers were also visualized in approximately 5% spikelets of the RNAi lines (Fig. 3J). The OsIG1-RNAi ovaries (Fig. 3K) were clearly swollen compared to WT (Fig. 3B). Compared with WT, besides the abnormal morphology of floral organs, OsIG1-RNAi plants displayed altered inflorescence shape with elongated and curved peduncles (Fig. 3G, L).
Fig. 3.
Phenotypes of WT and OsIG1-RNAi inflorescences and spiklets. (A, B) WT spiklet (A) and (B) with two lodicules, six stamens and one carpel. The two empty glumes, lemma and palea were removed for clarity in (B). (C–K) OsIG1-RNAi spiklet. The lemma and palea were removed in (I) and (J) for clarity. (C) A RNAi spikelet with elongated empty glume (arrow). (D) The number of empty glumes increased in the RNAi spiklet. (E) An extra glume-like organ formed in the RNAi spiklet. (F) A spikelet with velum-like organs. (G) A spikelet with degenerated or malformed palea in inflorescences at stage In9; arrowhead indicates peduncle. (H) The florets with severe phenotypes showing no inner floral organs. (I) Lodicules were fused at the base and transformed into carpel-like organs. (J) One stamen is transformed into a stigma-like organ in an OsIG1-RNAi spiklet. (K) The OsIG1-RNAi ovaries were clearly swollen compared with WT (B). (L) Inflorescences at stage In9 of WT; arrowhead indicates peduncle. ca, carpel; dpa, degenerated palea; eg, empty glume; est, ectopic stigma; fi, filament; gl, glume-like organ; le, lemma; lo, lodicule; pa, palea; st, stamen; sti, stigma; vl, velum-like organ. Scale bars, 1mm.
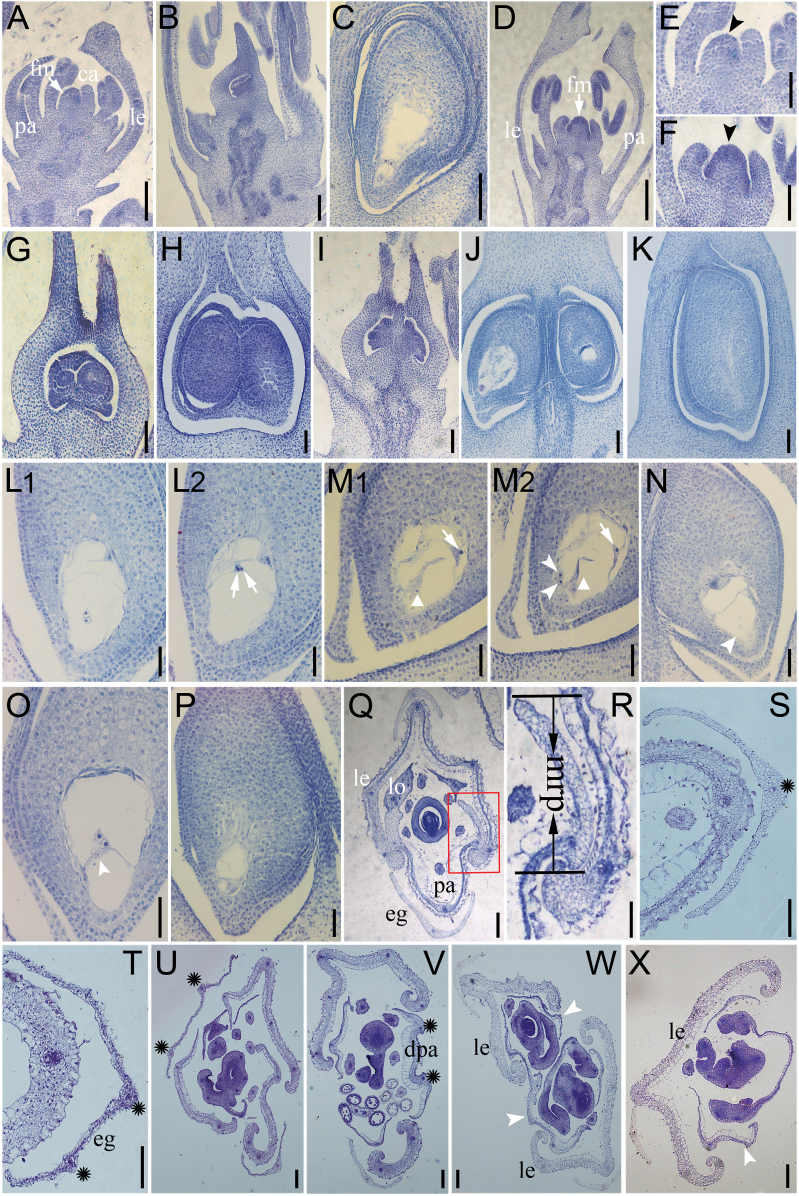
To characterize the observed morphological defect in more details, histological analyses were performed. In WT plants after the carpel primordium arises at the floral meristem near the lemma side, a lateral region of the floral meristem adjacent to the carpel initiation transformed into an ovule primordium (Fig. 4A) in which megaspore mother cell (Fig. 4B) occurs to form an embryo sac, the female gametophyte (Fig. 4C). By contrast, the size of the OsIG1-RNAi floral meristem at the early development stage of the pistil was larger than that of the WT (Fig. 4D–F). Also, the ovule primordia initiated on both the palea and the lemma side of floral meristem (Fig. 4D, G). Surprisingly, unusual double ovules, fused (Fig. 4G, H) or not fused (Fig. 4I, J), occurred in approximately 40% of the transgenic ovaries (Table 1). In addition, OsIG1-RNAi repression showed pleiotropic effects on female gametophyte differentiation, resulting in a variety of abnormally developed embryo sacs including embryo sac degeneration (Fig. 4K), an embryo sac with abnormal polar nuclei (Fig. 4L1, L2) and an embryo sac with an extra egg cell (Fig. 4M1, M2), as well as other abnormal embryo sacs, such as an embryo sac without a female germ unit (Fig. 4N), an embryo sac with abnormal nuclear migration (Fig. 4O) and an abnormally small embryo sac (Fig. 4P). The frequencies of various types of abnormalities are summarized in Table 1, with the RNAi lines showing 7–11% embryo sacs with abnormal polar nuclei, 27–29% embryo sac degeneration, 6–9% embryo sacs with an extra egg cell and 20–26% other abnormal embryo sacs.
Fig. 4.
Histological analyses of WT and OsIG1-RNAi flowers. (A–C) Pistil development in the WT. A carpel protruded from the lemma side of the floral meristem (A). When the carpel completely enclosed the ovule primordium, a megaspore mother cell became visible and an integument primordium was differentiated in the ovule primordium (B). In a mature ovule, a vacuolated embryo sac was visible (C). (D) When a carpel protruded, the floral meristem in OsIG1-RNAi (D) was larger than that of the WT (A). (E, F) Close-up of floral meristem shown in the WT (A) and OsIG1-RNAi (D), respectively. (G–J) Unusual double ovules fused (G, H) or not fused (I, J) occurred in the transgenic ovaries. (K) Embryo sac degeneration in OsIG1-RNAi ovule. (L1, L2) Serial section showing OsIG1-RNAi embryo sac with extra polar nuclei (arrows). (M1, M2) Serial section showing OsIG1-RNAi embryo sac with possible multiple egg cells (arrowheads), extra synergids (triangular arrows) and polar nuclei located in an abnormal position close to the chalazal end (arrows). (N) Embryo sac without female germ unit (arrowhead). (O) Embryo sac with abnormal nuclear migration (or abnormal position of egg cell and synergid cells; arrowhead). (P) Abnormally small embryo sac. (Q–S) Cross-sections of a WT spikelet. (R) Magnification of the area within the box on Q showing interlocking of lemma and palea. (S) The WT empty glume with one vascular bundles (asterisk). (T-X) Cross-sections of the OsIG1-RNAi spikelets. (T, U) Two vascular bundles in the OsIG1-RNAi empty glumes (asterisks in T), as opposed to one in WT (asterisks in S). Edges of lemma and palea are hooked in the WT (Q), but there is no interlock with the lemma in the OsIG1-RNAi spikelet (U-X). (U) An abnormal flower with hull-like organs and two vascular bundles of empty glume (asterisks) in an OsIG1-RNAi flower. (V) The OsIG1-RNAi spikelet has only two full-developed vascular bundles (asterisks) in degenerated palea. (W) Transverse section of twin-flower spikelet (one spikelet containing two separate carpels) of OsIG1-RNAi; arrowheads indicate the degenerated palea. (X) The degenerated palea (arrowhead) lacking a bop and retaining mrp-like structures in an OsIG1-RNAi spikelet. ca, primary carpel; dpa, degenerated palea; eg, empty glume; fm, floral meristem; le, lemma; lo, lodicule; mrp, marginal region of palea; pa, palea. Scale bars, 100 μm (A–D, Q–X), 50 μm (E–P).
Table 1.
Frequencies of various types of abnormal ovules and embryo sacs in the OsIG1-RNAi repression lines (60 spikelets examined for each line were collected at the mature stage)
| Genotype | One ovule | Double ovules | Total ovules examined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ESD | EESWA | OAT | ESE | NES | ESD | ESWA | OAT | ESE | NES | ||
| WT | 0 | 0 | 1 | 0 | 59 | 0 | 0 | 0 | 0 | 0 | 60 |
| RNAi-1 | 2 | 4 | 2 | 2 | 26 | 21 | 5 | 17 | 3 | 2 | 84 |
| RNAi-2 | 3 | 3 | 2 | 3 | 28 | 19 | 3 | 14 | 4 | 2 | 81 |
| RNAi-3 | 2 | 3 | 3 | 2 | 23 | 24 | 4 | 20 | 3 | 3 | 87 |
ESD, embryo sac degeneration; ESE, embryo sac with an extra egg cell; ESWA, embryo sac with abnormal polar nuclei; NES, normal embryo sac; OAT, other abnormal types.
Although the palea is similar to the lemma in its outer morphology in rice, a few critical differences exist: the lemma has five vascular bundles, whereas the palea has three vascular bundles and consists of two parts [the body of the palea (bop) and two marginal regions of the palea (mrps)]. The bop shares similar cellular morphology with lemma (Prasad et al., 2005), but the mrp has a distinctive smooth epidermis specific to palea (Fig. 4Q, R; Chen et al., 2006). In approximately 12% of the RNAi spikelets the elongated empty glumes had two vascular bundles as opposed to one in the WT (Fig. 4S–U). Interestingly, most of the degenerated palea (80%) exhibited reduced bop in size and nearly normal mrp (Fig. 4V). And about 6% degenerated palea lacked bop and retained only mrp-like structures with nonsilicified upper epidermis and without trichomes and protrusions (Fig. 4W, X), suggesting that the development of the bop of palea was severely affected in the RNAi lines. Additionally, all of RNAi lines displayed semi-sterile phenotype (the seed setting rate was approximately 38.6%).
To determine the nature of the reproductive defect found in the RNAi lines, we conducted reciprocal crosses and I2/KI staining assays for pollen from both WT and the RNAi lines. The results showed that offspring from the control cross (WT♀×WT♂) exhibited 86–91% seed-set fertility, while the pollination of WT pistils with pollen from RNAi plants produced seed-set rates ranging from 78 to 83% fertility. However, when using RNAi plants as the maternal recipient pollinated with WT pollen (RNAi♀×WT♂), the seed setting rates were scored as 46, 51 and 53% in OsIG1-RNAi repression panicles. Also, I2/KI assay showed that pollen from both WT (Supplementary Fig. 4A) and RNAi plants (Supplementary Fig. 4B–D) exhibited similar morphology and viability when fertilizing the emasculated rice spikelets. Taken together, these results suggest that the pollen from the RNAi plants was functional and that the sterility of the RNAi plants might be mainly due to defects in the female reproductive organs.
RNAi plants exhibited defects in meristem maintenance as well as floral organ development
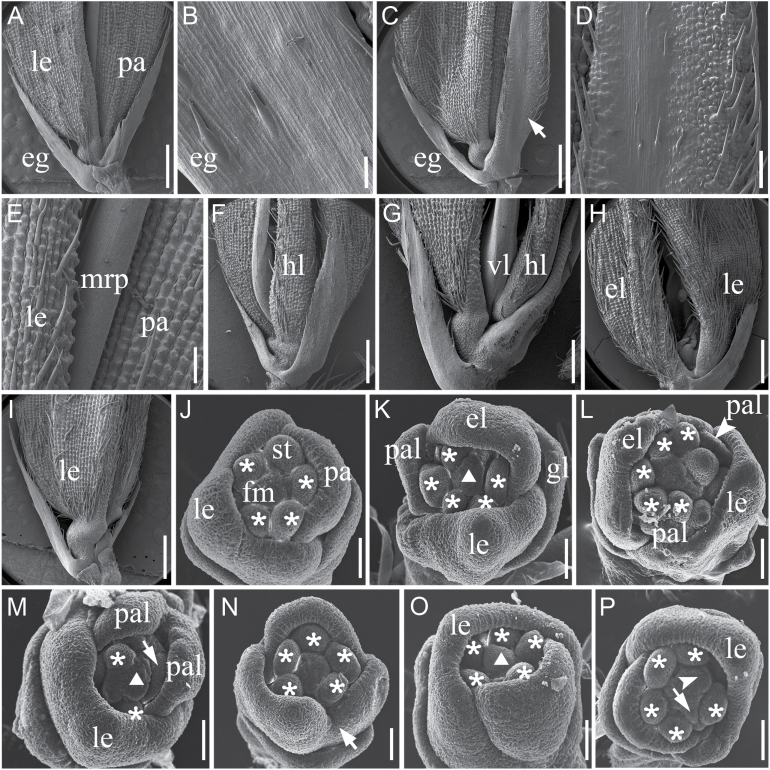
To study the identity of the abnormal organs observed in the RNAi lines, we examined spikelets at the heading and flowering stage by scanning electron microscopy. Compared with the smooth outer epidermal surface of WT empty glumes (Fig. 5A, B), epidermal cells of the elongated empty glumes in the OsIG1-RNAi lines displayed bulges and bristles that are similar to phenotype of eg1 mutation and ectopic expression of OsMADS1 in rice (Fig. 5C, D; Jeon et al., 2000; Prasad et al., 2001; Li et al., 2009). In OsIG1-RNAi lines, the extra hull-like organs showed a similar upper epidermis, resembling the WT lemma (Fig. 5E–G). In some cases, the palea was degenerated to form mrp-like structures (Fig. 5G). In other cases, an additional lemma-like organ occurred in place of OsIG1-RNAi palea (Fig. 5H), or OsIG1-RNAi spikelets lacked palea (Fig. 5I), suggesting that the development of palea was severely affected.
Fig. 5.
Scanning electron micrographs of WT and OsIG1-RNAi spikelets. (A) A WT spikelet. (B) Epidermal surface of an empty glume in the WT. (C) An OsIG1-RNAi spikelet with elongated empty glume (arrow). (D) Close-up view of the epidermal surface of the elongated empty glume in (C) showing a empty-glume-like surface in the central region and a lemma-/palea-like surface in the lateral region. (E) Epidermal surface of the lemma, palea and mrp in the WT. (F) An OsIG1-RNAi spikelet with an extra hull-like organ between the lemma and palea. (G) An OsIG1-RNAi spikelet with hull- or velum-like organs. (H) An OsIG1-RNAi spikelet with an extra lemma instead of the palea. (I) An OsIG1-RNAi spikelet with lacked palea. (J) WT flowers with six stamen primordia and a flat floral meristem. (K–P) OsIG1-RNAi flowers. (K) Showing one palea-like and one extra lemma primordium. (L) One abnormal flower with ectopic palea-like and extra lemma-like structures. (M) OsIG1-RNAi flower producing a stamen primordium ectopically in the centre of the floral meristem (triangular arrow), ectopic lodicule (arrow) and palea-like primordium. (N) One abnormal lemma shape. (O) At the late stage of stamen initiation, the floral meristem in OsIG1-RNAi lines remained bulged instead of being flat. (P) The floral meristem is uneven and bifurcated (arrow and arrowhead). eg, empty glume; el, extra lemma-like structure; fm, floral meristem; gl, glume-like organ; hl, hull-like organ; le, lemma; mrp, marginal regions of the palea; pa, palea; pal, palea-like organ; st, stamen; vl, velum-like organ. Asterisks indicate stamen primordia. Scale bars, 1mm (A, C, F–I), 100 µm (B), 200 µm (E, D), 50 µm (J–P).
To further characterize the developmental defects of the OsIG1-RNAi spikelet, we investigated the flower morphology of down-regulated OsIG1 transgenic lines at early stage by scanning electron microscopy. During the spikelet 6 stage (Sp6), the WT flower formed six stamen primordia, and it was apparent that the lemma with bumps at the top was larger than the palea (Fig. 5J). However, some OsIG1-RNAi spikelets developed extra lemma- or palea-like or glume-like structures (Fig. 5K–M). At the stage of stamen initiation, the floral meristem in WT flowers tended to be flat (Fig. 5J; Itoh et al., 2005). By contrast, the floral meristem in RNAi lines still bulged (Fig. 5N, O), suggesting that floral meristem determinacy was severely affected. Occasionally, floral meristem was doubled in one spikelet (Fig. 5P), suggesting defects in floral meristem determinacy. Together these observations indicated that down-regulated OsIG1 triggered abnormal spikelet development at an early stage, including the formation of ectopic floral organs, changes in organ number and alteration of floral meristem determinacy.
Alteration of vegetative development in OsIG1-RNAi plants
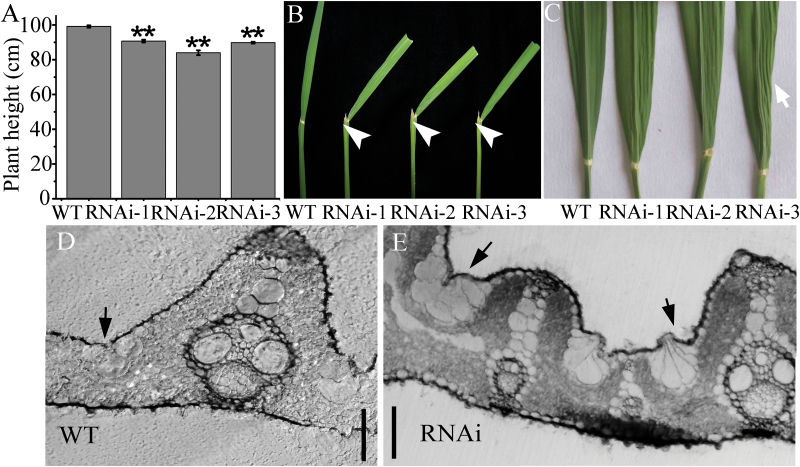
In addition to the altered morphology of reproductive organs, we also observed abnormal vegetative development by OsIG1-RNAi repression with reduced plant height and increased leaf inclination angle. Plant height for 20 field-grown individuals from each genotype was determined at the mature stage, and the average plant heights of the WT control, RNAi-1, RNAi-2 and RNAi-3 lines were 99.1, 90.7, 84.0 and 89.8cm, respectively (Fig. 6A), indicating a slightly reduced internodal elongation conferred by down-regulated OsIG1 expression. Distinctly, an increased flag leaf inclination angle resulting from OsIG1-RNAi repression was visualized at the heading stage. Leaf angle is the angle of inclination between the leaf blade and the vertical culm (Zhao et al., 2010). The lamina joint is a whitish region at the base of the blade, which joins the rice leaf blade and sheath and functions in bending the leaf blade toward the abaxial side. In the RNAi lines, the bending of the leaf blade occurred at the lamina joint. As shown in Fig. 6B, the leaf angle of the RNAi lines was much greater than that of the WT and the angle of leaves was increased and reached a maximum of approximately 44 °, whereas the leaf angle of WT plants appeared to plateau at approximately 16 °.
Fig. 6.
Vegetative phenotypic analyses of OsIG1-RNAi transgenic lines. (A) Plant height (cm). Student’s t test was used to analyse significant differences between WT and OsIG1-RNAi transgenic lines. Values are means ± SE from 20 plants. **P < 0.01 (t test). (B) Showing the increased leaf angle in OsIG1-RNAi transgenic lines. Arrows indicate the leaf lamina joint. (C) Some leaves of OsIG1-RNAi lines showing wrinkled blades, which was partially similar to that of plants overexpressing the class 1 KNOX gene OSH1. (D, E) Cross-sections of a WT (D) and OsIG1-RNAi (E) leaf blade. Arrows indicate bulliform cells. Scale bars, 50 µm. A colour version of this figure is available at JXB online.
It has been reported that maize ig1 mutant leaves have disrupted adaxial–abaxial polarity and fail to repress the expression of meristem-specific KNOX genes in leaf primordia, causing a proliferative, stem cell identity to persist in these cells (Evans, 2007). Occasionally, as shown in Fig. 6C, some leaves of OsIG1-RNAi lines exhibited wrinkled blades, resembling the phenotype observed in plants overexpressing the class 1 KNOX gene OSH1 (Sato et al., 1996), but no alteration of OSH1 expression was detected in the leaves with the mutant phenotype in the OsIG1-RNAi lines (data not shown). Compared with the WT (Fig. 6D), the number of bulliform cells located between the vasculatures was significantly increased in the RNAi lines (Fig. 6E). In addition, these cells did not show a balloon-like form as occurring in the WT (Fig. 6E). No obvious difference in other leaf cell types or alteration in adaxial–abaxial polarity was observed between the RNAi lines and WT (Fig. 6D, E), indicating that the wrinkled blade phenotype may be caused by the increase in bulliform cell number. Together these results suggested that the OsIG1 gene is involved in division and differentiation of bulliform cell and lateral growth during leaf development.
Down-regulation of the OsIG1 gene affects the expression of the EG1, OsMADS6 and OsMADS1 genes
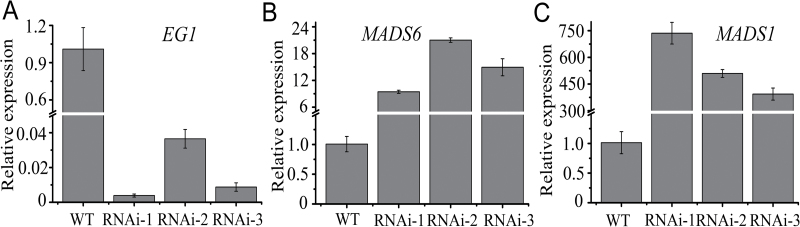
Recently, a number of plant genes involved in floral organ development have been identified. We asked whether the OsIG1 gene mediates floral development by regulating well-characterized floral-organ-associated genes. To this end, we investigated the expression of several genes which are profoundly involved in this process, including putative B function genes (OsMADS2 and OsMADS16), C function genes (OsMADS3, OsMADS58), a YABBY gene DROOPING LEAF (DL), a D function gene (OsMADS13), an E function gene (OsMADS1), an AGAMOUS-LIKE6 (AGL6)-like gene (OsMADS6) and KNOX genes (OSH1 and OSH6), as well as CLV1-homologue FON1 and jasmonic acid-synthesizing gene EG1 in the inflorescences of WT and the OsIG1-RNAi lines (less than 1, 5, 10, 15 and 20cm in length) using qRT-PCR analyses (Agrawal et al., 2005; Cai et al., 2014; Dreni et al., 2007; Jeon et al., 2000; Kater et al., 2006; Kyozuka et al., 2000; Li et al., 2009, 2010; Matsuoka et al., 1993; Moon et al., 1999; Nagasawa et al., 2003; Ohmori et al., 2009; Suzaki et al., 2004; Yamaguchi et al., 2004, 2006; Zhang et al., 2013). The results showed that in OsIG1-RNAi lines EG1 expression was dramatically reduced by 62-fold (Fig. 7A), while expression of OsMADS6 and OsMADS1 was up-regulated by 15- and 545-fold compared with the WT (Fig. 7B, C), respectively. Other than this, we did not observe consistent changes in the expression of other genes tested among the transgenic and WT plants (Supplementary Fig. 5). These results suggested that OsIG1 regulates the morphogenesis and development of floral organs, probably by regulating the expression of EG1 and the floral organ identity genes OsMADS1 and OsMADS6 as well as by unidentified pathways.
Fig. 7.
Comparison of gene expression levels of EG1, OsMADS1 and OsMADS6 in the inflorescences of OsIG1-RNAi transgenic lines and WT. qRT-PCR analysis of EG1, OsMADS1 and OsMADS6 transcripts in 5cm inflorescences in the WT and OsIG1-RNAi lines. Actin expression in each sample was used as a control. The expression level in the WT was set as 1. Error bars represent SD from three independent biological replicates.
Discussion
OsIG1 may function to maintain empty-glume identity
Regarding the origin and evolution of the sterile lemma, there are two controversial hypotheses. One hypothesis considers that an ancestral rice spikelet has three florets, including one fertile floret and the other two degenerated sterile florets with only the lemmas remaining (Kellogg, 2009). The empty glumes thus would seem to be just the remnants of the two lower reduced florets (Yoshida et al., 2009; Kobayashi et al., 2010). Another hypothesis states that the rice spikelet has only one floret, and the sterile lemma and rudimentary glume are generally regarded as severely metamorphic bract structures (Schmidt and Ambrose, 1998; Terrell et al., 2001; Hong et al., 2010). Recently, several mutants and their underlying genes related to the development of glume-like organs have been identified in rice, including eg1 (Li et al., 2009), long sterile lemma (g1; Yoshida et al., 2009)/elongated empty glume1 (ele1; Hong et al., 2010) and osmads34 (Gao et al., 2010). Among these, g1/ele1 and osmads34 mutants displayed the transformation of sterile lemmas into lemma-like structures (Yoshida et al., 2009; Hong et al., 2010; Gao et al., 2010), suggesting that G1/ELE1 and OsMADS34 repress lemma identity at the sterile lemma positions during rice spikelet development (Yoshida et al., 2009; Hong et al., 2010). Unlike g1/ele1 and osmads34 mutants, eg1 mutation severely affects its number as well as sterile lemmas size (Cai et al., 2014). These observations suggest that different genes are probably involved in the pathway of empty-glume development. In our OsIG1-RNAi lines, we observed an increased number of empty glumes, and in some cases the empty glumes were converted into glume-like organs and the increased number of empty glumes occurred (Fig. 3D). Similar to the studies on EG1, OSMADS34 and G1/ELE1, our data for OsIG1 seems supporting the first hypothesis.
Additionally, nuclear localization of IG1 and its expression at the early developmental stage of empty glumes suggests that IG1 may achieve this function by targeting downstream genes needed for empty-glume growth. As in the loss-of-function eg1 mutation, the spikelets of OsIG1-RNAi lines developed extra glume-like structures (Fig. 3E; Li et al., 2009). In addition, in OsIG1-RNAi lines empty glumes were elongated and widened, resembling the lemma/palea in shape and size, a phenotype similar to that reported by Prasad et al. (2001, 2005), where OsMADS1 over-expression created a lemma-like epidermis in the transformed glumes. Intriguingly, these phenotypic alterations appear to be consistent with EG1 down-regulation and OsMADS1 up-regulation observed in OsIG1-RNAi repression lines (Fig. 7A, C). Our data showed that OsIG1-RNAi repression (by 35 S promoter) could greatly enhance expression of OsMADS6 and OsMADS1 (Fig. 7B, C), and we believe that the abnormal organ development occurring in OsIG1-RNAi lines could be ascribed to, at least in part, the constitutive/ectopic expression of these MADS genes (Fig. 3C, I, J). Also, our qRT-PCR analysis showed that the expression level of EG1 was remarkably decreased in all of three OsIG1-RNAi repression lines relative to WT (Fig. 7A), indicating that EG1 might function downstream of OsIG1. Nevertheless, Li et al. (2009) and Cai et al. (2014) reported that EG1 positively regulates expression of the floral meristem and organ identity gene OsMADS1, seemingly contradictory to our expression data for EG1 and OsMADS1 in the OsIG1-RNAi lines. However, this discrepancy implicates the complexity of floral organ gene expression and regulation under various concerts of background. Actually, Cai et al. (2014) observed a significant reduction of OsMADS1 expression in 2mm inflorescence of EG1-defective lines, but a large portion of this reduction disappeared in the 3–4mm inflorescence, suggesting their interaction is spatially and temporarily regulated. Different from these observations, opposite expression alteration of OsMADS1 and EG1 was visualized in the background of OsIG1-RNAi lines (Fig. 7A, C). We thought it is probably the different background that should be responsible for triggering the unusual activation/repression of gene expression. Although OsMADS1 has been shown as a positive target of EG1, it can not be ruled out that OsMADS1 acts as a target protein directly or indirectly regulated by other regulatory proteins, such as OsIG1 in the current study or other unidentified regulators. Therefore, our results point out that the effects of OsIG1-RNAi repression are probably not limited to EG1 function and that OsMADS1 as well as its unknown upstream genes also could be activated/repressed in an EG1-independent pathway in OsIG1-RNAi lines. It is indeed interesting that down-regulation of OsIG1 results in up-regulation of OsMADS1 expression in these RNAi lines, suggesting that there may be interplay between OsIG1 and the MADS box to mediate certain developmental cascades in an EG1-independent pathway. Nevertheless, further investigations are required to examine whether this interaction genuinely exists in the regulation of rice floral development. In addition, it is worth mentioning that another characteristic phenotype of the OsIG1-RNAi lines—that is, double ovules—does not occur in loss-of-function eg1 mutant (Li et al., 2009; Cai et al., 2014), suggesting that unidentified downstream genes of OsIG1 other than EG1 could be responsible for the initiation of ovule number control. All these hypotheses need to be further tested in the future.
OsIG1 regulates palea development
At present, several important genes are known to be involved in specifying the identity of the palea. In the mfo1 mutant mrp is absent and recent studies revealed that MFO1/OsMADS6 is expressed in the mrp (Ohmori et al., 2009; Li et al., 2010). Like the mfo1 mutant, in RNAi lines of LHS1/OsMADS1, mrp is absent in the abnormal palea (Prasad et al., 2005). These results imply that OsMADS1 and OsMADS6 specify palea identity by promoting mrp development. In contrast, dp1, sdp1/rep1 and osmads15 mutants all exhibited changes in the bop rather than the mrp (Li et al., 2008; Wang et al., 2010; Yuan et al., 2009). Our findings show that in most of the degenerated palea the development of mrp was normal while the size of bop was reduced in OsIG1-deficient lines in which the E function gene OsMADS1 and the functionally similar E function gene OsMADS6 were up-regulated. Apparently, the altered expression levels of EG1, OsMADS1 and OsMADS6 in OsIG1-RNAi lines contributed to the abnormal formation of the palea. Since the bop tissue alone was affected by OsIG1-RNAi repression, it is possible that the bop and mrp of the palea are controlled by different pathways. Indeed, several genes, including DP1, SDP1/REP1 and OsMADS15, have been reported to determine bop identity (Li et al., 2008; Yuan et al., 2009; Wang et al., 2010). Furthermore, LHS1/MADS1 and MFO1/MADS6 have been demonstrated to be involved in the regulation of mrp identity (Prasad et al., 2005; Ohmori et al., 2009). Additionally, DEGENRATED HULL1 (DH1), a LOB family transcription factor gene, was found to affect both palea and lemma formation (Li et al., 2008; Wang et al., 2011). Thus, it is possible that various genes, such as E function-like genes (OsMADS1 and OsMADS6), palea-specific genes (OsMADS15, DP1 and SDP1/REP1), as well as the LBD gene OsIG1 are required for the formation of palea.
OsIG1 is required for patterning and growth during ovule development
In the maize ig1 mutant, embryo sacs undergo extra rounds of free nuclear division that result in extra egg cells, extra central cells and extra polar nuclei, and also the mutation shows disrupted adaxial–abaxial polarity of leaf development (Huck et al., 2003). Our work revealed that the down-regulated OsIG1 had a similar disruption to embryo sac development (Table 1, Fig. 4L–P), but did not obviously influence the leaf adaxial–abaxial patterning as in the maize ig1 mutant (Evans, 2007), in spite of the altered inclination angle observed in the RNAi transgenic flag leaves (Fig. 6B). The phenotypic variations might be associated with unchanged meristem-specific KNOX gene expression between the transgenic rice lines and WT (data not shown), compared to the fact that alteration of KNOX gene expression is associated with adaxial–abaxial polarity in maize (Evans, 2007). Alternatively, it is possible that the rice RNAi lines retaining relative high residual OsIG1 transcript activities compared with the maize ig1 loss-of-function mutant might be the cause of the unaffected leaf adaxial–abaxial patterning. Interestingly, besides glume-like features, other unique characteristics occurring in OsIG1-RNAi lines has not yet been reported in rice eg1 and maize ig1 mutants, including abnormal carpels and unusual fused double ovules, suggesting that IG1 functions have diversified between the two species. Although our experiments suggest there is less possibility of incorrect targeting caused by OsIG1-RNAi, we could not completely exclude the possibility that the potential incorrect targeting might have effects on vegetative and reproductive processes. Hence, specific mutated lines (e.g. T-DNA insertion lines) would be a useful tool to confirm the phenotypic abnormalities triggered by OsIG1-RNAi repression.
Meanwhile, alteration of floral organ number has been demonstrated to be closely associated with a change in meristem size (Suzaki et al., 2004). In this study, silencing OsIG1 increased floral meristem size at the stage of carpel protrusion (Fig. 4D, F), resulting in increased ovule number (Fig. 4F, G). Additionally, ovule development was indistinguishable from that of the WT at a very early developmental stage. As shown in Fig. 4G, the change in ovule primordia size was accompanied by shape modification, evidenced by the unusual ovule primordia initiation existing on both sides of the palea and lemma of the floral meristem (Fig. 4D, G). This observation raises the possibility that the initiation of ovule primordia is disrupted by OsIG1-RNAi repression. Therefore, the OsIG1 protein may play an important role in inhibiting ovule primordium formation on the palea side in rice. Although it is well known that establishment of adaxial–abaxial polarity is essential for lateral organ development, little is known about the mechanisms underlying the polarity establishment in the carpel and ovule. On the basis of these observations, we speculate that although adaxial–abaxial polarity was less affected in the leaves of OsIG1-RNAi plants, the unusual ovule patterning of transgenic lines is presumably due to defects in the regulation of polarity. The polarity of the gynoecium (the lemmal versus paleal side) may be lost and the gynoecium forms two ovules instead of one. We propose that the gynoecium might contain two ovaries if the transformation is complete (Fig. 4H, I); however, incomplete transformation may result in the fused ovule phenotype (Fig. 4G, H).
In addition, given that aberrant shapes of ovule primordia were initiated by down-regulation of OsIG1 (Supplementary Fig. 2B), we postulated that OsIG1 might have an important role in maintaining the proper organization of the floral meristem, which is essential for the initiation of ovule primordium. In other words, OsIG1 may regulate carpel and ovule primordium initiation by maintaining the proper floral meristem organization during the early developmental phase in rice. Despite the fact that an increased size of the floral meristem/ovule primordial occurred in OsIG1-RNAi repression, the key genes which regulate ovule primordial initiation need to be identified. On the other hand, in the maize ig1 mutant, the proliferative phase is prolonged, suggesting that the IG1 protein restricts the proliferative phase of female gametophyte development. According to this opinion, it is possible that OsIG1 regulates carpel and ovule numbers by preventing overproliferation of their primordia in rice.
In this study, our results revealed that OsIG1 acts as a key regulator in controlling rice spikelet development. It was reported that IG1 interacts with ROUGH SHEATH2 (RS2), which is the maize orthologue of AtAS1 (Evans, 2009). Furthermore, it was shown that members of the basic helix-loop-helix (bHLH) family of transcription factors are capable of interacting with LOB (Husbands et al., 2007). Thus, it would be interesting to elucidate whether these genes are the targets of OsIG1 that may be involved in ovule development. Future identification of the specific interacting factors will shed new light on the molecular mechanism of ovule development in grasses.
Supplementary data
Supplementary material is available at JXB online.
Supplementary Fig. 1. Structure of OsIG1 and sequence analysis of OsIG1.
Supplementary Fig. 2. Constructs and molecular analysis of normal and transgenic plants.
Supplementary Fig. 3. Expression patterns of OsIAL1 and the expression levels of OsIAL1 and Os02g0318851 in the young panicles of OsIG1-RNAi transgenic lines and WT plants.
Supplementary Fig. 4. The I2/KI staining of pollen grains of the WT (A) and OsIG1-RNAi transgenic lines (B–D).
Supplementary Fig. 5. Expression-level comparison of flower-development-related genes between WT and OsIG1-RNAi lines in 1, 5, 10, 15 and 20cm inflorescences.
Supplementary Table 1. Primers used in this study.
Acknowledgements
This work is supported by the National Science Fund for Distinguished Young Scholars (no. 30825030) and the National Science and Technology Key Project of China (no. 2011CB100401).
Glossary
Abbreviations:
- bop
body of the palea
- bp
base pairs
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- LBD
LATERAL ORGAN BOUNDARIES DOMAIN/ASYMMETRIC LEAVES2-LIKE
- LOB
LATERAL ORGAN BOUNDARIES
- mrp
marginal regions of the palea
- qRT-PCR
real-time quantitative PCR
- WT
wild type.
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. 2005. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture induced loss-of-function mutants of the OsMADS1 gene. Plant Molecular Biology 59, 125–135. [DOI] [PubMed] [Google Scholar]
- Borghi L, Bureau M, Simon R. 2007. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. The Plant Cell 19, 1795–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri E, Chuck G, Vollbrecht E, Rocheford T, Martienssen R, Hake S. 2006. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. The Plant Cell 18, 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Yuan Z, Chen MJ, Yin CS, Luo ZJ, Zhao XX, Liang WQ, Hu JP, Zhang DB. 2014. Jasmonic acid regulates spikelet development in rice. Nature Communications 5, article number 3476; 10.1038/ncomms4476. [DOI] [PubMed] [Google Scholar]
- Chalfun-Junior A, Franken J, Mes JJ, Marsch-Martinez N, Pereira A, Angenent GC. 2005. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Molecular Biology 57, 559–575. [DOI] [PubMed] [Google Scholar]
- Chen L, Huang C, Yuan Z, Pan A, Liang W, Huang H, Shen M, Zhang D. 2006. Isolation and genetic analysis for rice mutants treated with 60 Co γ-Ray. Journal of Xiamen University (Natural Science) 45, 82–85. [Google Scholar]
- Dai M, Hu Y, Zhao Y, Liu H, Zhou DX. 2007. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiology 144, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreni L, Jacchia S, Fornara F, Fornari M, Ouwerkerk PB, An G, Colombo L, Kater MM. 2007. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. The Plant Journal 52, 690–699. [DOI] [PubMed] [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM. 2011. Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. The Plant Cell 23, 2850–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MM. 2007. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. The Plant Cell 19, 46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MM. 2009. Indeterminate gametophyte1 (ig1), mutations of ig1, orthologs of ig1, and uses. US Patent Application 20090151025. [Google Scholar]
- Gao XC, Liang WQ, Yin CS, Ji SM, Wang HM, Su X, Guo C, Kong HZ, Xue HW, Zhang DB. 2010. The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiology 153, 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Hong L, Qian Q, Zhu K, Tang D, Huang Z, Gao L, Li M, Gu M, Cheng Z. 2010. ELE restrains empty glumes from developing into lemmas. Journal of Genetics and Genomics 37, 101–115. [DOI] [PubMed] [Google Scholar]
- Huck N, Moore JM, Federer M, Grossniklaus U. 2003. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development 130, 2149–59. [DOI] [PubMed] [Google Scholar]
- Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. 2007. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Research 35, 6663–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. 2005. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. The Plant Cell 17, 1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. 2005. Rice plant development: from zygote to spikelet. Plant Cell Physiology 46, 23–47. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Ueno Y, Semiarti E, et al. 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiology 43, 467–478. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, et al. 2000. leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. The Plant Cell 12, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kater MM, Dreni L, Colombo L. 2006. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. Journal of Experimental Botany 57, 3433–3444. [DOI] [PubMed] [Google Scholar]
- Kellogg EA. 2009. The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza. Rice 2, 1–14. [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. 2010. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiology 51, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Kobayashi T, Morita M, Shimamoto K. 2000. Spatially and temporally regulated expression of rice MADS box genes with similarity to Arabidopsis class A, B and C genes. Plant Cell Physiology 41, 710–718. [DOI] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J. 2009. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiology 151, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Zhang Y, Wu X, Tang W, Wu R, Dai Z, Liu G, Zhang H, Wu C, Chen G, Pan X. 2008. DH1, a LOB domain-like protein required for glume formation in rice. Plant Molecular Biology 66, 491–502. [DOI] [PubMed] [Google Scholar]
- Li HF, Liang WQ, Jia RD, Yin CS, Zong J, Kong HZ, Zhang DB. 2010. The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Research 20, 299–313. [DOI] [PubMed] [Google Scholar]
- Li HG, Xue DW, Gao ZY, Yan MX, Xu WY, Xing Z, Huang DN, Qian Q, Xue YB. 2009. A putative lipase gene EXTRA GLUME1 regulates both empty-glume fate and spikelet development in rice. The Plant Journal 57, 593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HF, Liang WQ, Yin CS, Zhu L, Zhang DB. 2011. a Genetic Interaction of OsMADS3, DROOPING LEAF, and OsMADS13 in Specifying Rice Floral Organ Identities and Meristem Determinacy. Plant Physiology 156, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liang W, Hu Y, Zhu L, Yin C, Xu J, Dreni L, Kater MM, Zhang D. 2011. b Rice MADS6 interacts with the floral homeotic genes SUPERWOMAN1, MADS3, MADS58, MADS13, and DROOPING LEAF in specifying floral organ identities and meristem fate. The Plant Cell 23, 2536–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Shuai B, Springer PS. 2003. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. The Plant Cell 15, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang S, Yu X, Yu J, He X, Zhang S, Shou H, Wu P. 2005. ARL1, a LOB-domain protein required for adventitious root formation in rice. The Plant Journal 43, 47–56. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lopez-Dee ZP, Wittich P, Enrico Pè M, Rigola D, Del Buono I, Gorla MS, Kater MM, Colombo L. 1999. OsMADS13, a novel rice MADS-box gene expressed during ovule development. Journal of Genetics and Development 25, 237–244. [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Ichikawa H, Saito A, Tada Y, Fujimura T, Kano Murakami Y. 1993. Expression of a rice homeobox gene causes altered morphology of transgenic plants. The Plant Cell 5, 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YH, Kang HG, Jung JY, Jeon JS, Sung SK, An G. 1999. Determination of the motif responsible for interaction between the rice APETALA1/AGAMOUS-LIKE9 family proteins using a yeast two-hybrid system. Plant Physiology 120, 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. 2003. SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. [DOI] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. 2009. MOSAIC FLORAL ORGANS1, an AGL6- like MADS box gene, regulates floral organ identity and meristem fate in rice. The Plant Cell 21, 3008–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M. 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis . The Plant Cell 19, 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532. [DOI] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L, Ye D, Sundaresan V. 2004. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis . Development 132, 603–614. [DOI] [PubMed] [Google Scholar]
- Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF. 2003. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U. 2001. Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Development Genes and Evolution 211, 281–290. [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U. 2005. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. The Plant Journal 43, 915–928. [DOI] [PubMed] [Google Scholar]
- Sato Y, Hong SK, Tagiri A, Kitano H, Yamamoto N, Nagato Y, Matssska M. 1996. A rice homeobox gene, OSH1, is expressed before organ differentiation in a specific region during early embryogenesis. Proceedings of the National Academy of Science, USA 93, 8117–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt RJ, Ambrose BA. 1998. The blooming of grass flower development. Current Opinion in Plant Biology 1, 60–67. [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. 2001. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128, 1771–1783. [DOI] [PubMed] [Google Scholar]
- Shuai B, Reynaga-Peña CG, Springer PS. 2002. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiology 129, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. 1997. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located lntragenically. The Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DJ, Hill TA, Gasser CS. 2004. Regulation of ovule development. The Plant Cell 16, S32–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Sato M, Ashikari M, Miyoshi M, Nagato Y, Hirano HY. 2004. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat recetor kinase orthologous to Arabidopsis CLAVATA1 . Development 131, 5649–5657. [DOI] [PubMed] [Google Scholar]
- Terrell EE, Peterson PM, Wergin WP. 2001. Epidermal features and spikelet micromorphology in Oryza and related genera (Poaceae: Oryzeae). Smithsonian Contributions to Botany 91, 1–50. [Google Scholar]
- Wang KJ, Tang D, Hong LL, Xu WY, Huang J, Li M, Gu MH, Xue YB, Cheng ZK. 2010. DEP and AFO regulate peproductive habit in rice. PLoS Genetics 6, e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Wang CS, Tseng TH, Hou YL, Chen KY. 2011. High-resolution genetic mapping and candidate gene identification of the SLP1 locus that controls glume development in rice. Theoretical and Applied Genetics 122, 1489–1496. [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. 2003. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130, 4097–4107. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. 2006. Functional diversification of the two C-Class MADS box genes OsMADS3 and OsMADS58 in Oryza sativa . The Plant Cell 18, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nagasawa N, Kawasaki S, Matsuoka M, Nagato Y, Hirano HY. 2004. The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. The Plant Cell 16, 500–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaki S, Nagato Y, Kurata N, Nonomura K. 2011. Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Developmental Biology 351, 208–216. [DOI] [PubMed] [Google Scholar]
- Yang Y, Yu X, Wu P. 2006. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Molecular Phylogenetic Evolution 39, 248–262. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Suzaki T, Tanaka W, Hirano HY. 2009. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proceedings of the National Academy of Sciences, USA 106, 20103–20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y. 2011. Flower development in rice. Journal of Experimental Botany 62, 4719–4730. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gao S, Xue DW, Luo D, Li LT, Ding SY, Yao X, Wilson ZA, Qian Q, Zhang DB. 2009. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiology 149, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DB, Yuan Z, An G, Dreni L, Hu JP, Kater MM. 2013. Panicle development. In: Zhang QF, Wing RA, eds. Genetics and genomics of rice . Plant Genetics and Genomics: Crops and Models, vol. 5. Heidelberg: Springer, 279–295. [Google Scholar]
- Zhao S, Hu J, Guo L, Qian Q, Xue H. 2010. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Research 20, 935–947. [DOI] [PubMed] [Google Scholar]
- Zhu QH, Guo AY, Gao G, Zhong YF, Xu M, Huang M, Luo J. 2007. DPTF: A database of poplar transcription factors. Bioinformatics 23, 1307–1308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.