Summary
A reversible Renilla luciferase protein complementation assay for rapid identification of protein–protein interactions revealed the existence of an interaction network involved in xyloglucan biosynthesis in the Golgi apparatus in plants.
Key words: Arabidopsis thaliana, glycosyltransferase, Golgi apparatus, Nicotiana benthamiana, plant cell wall, polysaccharides, protein–protein interaction, Renilla luciferase, type II membrane protein, xyloglucan.
Abstract
A growing body of evidence suggests that protein–protein interactions (PPIs) occur amongst glycosyltransferases (GTs) required for plant glycan biosynthesis (e.g. cell wall polysaccharides and N-glycans) in the Golgi apparatus, and may control the functions of these enzymes. However, identification of PPIs in the endomembrane system in a relatively fast and simple fashion is technically challenging, hampering the progress in understanding the functional coordination of the enzymes in Golgi glycan biosynthesis. To solve the challenges, we adapted and streamlined a reversible Renilla luciferase protein complementation assay (Rluc-PCA), originally reported for use in human cells, for transient expression in Nicotiana benthamiana. We tested Rluc-PCA and successfully identified luminescence complementation amongst Golgi-localizing GTs known to form a heterodimer (GAUT1 and GAUT7) and those which homooligomerize (ARAD1). In contrast, no interaction was shown between negative controls (e.g. GAUT7, ARAD1, IRX9). Rluc-PCA was used to investigate PPIs amongst Golgi-localizing GTs involved in biosynthesis of hemicelluloses. Although no PPI was identified among six GTs involved in xylan biosynthesis, Rluc-PCA confirmed three previously proposed interactions and identified seven novel PPIs amongst GTs involved in xyloglucan biosynthesis. Notably, three of the novel PPIs were confirmed by a yeast-based split-ubiquitin assay. Finally, Gateway-enabled expression vectors were generated, allowing rapid construction of fusion proteins to the Rluc reporters and epitope tags. Our results show that Rluc-PCA coupled with transient expression in N. benthamiana is a fast and versatile method suitable for analysis of PPIs between Golgi resident proteins in an easy and mid-throughput fashion in planta.
Introduction
In eukaryotes, biosynthetic machineries of extracellular matrix polysaccharides (e.g. cell wall polysaccharides, proteoglycans) and N- and O-protein glycosylation are located in the endomembrane system, particularly in the Golgi apparatus. It is known in plants, yeast, and mammals that the Golgi-localization and function of some of glycan biosynthetic enzymes, including glycosyltransferases (GTs), are regulated through protein–protein interactions (PPIs) (de Graffenried and Bertozzi, 2004; Jungmann and Munro, 1998; McCormick et al., 2000; Schoberer et al., 2013; Tu and Banfield, 2010). A growing body of evidence suggests that functions of some GTs and other proteins involved in plant cell wall biosynthesis are also controlled by PPIs (Oikawa et al., 2013). In dicots, pectins and hemicelluloses are two main classes of plant cell wall polysaccharides, the biosynthesis of which occurs in the Golgi apparatus (Driouich et al., 2012; Dupree and Sherrier, 1998), and which play pivotal roles in providing physical support to the cell wall and during plant growth, development, reproduction, and in response to environmental stimuli (Bellincampi et al., 2014; Scheller and Ulvskov, 2010; Wolf et al., 2009). These polysaccharides also present a rich renewable source of biomass for a wide range of industrial processes including production of food, fuel, and fibres (Carpita, 2012; Pauly and Keegstra, 2008). Impressive progress has been made during the past decade in the identification of genes and enzymes involved in pectin and hemicellulose biosynthesis; however, mechanisms of their functional control by PPIs remain largely elusive, owing in part to technical limitations as described below.
In Arabidopsis thaliana, the following PPIs have been experimentally demonstrated for GTs involved in cell wall biosynthesis. The term ‘GT’ is used for proteins that are classified as GTs by the Carbohydrate Active Enzyme database (http://www.cazy.org) based on phylogenetic relatedness even though some of these proteins, such as GALACTURONOSYLTRANSFERASE7 (GAUT7) may not have glycosyltransferase activity (Atmodjo et al., 2011). Of note, heterooligomerization of GAUT1 and its homologue, GAUT7, in biosynthesis of pectic homogalacturonan (Atmodjo et al., 2011); hetero- and homo-oligomerization of the putative arabinosyltransferases ARABINAN DEFICIENT1 (ARAD1) and ARAD2 in pectic arabinan biosynthesis (Harholt et al., 2012; Søgaard et al., 2012); and homo- and heterooligomerization of XYLOSYLTRANSFERASE1 (XXT1), XXT2, XXT5, and CELLULOSE-SYNTHASE LIKE C4 (CSLC4) in hemicellulosic xyloglucan (XyG) biosynthesis (Chou et al., 2012) have been demonstrated. In wheat, a high-molecular weight complex involving GTs belonging to the GT families 43, 47, and 75 has been identified in glucurono(arabino)xylan biosynthesis (Zeng et al., 2010). In addition, it has been proposed that UDP-arabinose mutases, also known as REVERSIBLY GLYCOSYLATED POLYPEPTIDEs (RGPs), required for the interconversion of UDP-arabinopyranose to UDP-arabinofuranose, are retained at the cytosolic face of the Golgi membrane by PPIs for efficient supply of the substrate to the Golgi lumen (Konishi et al., 2007; Rautengarten et al., 2011). Precise biological/biochemical functions of these PPIs are only beginning to be understood. For instance, GAUT1 undergoes proteolytic processing in the endomembrane system, which renders the C-terminal catalytic domain of GAUT1 secretable, whereas GAUT7, which does not seem to undergo proteolytic processing, anchors the secretable C-terminal domain of GAUT1 in the Golgi lumen where homogalacturonan biosynthesis occurs (Atmodjo et al., 2011). Given the structural complexity of polysaccharides, PPIs among GTs, modifying enzymes (e.g. methyl- and acetyltransferases, nucleotide-sugar converting enzymes) and other functional proteins (e.g. transporters and chaperones) are likely to occur and may play pivotal roles in the regulation of biosynthesis of these polymers (Oikawa et al., 2013). Analysis of PPIs among integral membrane proteins in the Golgi apparatus is technically challenging because many of these proteins are often in low abundance and isolation of protein complexes requires careful choice and optimization of conditions (e.g. detergents, buffers, chromatographic resins) for each individual PPI investigated. In situ binary PPI assays, such as bimolecular fluorescence complementation (BiFC), and Förster resonance energy transfer (FRET) are non-invasive and can facilitate identification of PPIs in living tissues, and in addition can provide information about the subcellular localization of PPIs in cells. However, these techniques have their caveats. In BiFC, the split halves of the fluorophores irreversibly assemble, causing a high rate of false-positive interactions (Magliery et al., 2005; Søgaard et al., 2012; Zamyatnin et al., 2006). Although BiFC has been successfully used to aid identification of PPIs among the Golgi-localizing GTs in plants and mammals (Chou et al., 2012; Hassinen et al., 2010), it also requires use of a flow cytometer, an expensive and maintenance-intensive instrument, for high-confidence identification of PPIs. Whereas FRET offers a lower false positive rate, it has a low signal-to-noise ratio and requires additional data processing and an expensive instrumental setup (Piehler, 2005). The split-ubiquitin assay in yeast (Stagljar et al., 1998) is widely used for PPIs among membrane proteins. Ubiquitin is split into two fragments, the N-terminal ubiquitin fragment (Nub) and the C-terminal ubiquitin fragment (Cub). The native NubI, with “I” being isoleucine at position 13, interacts irreversibly with Cub and is used as a positive control, whereas NubG, with “G” being glycine replacing the isoleucine, interacts reversibly with Cub and is used for the interaction assay (Johnsson and Varshavsky, 1994). Cub is fused to a synthetic transcription factor (TF) (protein A–LexA–VP16), and when reconstituted with NubI or NubG the C-terminus of Cub is cleaved by cytosolic ubiquitin-specific proteases releasing the synthetic transcriptional factor that subsequently initiates transcription of reporter genes. The split-ubiquitin assay is powerful as it allows high-throughput screening of PPIs amongst membrane-bound proteins, and has been successfully used in characterisation of the cellulose synthase complex in Arabidopsis (Timmers et al., 2009). However, as plant proteins are expressed in a non-native system, misfolding and mislocalization can result in a relatively high rate of false-negative interactions (Oikawa et al., 2013).
In this article, we present a successful adaptation of a reversible Renilla luciferase complementation assay (Rluc-PCA), previously reported in human cells (Stefan et al., 2007), for screening of PPIs among Golgi-localizing proteins in planta. Luciferase-based PCA offers a superb signal-to-noise ratio and maintains reversibility of PPIs (Stefan et al., 2007). Agrobacterium tumefaciens-mediated transient transfection of Nicotiana benthamiana was used to express proteins of interest (POI) fused with the N- and C-terminal human-codon optimized Renilla luciferase (hRluc) fragments in Gateway-enabled expression vectors. Co-transfection of Agrobacterial strains carrying different POI-hRluc constructs allowed versatility in choice of binary interaction assay to be performed. To strengthen the versatility of the system, compatible Gateway expression vectors for the yeast split-ubiquitin assay were generated. The assay is easy, robust, and requires standard laboratory equipment. Furthermore, using Rluc-PCA enabled successful identification of novel candidates for PPIs amongst XyG biosynthetic enzymes.
Materials and methods
Generation of hRluc and ST–hRluc
hRluc was reconstituted from the two hRluc fragments (a gift from S.W. Michnick, University of Montreal) by USER fusion (Geu-Flores et al., 2007; Nour-Eldin et al., 2006). hRluc fragment [F1] was amplified from PKACat.hRluc-F[1] (Stefan et al., 2007) using primers USERF1 F and USERF1 R. Fragment [F2] was amplified from PKACat.hRluc-F[2] (Stefan et al., 2007) using USERF2 F and USERF2 R. The two products were combined and inserted into pCambia3300u (Geu-Flores et al., 2007; Nour-Eldin et al., 2006) by USER fusion. Gateway entry clones were produced for both the N-terminus of rat sialyltransferase (ST) combined with hRluc (ST–hRluc) and hRluc in pDONRTM/Zeo by BP recombination. An attB flanked hRluc was amplified using primers attB1Luc F and attB2Luc R. An attB flanked ST–hRluc was produced by overlap PCR. ST was amplified from Yn-TMD (Søgaard et al., 2012) using primers attB1ST F and LucST R. hRluc was amplified using primers STLuc F and attB2Luc R. Products were combined and attB1ST F and attB1Luc R primers used to amplify the ST–hRluc chimera. Primer sequences are detailed in Supplementary Table S1. Constructs for measurement of activity of the ST–hRluc fusion protein and hRluc were produced without C-terminal epitope fusions by recombination with pEarleygate100 (Earley et al., 2006). Constructs for localization of the ST–hRluc fusion protein and hRluc were produced by LR recombination with pEarleygate101 to produce C-terminal YFP fusions.
Transient expression in N. benthamiana
Transient expression in N. benthamiana was performed as described by Sakuragi et al. (2011) using Agrobacterium tumefaciens GV3101 as a bacterial host and included the co-infiltration of the viral silencing suppressor p19 (Voinnet et al., 2003). Transient expression of fusion proteins was carried out in 4-week-old N. benthamiana plants grown under a 16h photoperiod at 26/24°C (day/night), 60% humidity and light intensities of 115–150 µE m–2 s–1. Each A. tumefaciens strain was infiltrated at a final OD600nm of 0.2, unless stated otherwise, and that harbouring p19 at OD600 0.05. Infiltrated plants were returned to the same growth conditions for 72h before harvest of material.
Fluorescence confocal microscopy
ST–hRluc–YFP and the Golgi marker α-mannosidase–CFP (Nelson et al., 2007) were co-infiltrated into N. benthamiana to confirm targeting of ST–hRluc to the Golgi apparatus. Abaxial epidermal sections from leaves 72h post infiltration were prepared. A Zeiss LSM 710 confocal microscope equipped with Argon and InTune lasers was used for confocal laser-scanning microscopy. All images were obtained with a 0.9NA 40X air objective using the Zen software package (Carl Zeiss Inc., Oberkochen, Germany). Emission was collected at 463 to 484nm (CFP) and 521 to 572nm (YFP), laser lines 405nm and 514nm. The pinhole diameter was set at 1 airy unit. Image analysis and processing (scale bar, brightness, and contrast) used ImageJ (Version 1.6r).
Construction of phRluc[F1] and phRluc[F2] vectors
Gateway compatible phRluc[F1] and phRluc[F2] vectors were generated by USER cloning of overlap PCR amplicons into pCambia3300u downstream of a CaMV 35S promoter. pEarleygate101 was used as a template to amplify the Gateway cassette, PKACat.hRluc-F[1] for hRluc [F1] and PKACat.hRluc-F[2] for hRluc [F2]. In addition, a C-terminal HA epitope was added to hRluc-[F1] and a FLAG epitope to hRluc [F2] by inclusion in the reverse primer sequence. All primers used are detailed in Supplementary Table S1. The amplicons for fusion were amplified using the products of USER GW F and LucF1GW R, and GWLucF1 F and USERLucF1HA R for phRluc[F1] and USERGW F and LucF2GW R combined with GWLucF2 F and USERLucF2FL R for phRluc[F2]. The final cassettes were amplified from a mixture of the two fragments with USER site flanked primers.
Gateway enabled DUALmembrane system vectors
pPR3-N and pBT3-N (Dualsystems Biotech AG, Schlieren, Switzerland) were used to create Gateway destination vectors by ligation of a Gateway cassette by the SfiI site downstream of the NubG and Cub, thereby generating pPR3-GW and pBT3-GW, respectively. The Gateway cassette was amplified from pEarleygate101 using primers SfiGW F and SfiGWStop R to include a stop codon in frame to the 3’ of the attR2 site.
Generation of HG, XyG, and xylan-related GT Rluc-PCA constructs
Coding sequences lacking stop codons of GAUT1 (At3g61130.1), GAUT7 (At2g38650.1), ARAD1 (At2g35100.1), IRREGULAR XYLEM9 (IRX9, At2g37090.1), XXT5 (At1g74380.1), MURUS3/KA- TAMARI1 (MUR3) (At2g20370.1) and FUCOSYLTRANSFERA- SE1 (FUT1, At2g03220.1) were inserted into pDONRTM/Zeo by BP recombination (Invitrogen, Carlsbad, CA, USA), primers detailed in Supplementary Table S1. Coding sequences of XXT1 (At3g62720.1), XXT2 (At4g02500.1), CSLC4 (At3g28180.1), IRX9-LIKE (IRX9-L, At1g27600.1), IRX10 (At1g27440.1), IRX10-L (At5g61840.1), IRX14 (At4g36890.1) and IRX14-L (At5g67230.1) in pDONR223 lacking stop codons were obtained from the JBEI GT collection (Lao et al., 2014). In-frame fusions into phRluc[F1] and phRluc[F2] vectors were made by LR recombination (Invitrogen, Carlsbad, CA, USA). Additionally, the 35S–ARAD1–cMyc construct, a gift from J. K. Jensen (Michigan State University, USA) was used as the competitor in the competition assay.
Reversible Renilla luciferase protein complementation assay
Three leaf discs (Ø 0.5cm), one from each of three infiltrated leaves, were punched out and pooled into tubes containing 200 µl ice cold assay buffer [0.5M NaCl, 0.1M potassium phosphate pH 7.4, 1mM EDTA, 0.02% (w/v) BSA supplemented with protease inhibitors (cOmplete EDTA-free protease inhibitor, Roche, Basel, Switzerland)] and a chrome ball (Ø 3mm). The plant material was macerated in a mixer mill (Retsch MM301, Haan, Germany) at 25–30 Hz for 1min. Samples were kept on ice whenever possible. Of each sample, 100 µl was transferred to a Nunc black 96-well plate (Thermo Scientific, Rockford, IL, USA). Coelenterazine-h (Biosynth AG, Staad, Switzerland) was added to a final concentration of 10 µM to each well by an automated injector and bioluminescence measured for 30 s immediately after addition using a luminometer (Berthold TriStar2 LB 942, Berthold, Bad Wildbad, Germany). For each PPI tested, three independent samples, each comprised of a pool of three independent leaf discs, were assayed. The experiment was repeated three times with independent transfection of N. benthamiana. Means of the RLU values derived from the three independent experiments were transformed to the Log10 scale, which were used for statistical evaluation by Student′s t-test (independent test with two tails) for evaluation of the difference from the Log10-transformed RLU value obtained for samples expressing p19 alone.
Immunoblotting
Pooled leaf discs as described above were either homogenised directly in 100 µl Laemmli buffer or were macerated in the Rluc-PCA assay buffer and Laemmli buffer added. The samples were boiled for 5min and cooled on ice. Ten microliters of the homogenate were separated on a 12% 1-mm thick polyacrylamide gel (Criterion™ XT Bis-Tris precast polyacrylamide gel, Biorad, Hercules, CA, USA) in 1× XT-MOPS buffer (Biorad, Hercules, CA, USA). Proteins were transferred to a nitrocellulose membrane and probed with primary and secondary antibodies. Antibodies were diluted in PBS-T 1% (w/v) skimmed milk powder as the following: rabbit α-HA (Sigma-Aldrich, St. Louis, MO, USA), 1:500; swine α-rabbit HRP-conjugate (Dako, Glostrup, Denmark), 1:1700; mouse α-FLAG M2 (Sigma-Aldrich), 1:1000; rabbit α-mouse HRP-conjugate (Dako, Glostrup, Denmark), 1:2000; mouse α-cMyc 9E10 (Sigma-Aldrich), 1:1000, where skimmed milk powder was omitted. Detection was performed with SuperSignal West Dura chemiluminescent substrate (Thermo Scientific, Rockford, IL, USA).
Yeast split-ubiquitin assay
The split-ubiquitin assay was performed in yeast strain NMY51 using pBT3-N/pBT3-GW and pPR3-N/pPR3-GW vectors (Dualsystems Biotech AG, Schlieren, Switzerland). The coding sequences of tested GTs were PCR amplified using primers detailed in Supplementary Table S1, and ligated into pBT3-N and pPR3-N (Dualsystems Biotech AG, Schlieren, Switzerland) at the SfiI restriction site. The coding sequence of FUT1 was inserted in-frame into pBT3-GW and pPR3-GW by LR recombination. The plasmids were introduced in pairs into NMY51 by LiAc transformation (Gietz and Woods, 2002). Transformants were selected on SD-Leu-Trp and strains carrying both vectors were grown to OD546 of 1.5. Serial dilutions (from 1–1000 fold) were spotted on SD-His-Leu, SD-His-Leu-Trp and SD-His-Leu-Trp-Ade plates. Growth on SD-His-Leu-Trp-Ade plates was scored as an indication of interaction. Yeast growing on SD-His-Leu plates were tested for β-galactosidase activity using the X-gal overlay assay (Obrdlik et al., 2004).
Results and discussion
The choice of luciferase-based PCA system for analysis of PPIs in the Golgi lumen
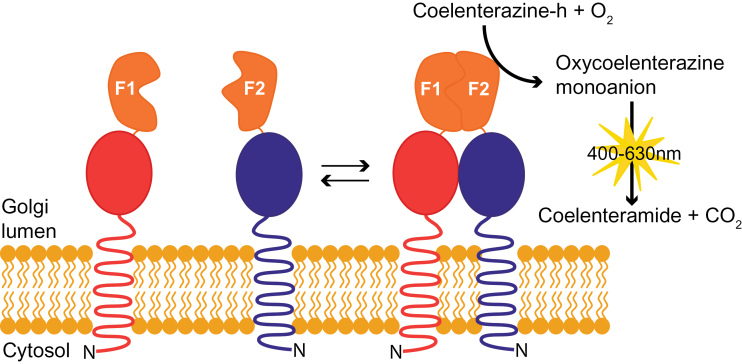
Before this study, two versions of luciferase-based PCA had been developed for study of PPIs among cytosolic proteins in planta. Firefly luciferase was used to successfully detect PPIs amongst cytosolic proteins in N. benthamiana (Gehl et al., 2011). However, this system is unsuitable for PPI assays in the Golgi lumen because of the absence of ATP in this compartment. A luciferase from sea pansy (Renilla reniformis; Rluc) does not require ATP for its catalytic action and has been successfully used for in vivo detection of PPIs amongst cytosolic proteins in Arabidopsis protoplasts (Fujikawa and Kato, 2007; Kato and Jones, 2010). This system also integrated a Gateway- and Cre-loxP-enabled vector cloning system, allowing high-throughput cloning and screening of PPIs in planta. However, reversibility of the association between the two Rluc fragments (amino acid residues 1–299, N-terminal fragment; residues 299–310, C-terminal fragment) has not yet been experimentally demonstrated. A human-codon optimized Rluc PCA with structure-based design of fragments (amino acid residues 1–110, N-terminal fragment [F1]; residues 111–310, C-terminal fragment [F2]) has been developed for use in human cell line HEK293T and Chinese hamster ovary cells (Stefan et al., 2007). Notably, the reversible reconstitution of the two fragments has been experimentally demonstrated. The reversibility of the system is particularly important for an assay system dealing with endomembrane proteins because their diffusion is limited in a restricted two-dimensional space. As a consequence there would be a considerably higher frequency of false-positive interaction should the two fragments irreversibly assemble. Therefore, we have used Rluc-PCA for the subsequent experiments and used N. benthamiana as expression host owing to its ease of transfection and efficient expression of transient proteins with minimal handling compared with Arabidopsis protoplast based assays. A schematic representation of Rluc-PCA adapted for a Golgi PPI assay is shown in Fig. 1.
Fig. 1.
Schematic representation of the reversible Renilla luciferase protein complementation assay (Rluc-PCA) to study Golgi lumenal protein interactions. Membrane proteins with a type II membrane topology, spanning the membrane once with the N-terminus (N) in the cytosol and a lumenal C-terminus, are shown fused to the N-terminal domain (F1) and C-terminal domain (F2) of human-codon optimized Renilla luciferase (hRluc). Arrows denote the dynamics of the protein interaction, the coupling and decoupling of the two domains of hRluc. (This figure is available in colour at JXB online.)
Assay of hRluc activity in the Golgi apparatus of N. benthamiana
We placed the hRluc fragments on the carboxy (C) termini of the POIs because amino (N) terminal tagging of integral membrane proteins may affect their membrane protein topologies (Søgaard et al., 2012). Furthermore, there is precedence for post-translational proteolytic processing that cleaves the N-terminal domain from the C-terminal domain that contains features required for PPIs (Atmodjo et al., 2011).
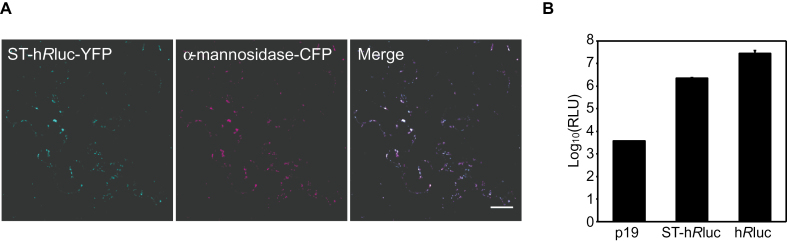
The functionality of hRluc within the Golgi lumen, never previously demonstrated in planta, was determined. The 52 amino acid residues from the N-terminus of ST, including the 17 residue trans-membrane region and 9 cytosolic residues, have previously been shown to be sufficient for Golgi localization (Boevink et al., 1998). Notably the C-terminus localizes to the Golgi lumen (Søgaard et al., 2012). A fusion protein between the N-terminal domain of ST and hRluc was generated. Transfection of ST–hRluc with a C-terminal YFP fusion alongside the Golgi marker α-mannosidase (Nelson et al., 2007), with C-terminal CFP fusion, into N. benthamiana confirmed the targeting of hRluc to the Golgi apparatus (Fig. 2A). ST–hRluc and hRluc were transiently expressed in N. benthamiana and activity assayed by the conversion of coelenterazine-h to coelenteramide, which undergoes relaxation from an electronically excited state, emitting a photon of blue light. Leaf discs were excised from the infiltrated areas and macerated in an assay buffer (see materials and methods) and this macerate was directly used for the luciferase assay. Robust bioluminescence significantly above background was observed when assaying both ST–hRluc and hRluc (Fig. 2B). These results demonstrate the suitability of hRluc as a reporter in the Golgi lumen. The signal intensity derived from ST–hRluc was an order of magnitude lower than that from hRluc, which is expected either owing to the generally low abundance of Golgi localized proteins due to the smaller compartment volume of the Golgi apparatus as compared to the cytosol and/or owing to different extractability of the proteins in these compartments.
Fig. 2.
Localization and activity of Golgi lumenal localized human-codon optimized Renilla luciferase (hRluc). Rat sialyltransferase transmembrane domain (ST) fused to hRluc was used to target hRluc to the Golgi apparatus. (A) ST–hRluc-YFP co-localized with the cis-Golgi marker α-mannosidase-CFP. Scale bar=20 µm. (B) ST–hRluc and hRluc activities in transiently expressing N. benthamiana crude leaf protein extracts. The silencing suppressor p19 was co-expressed with ST–hRluc and hRluc constructs and alone is used as a negative control for background luminescence. Error bars represent 95% confidence interval, n=3. Log10(RLU); relative luminescence units transformed to Log10. (This figure is available in colour at JXB online.)
Construction of a Gateway-compatible hRluc binary vector system
To facilitate the combination of the advantages of Rluc-PCA with the requirement to examine a multitude of candidate protein interactions within the Golgi apparatus, we sought to create Rluc-PCA vectors that use Gateway® cloning technology (Life Technologies). Gateway-compatible destination vectors phRluc[F1] (HA-tag) and phRluc[F2] (FLAG-tag) (Supplementary Fig. S1A) were generated that allowed rapid recombination with libraries of genes contained in entry vectors (Lao et al., 2014) and fusion with epitope tags (HA, hemagglutinin; FLAG, the octapeptide DYKDDDDK) for detection of expressed proteins. As the system is Gateway-compatible, genes of interest can easily be cloned and tested in several Gateway-enabled PPI systems including bioluminescence resonance energy transfer (BRET) (Subramanian et al., 2006), FRET (Miyawaki and Tsien, 2000; Siegel et al., 2000), BiFC (Gehl et al., 2009), and the BiFC-based membrane topology analysis (Søgaard et al., 2012). In addition to the Gateway-compatible systems already available, a commercially available split-ubiquitin assay system (DUALmembrane system, Dualsystems Biotech AG, Schlieren, Switzerland) (see details below) was Gateway-enabled for testing membrane-localized PPIs in yeast (Supplementary Fig. S1B).
Optimization of the Rluc-PCA system in transient expression in N. benthamiana
Initially, highly variable signals of hRluc were seen between different infiltrated areas and leaves. As N. benthamiana leaves are known to express proteins to different degrees depending on growth stage of the leaves (Cazzonelli and Velten, 2006), the activity of complemented hRluc in tissue macerated from manually infiltrated leaves of different ages on the same plant was determined and compared with tissue pooled from the same three leaves (Supplementary Fig. S2A). Expression between leaves was found to be variable within the same plant and therefore the method was refined to pool tissue to reduce variability and ensure reproducibility. Cazzonelli and Velten (2006) found that optimal protein expression occurs between 44–96h post infiltration. To ensure optimal expression, a 72h period was chosen. To determine the optimal integration time for measurement of complemented hRluc activity, relative luminescence units (RLU) were measured at half-second intervals for 10 s before and 300 s after addition of coelenterazine-h (Supplementary Fig. S2B). An integration time of 30 s was applied to maximize the integrated signal intensity while minimizing protein degradation. Vacuum infiltration with Agrobacterium was also tested, although this resulted in very poor signals, and therefore manual infiltration, pooling of three leaf discs and measurement after 3 d were used for the subsequent experiments.
An overview of the developed assay procedure is shown in Fig. 3. Combinations of Agrobacterial strains containing constructs of interest can be made in 24-well plates for convenient handling, with each combination requiring not more than 1ml in volume. In our hands, manual infiltration of 50 combinations, each infiltrated in three different leaves, by one person takes approximately 1–2h, allowing a mid-throughput analysis. This processing time was comparable to that required for a manually performed split-ubiquitin assay in yeast with the same number of samples. After 72h, three leaf discs, each derived from independent infiltrated areas, were excised, pooled, and macerated in 96-well plates with a ball mixer mill. The macerates were then transferred to fresh 96-well plates for luminescence measurement. A microplate reader with high-sensitivity luminescence detector equipped with auto-reagent-injectors was used in this study.
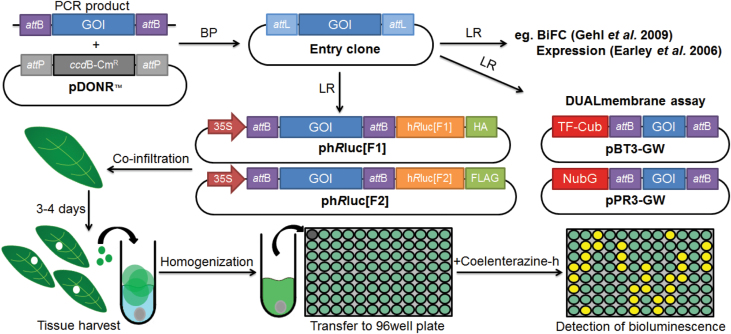
Fig. 3.
Schematic protocol for Rluc-PCA. Genes of interest (GOIs) are PCR amplified and recombined into pDONRTM/Zeo vector by BP cloning (BP). The GOI-containing entry clone can be recombined by LR cloning (LR) to insert GOI into desired destination vectors, which here is the Gateway-compatible phRluc[F1] and phRluc[F2] belonging to Rluc-PCA. Alternatively, GOI can be recombined into other destination vectors, e.g. Gateway-compatible DUALmembrane vectors containing the amino terminal ubiquitin fragment (NubG) and the carboxy terminal ubiquitin fragment fused to an artificial transcription factor (TF–Cub). In Rluc-PCA, phRluc[F1]- and phRluc[F2]-fused GOIs are individually introduced into different Agrobacterium and then co-infiltrated into the leaves of N. benthamiana in desired combinations to test PPIs. The assay is performed 3–4 d post infiltration by harvesting three leaf discs and transferring to tubes containing 200 µl assay buffer and a chrome ball. Leaf discs are macerated and 100 µl is transferred into a black 96-well plate. Bioluminescence upon addition of coelenterazine-h is monitored in a plate luminometer. (This figure is available in colour at JXB online.)
Establishing a proof of concept for identifying PPIs in the Golgi lumen
To confirm that the Rluc-PCA was functional in reporting PPIs within the Golgi lumen, cell wall biosynthetic GTs that were previously shown to form complexes in the Golgi apparatus were selected to tested with Rluc-PCA. GAUT, GAUT7, and ARAD1 were selected as these proteins form hetero- and homodimeric complexes, respectively (Atmodjo et al., 2011; Harholt et al., 2012). These proteins exhibit the canonical type II membrane topology and the C-termini of these proteins are known to be lumenal (Atmodjo et al., 2011; Søgaard et al., 2012). The GAUT1–GAUT7 PPI also allowed the resilience of the system in assaying interactions between membrane-associated proteins and soluble lumenal proteins. Additionally, IRX9, also a Golgi-localizing GT (Brown et al., 2007; Peña et al., 2007), was included as a control to test for non-specific interactions. As IRX9 is known to be involved in hemicellulosic glucuronoxylan synthesis in the secondary cell wall (Brown et al., 2007; Peña et al., 2007), whereas the GAUT1–GAUT7 heterodimer and the ARAD1 homodimer are involved in pectic homogalacturonan (Atmodjo et al., 2011) and arabinan synthesis (Harholt et al., 2006) in the primary cell wall, respectively, we hypothesised that these proteins would not interact. Analysis of the amino acid sequence of IRX9 shows that it has a typical type II membrane topology, indicating that the IRX9 C-terminus is also lumenal (Supplementary Fig. S3).
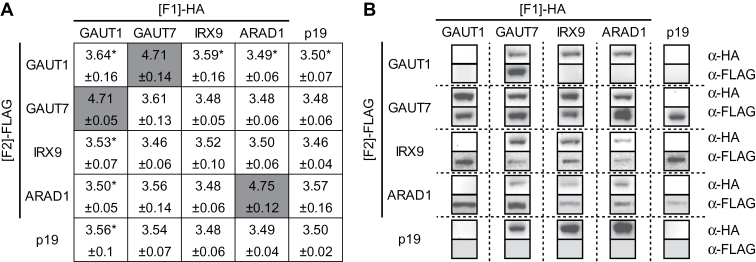
Agrobacterial strains harbouring GAUT1, GAUT7, ARAD1, and IRX9 recombined into phRluc[F1] and phRluc[F2] were co-infiltrated into N. benthamiana leaves alongside the silencing suppressor p19 (Voinnet et al., 2003). The OD value of each strain was 0.2 whereas that for p19 was 0.05. Binary PPI assays among GAUT1, GAUT7, ARAD1, and IRX9 showed that, after 72h incubation, only tissues expressing the combinations GAUT1-[F1] and GAUT7-[F2] (Log10 value: 4.71), GAUT7-[F1] and GAUT1-[F2] (Log10 value: 4.71), and ARAD1-[F1] and ARAD1-[F2] (Log10 value: 4.75) showed complemented luciferase activity with an order of magnitude higher RLU than those expressing the single halves of hRluc or p19 alone (Log10 value: 3.50) (Fig. 4A). Immunoblots confirmed that, as expected, soluble GAUT1 was only retained in leaves also expressing its anchor GAUT7. In all other cases, immunoblots confirmed expression of two proteins in extracts testing both positive and negative interactions by Rluc-PCA, indicating a negative measurement was due to lack of interaction and not lack of expression (Fig. 4B). Measured RLUs of positive complementations were between approximately 16–18 fold higher than that of background demonstrating the robustness of the Rluc-PCA in discerning positive interactions in the Golgi lumen above non-specific noise. The average RLU from the positive interactions was 2.3%±0.12 of the RLU obtained for the Golgi-localized hRluc. Taken together, these results demonstrate that Rluc-PCA can successfully identify known Golgi PPIs and can distinguish positive PPIs from the background.
Fig. 4.
Rluc-PCA identifies the GAUT1–GAUT7 core-complex and ARAD1–ARAD1 homodimer. (A) Heat map of Log10 values of RLU where dark grey denotes statistically significant higher Log10 values of RLU above the background level (p19). Statistical analysis was performed on the averages derived from three independent experiments, each consisting of three biological replicates (pools) (see materials and methods). A vector containing the silencing suppressor p19 was co-transfected along with GOI–hRluc[F1] and GOI–hRluc[F2]. Error represents 95% confidence interval, n=3. Asterisk represents extracts where GAUT1 was not detected by immunoblot owing to proteolytic processing and possible degradation (Atmodjo et al., 2011). (B) Immunoblot of expressed proteins probed with anti-HA and anti-FLAG primary antibodies.
Impact of protein overexpression on the bioluminescence complementation was analysed. ARAD1-[F1] and ARAD1-[F2] were co-expressed at equal Agrobacterial OD values ranging from 0.025–0.2. This OD range was chosen because ARAD1 fused to GFP localizes to the Golgi apparatus when infiltrated at the OD value of 0.05, whereas increasing ODs caused mistargeting to the endoplasmic reticulum (Sakuragi et al., 2011). Log10 RLU values obtained for all the samples were significantly higher than that of the negative control (p19 only), whereas no significant difference was observed among the samples within the tested OD range (Supplementary Table S2). These results indicate that overexpression of ARAD1 does not increase the bioluminescence signal. Targeting of glycosyltransferases to sub-Golgi compartments can be mediated by protein complex formation, known as “kin recognition”, which functions by forming protein aggregates that are too large to enter transport vesicles (Nilsson et al., 1993). It is plausible that ARAD1 forms homomeric complexes to remain in the Golgi apparatus or in a sub-Golgi compartment and those proteins that were mistargeted to the endoplasmic reticulum owing to overexpression do not form complexes and thus do not contribute to bioluminescence complementation.
In addition, higher OD values (0.2 and 0.1) for ARAD1-[F1] were infiltrated alongside a lower OD value (0.05) for ARAD-[F2] and bioluminescence measured. Log10 RLU values of both combinations were significantly higher than that of the negative control but were not significantly different in comparison to the sample where the OD value for the both proteins was 0.2 (P-value>0.05) (Supplementary Table S2). This result suggests that the bioluminescence complementation of ARAD1-[F1] and ARAD1-[F2] is independent of the ratio of the expressed protein levels within the range tested.
Finally, a competition assay was performed in which ARAD1-[F1] and ARAD1-[F2] (OD value of 0.1 for each) were co-expressed with a cMyc-tagged ARAD1 as the competitor (OD values of 0, 0.2, and 0.4) (Table 1 and Supplementary Fig. S4). The complemented bioluminescence diminished with increasing concentration of the competitor, demonstrating that the observed bioluminescence complementation is not due to a false positive effect.
Table 1.
Competition assayARAD1-[F1] and ARAD1-[F2] were co-expressed with ARAD1–cMyc as the competitor at increasing Agrobacterial ODs. p19 was co-expressed in all samples (OD 600nm=0.05). Log10 values of RLU were obtained from three biological replicates, errors represents the 95% confidence interval.
| Construct | OD | ||
|---|---|---|---|
| ARAD1-[F1] | 0.1 | 0.1 | 0.1 |
| ARAD1-[F2] | 0.1 | 0.1 | 0.1 |
| 35S–ARAD1–cMyc | 0 | 0.2 | 0.4 |
| Log10(RLU) | 4.20 ±0.059 |
4.07 ±0.019 |
3.96 ±0.011 |
Rluc-PCA among hemicellulosic xyloglucan and xylan biosynthetic enzymes
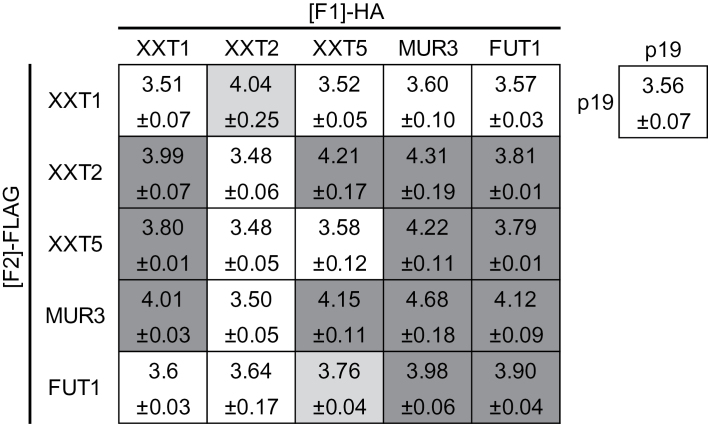
Rluc-PCA coupled with transient expression in N. benthamiana was applied to test binary interactions among XyG biosynthetic enzymes: XXT1 (Cavalier and Keegstra, 2006), XXT2 (Cavalier and Keegstra, 2006), XXT5 (Zabotina et al., 2008), MUR3 (Madson et al., 2003; Tamura et al., 2005), FUT1 (Perrin et al., 1999; Perrin et al., 2003), and CSLC4 (Cocuron et al., 2007). Expression of fusion proteins was confirmed by immunoblot analysis (Supplementary Fig. S5), with the exception of CSLC4-[F1] and -[F2], which were not detectable. The background RLU level of N. benthamiana expressing p19 was Log10 value of 3.56. The lower and upper limits of the range of detected RLU found to be significantly higher than background (p19) were XXT5-[F1] and FUT1-[F2] with a Log10 value of 3.76, and MUR3-[F1] and MUR3-[F2] with a Log10 value of 4.75, which are approximately 5800 RLU and 56000 RLU, respectively. The tested combination consisting of XXT1 and XXT2, XXT5 and FUT1, XXT5 and MUR3, MUR3 and FUT1, MUR3 and MUR3, FUT1 and FUT1 (Fig. 5) all had statistically significant (P<0.05) RLUs above the background level. These positive PPIs were detectable when the [F1] and [F2] tags were swapped in at least five out of six biological replications and therefore were considered as genuine PPIs with high confidence. XXT1 and XXT2 formed PPIs with many other XyG enzymes when they were fused to the [F2] and [F1] tags, respectively, whereas no PPI was formed when the tags were swapped (Fig. 5). XXT1-[F2] and XXT2-[F1] may be improperly folded or the hRluc tags are not oriented properly to allow complementation of the luciferase activity. Therefore, the results were interpreted as an indication of PPIs with lower confidence among the following XyG enzymes: XXT1 and XXT5, XXT1 and MUR3, XXT2 and XXT5, XXT2 and MUR3, XXT2 and FUT1. CSLC4 has a topology locating both N- and C-termini to the cytosolic side of the Golgi membrane (Davis et al., 2010), whereas the other tested proteins are Golgi-localized type II membrane proteins that have their C-termini within the Golgi lumen (Søgaard et al., 2012). This caused the split hRluc tags to be located on opposite faces of the membrane rendering complementation of hRluc impossible when testing CSLC4 against Golgi-localized type II membrane proteins, and such were not tested.
Fig. 5.
Application of the Rluc-PCA to test PPIs among XyG biosynthetic enzymes. Three independent experiments, each consisting of three biological replicates (pools) were made (see materials and methods). Generated results are shown as heat map of Log10 values of RLU where dark grey denotes samples with all experiments being significantly higher than the background level, whereas light grey denotes samples with two out of three experiments being significantly higher than the background level and white denotes Log10 values of RLU of the background p19 infiltrated control in a complementation assay. Statistical analysis was performed on the averages derived from three independent experiments. Plants were co-transfected with Agrobacteria carrying vectors containing silencing suppressor p19, GOI–hRluc[F1], and GOI–hRluc[F2]. Error represents 95% confidence interval, n=3.
There is evidence from wheat that proteins from GT43, GT47, and GT75 form a higher order complex in arabinoxylan synthesis (Zeng et al., 2010). It has previously been speculated that the enzymes involved in synthesis of the β-1,4-linked xylan backbone, namely IRX9 and IRX14 of GT43, and IRX10 of GT47, may too form PPIs in Arabidopsis (Brown et al., 2009; Faik et al., 2014; Oikawa et al., 2013). We carried out Rluc-PCA amongst these proteins and their homologues IRX9-L, IRX14-L, and IRX10-L. Luminescence above background was not detected for any combination of these enzymes, indicating no direct PPIs occurring amongst the xylan biosynthetic GTs under the conditions tested (Supplementary Fig. S6).
Split-ubiquitin assay of XyG and xylan biosynthetic enzymes
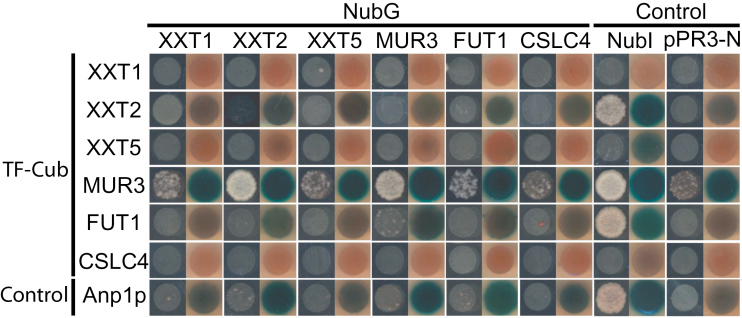
For comparison, the split-ubiquitin assay in yeast was used to test PPIs among the XyG and xylan biosynthetic enzymes. The original split-ubiquitin system requires the synthetic TF to be fused to the C-terminus of Cub and to be localized to the cytosol to be cleaved off by ubiquitin-specific proteases and to initiate transcriptional activation of reporter genes in the nucleus (Stagljar et al., 1998). Because many GTs, including those described in this study, have type II membrane topology, C-terminal tagging would result in the Cub-TF to localize to the Golgi lumen; thus no complementation can occur. Recently a modified yeast split-ubiquitin assay system was made commercially available (Dualsystems Biotech AG, Schlieren, Switzerland), allowing the fusion of the TF at the N-terminus of Cub (TF–Cub); thus the cytosol-localized N-termini of type II membrane proteins. Here, the modified split-ubiquitin assay integrating the Gateway cloning system (see above) was used with a series of controls. Ost1p–NubI was used, where Ost1p, a yeast type I membrane protein localized to the endomembrane system, is fused to NubI, a wild-type variant of Nub that forms stable interaction with Cub. This fusion protein is routinely used to test the functionality of the Cub fusion proteins as a positive control (Stagljar et al., 1998). The empty plasmid, pPR3-N, was used as a negative control to assess auto-activation by the Cub fusion proteins. Anp1p is a Golgi-localized enzyme of yeast involved in N-glycan biosynthesis and was fused to TF–Cub in this study to generate Cub–Anp1p (TF–Cub–Anp1p) as a control to assess random interaction by NubG-fused proteins of interest. TF-Cub-fused XXT1, XXT5, and CSLC4 were found to be non-functional because no growth was found when paired with NubI-fused Ost1p (Fig. 6). Consistently, no PPI involving these proteins was detected. TF-Cub-fused XXT2 was functional but did not form reproducibly detectable PPIs (Fig. 6). In contrast, the TF-Cub-fused MUR3 was found to be functional with only a limited degree of auto-activation (Supplementary Fig. S7), and it showed a significantly high degree of growth when paired with NubG-fused XXT2 and MUR3. TF-Cub-fused FUT1 was also functional but it only showed a limited growth and β-galactosidase activity when paired with NubG-fused MUR3, suggesting an interaction with low confidence. These results indicate that under the conditions tested the split-ubiquitin assay detected PPIs between MUR3 and XXT2, MUR3 and MUR3, and with MUR3 and FUT1.
Fig. 6.
Split-ubiquitin assay used to detect PPIs among XyG biosynthetic proteins. Transformed yeast containing the indicated combinations of TF–Cub and NubG fused proteins were spotted in an OD546 of 1.5 and up to 1000× dilution on SD-His-Leu and SD-His-Leu-Trp-Ade plates. Growth on SD-His-Leu-Trp-Ade plates indicates a positive interaction. X-Gal assay performed on growing yeast on SD-His-Leu is a test for β-galactosidase activity, a reporter for interaction upon blue colour formation, Ost1p–NubI (NubI) and pPR3-N test for the functionality and random interaction of the Cub-fused proteins, respectively. The type II membrane protein TF–Cub–Anp1p tests for random interaction among NubG-fused proteins. Consensus of three biological replicates is shown. (This figure is available in colour at JXB online.)
Of the xylan biosynthetic proteins, when expressed alongside NubI-fused Ost1p, only IRX14 and IRX14-L were demonstrated to be functional TF-Cub fusions (data not shown). The lack of functionality of the majority of the xylan backbone-related GTs under test meant that this line of investigation was not furthered.
Comparison of XyG and xylan PPIs detected by Rluc-PCA, the split-ubiquitin assay, and BiFC.
Results obtained in this study and in the previous study by Chou et al. (2012), which applied BiFC in Arabidopsis protoplasts combined with co-immunoprecipitation of recombinant proteins expressed in E. coli, were used for a comparison of the three binary PPI assays for the plant Golgi-localizing proteins involved in XyG biosynthesis (Table 2). Of 10 combinations tested by BiFC in Arabidopsis protoplasts and by co-immunoprecipitation of recombinant proteins expressed in E. coli, 7 PPIs were observed (Chou et al., 2012). In the study presented here, of the 21 tested, Rluc-PCA detected 11 PPIs. Rluc-PCA successfully confirmed three of the PPIs previously detected by Chou et al. (2012), XXT1 and XXT2, XXT1 and XXT5, and XXT2 and XXT5, whereas it did not detect homooligomerization of XXT2 and XXT5. The lack of homomeric complementation by XXT2 is likely to be due to the aforementioned improper function of XXT2-[F1], whereas the lack of homomeric complementation by XXT5 is not readily reconciled. It is possible that XXT5 forms a transient interaction that occurs in a kiss-and-go manner, where the proteins are mainly in monomeric form and the complexes form only in a small fraction of time and/or with forces that are too weak to maintain the complex during the sample preparation. Similarly to co-immunoprecipitation, Rluc-PCA would not be able to generate sufficiently high signals for such interactions. Alternatively, XXT5 may form a transient homomeric association when overexpressed, which could be detectable by BiFC owing to irreversibility of the reporter reconstitution. Aside from the previously reported interactions, Rluc-PCA identified seven novel PPIs among XyG biosynthetic enzymes: XXT1 and MUR3, XXT2 and MUR3, XXT2 and FUT1, XXT5 and MUR3, XXT5 and FUT1, MUR3 and MUR3, and FUT1 and FUT1. Heterooligomerization of XXT2 and MUR3, and XXT2 and FUT1 have previously been implicated by Zabotina (2012). During the preparation of this manuscript, Zabotina and colleagues have identified heterooligomerization of XXT2 and FUT1, XXT5 and FUT1, MUR3 and FUT1 and homooligomerization of FUT1 by using BiFC and co-immunoprecipitation (personal communication), corroborating our results. Furthermore, PPIs between XXT2 and MUR3, MUR3 and FUT1, and MUR3 itself were verified by split-ubiquitin assay in yeast as described below.
Table 2.
Comparison of the results obtained by Rluc-PCA, the split-ubiquitin assay (Split-Ub), and bimolecular fluorescence complementation (BiFC)/co-immunoprecipitation (Co-IP) (Chou et al., 2012)0, 1, and 2, indicate no PPI, a PPI with low confidence, and a PPI with high confidence, respectively. nt indicates not tested or not testable owing to non-functional or non-expressed proteins. POI, protein of interest.
| Combination | Rluc-PCA | Split-Ub | BiFC/Co-IP | |
|---|---|---|---|---|
| POI 1 | POI 2 | |||
| XXT1 | XXT1 | 0 | Nt | 0 |
| XXT2 | 2 | Nt | 2 | |
| XXT5 | 1 | Nt | 1 | |
| MUR3 | 1 | 0 | nt | |
| FUT1 | 0 | 0 | nt | |
| CSLC4 | nt | Nt | 0 | |
| XXT2 | XXT2 | 0 | Nt | 2 |
| XXT5 | 1 | 0 | 2 | |
| MUR3 | 1 | 2 | nt | |
| FUT1 | 1 | 0 | nt | |
| CSLC4 | nt | 0 | 0 | |
| XXT5 | XXT5 | 0 | Nt | 1 |
| MUR3 | 2 | 0 | nt | |
| FUT1 | 2 | 0 | nt | |
| CSLC4 | nt | Nt | 2 | |
| MUR3 | MUR3 | 2 | 2 | nt |
| FUT1 | 2 | 1 | nt | |
| CSLC4 | nt | 0 | nt | |
| FUT1 | FUT1 | 2 | 0 | nt |
| CSLC4 | nt | 0 | nt | |
| CSLC4 | CSLC4 | nt | Nt | 2 |
Recently, binary interactome analysis among 3286 membrane and signalling proteins from Arabidopsis were carried out (Jones et al., 2014) using the mating-based split-ubiquitin system (Obrdlik et al., 2004), wherein the reporters (Cub–TF and NubG) were fused at the C-termini of the tested proteins. As mentioned above, C-terminal tagging of type II membrane proteins renders the Cub and NubG fragments to be situated inside the Golgi lumen, thereby making them non-functional and this is reflected in the analysis; XXT5 and FUT1, fused to Cub–TF were initially represented in the interactome analysis but were excluded from the analysis owing to “bad topology”, whereas NubG-fusions of XXT5 and FUT1 were still included in the screen, but no PPI involving these proteins was identified. The yeast two-hybrid system was also used to construct an Arabidopsis interactome map ( Arabidopsis Interactome Mapping Consortium, 2011). The yeast two-hybrid system relies on reconstitution of a functional TF followed by transcriptional activation of reporter gene expression in the nucleus. Poor representation of membrane integrated GTs in the interactome by the yeast two-hybrid system is expected, because the system requires the relocation of the assemblage of the reconstituted TF fused to POIs into the nucleus for reporter genes to be transcriptionally activated. GAUT1, GAUT7, XXT2, FUT1, and IRX10-L were represented in the total of approximately 8000 open reading frames screened. The only PPI detected involving these proteins was between FUT1 and At4G19030, a putative aquaporin (AT-NLM1) that formed PPI with 127 other proteins, suggesting false-positive PPIs. The split-ubiquitin assay conducted in this study, using N-terminal tagging, successfully identified heterooligomerization of XXT2 and MUR3, and MUR3 and FUT1, and homooligomerization of MUR3, lending further support to the newly identified PPIs in XyG biosynthesis by Rluc- PCA. Yet, it is evident that the same assay grossly underestimated the XyG PPIs, and only detected three PPIs in total. This is in large part due to non-functionality of the expressed proteins.
Xylan biosynthetic enzymes of GT43 and GT47 did not form detectable PPIs (Supplementary Fig. S6). This was unexpected as it has been demonstrated that a higher order complex involving members of GT47 and GT43 is formed in wheat (arabino)xylan synthesis which promotes synthesis of the β(1,4) xylan backbone (Zeng et al., 2010) and because indirect evidence support the existence of xylan synthase complexes in Arabidopsis (Brown et al., 2009; Faik et al., 2014). It remains possible that these proteins do not form complexes and act independently on the oligosaccharide substrates. It is also possible that additional proteins are required to form complexes including these enzymes. Nevertheless, the lack of interactions in xylan biosynthesis underpins the specificity of PPIs detected by Rluc-PCA.
Based on Table 2, we hypothesize that there exists a XyG biosynthetic PPI network. In Arabidopsis XyG biosynthesis, CSLC4 is thought to catalyse the synthesis of β-1,4-linked glucan backbone (Cocuron et al., 2007), which is decorated with the side-chain α-1,6-xylosyl residues by XXT1, XXT2, and XXT5 (Cavalier and Keegstra, 2006; Cavalier et al., 2008; Zabotina et al., 2008) and can be further substituted with β-1,2-galactosyl residue by MUR3 and XYLOGLUCAN L-SIDE CHAIN GALACTOSYLTRANSFERASE POSITION2 (XLT2) and α-1,2-fucosyl residues by FUT1 (Faik et al., 2000; Jensen et al., 2012; Madson et al., 2003; Scheller and Ulvskov, 2010; Zabotina et al., 2008). In this study, PPIs among XXT1, XXT2, XXT5, MUR3, and FUT1 were tested and nearly all these side-chain forming enzymes were found capable of forming PPIs with each other, which raises a possibility that XXT1, XXT2, XXT5, MUR3, and FUT1 form a side-chain forming complex(es). It is noteworthy that XXT5 is thus far the only side-chain forming enzyme that has been shown to form PPI with CSLC4 (Chou et al., 2012). Also noteworthy is that MUR3 was previously identified as KATAMARI1, which is required for the proper functioning of actin in maintaining the endomembrane organization in Arabidopsis through protein complex formation with actin (Tamura et al., 2005). Based on these previous observations and the results obtained in the present study, we speculate (i) that XXT5 tethers the side-chain forming complex(es) to the backbone synthesis by CSLC4 to form a XyG biosynthetic complex(es) and (ii) that this complex(es) is further anchored to actin filaments via MUR3 to ensure proper actin organization that is required for the secretion of polysaccharides from the Golgi stacks to the cell wall (Blancaflor, 2002; Hu et al., 2003; Tamura et al., 2005). The hypothesis of XXT5 tethering is consistent with the phenotypes of various xyloglucan mutants as previously reported (Zabotina et al., 2012). The authors concluded that XXT5 cannot add the xylose residues on its own and raised a possibility that the function of XXT5 is to maintain the integrity of a synthetic complex involved in xyloglucan biosynthesis rather than to function as a xylosyltransferase. Although this possibility is yet to be substantiated, our results lend support to it. Based on the physiological data by Zabotina et al. (2012) and our results, we speculate that the exact protein composition of xyloglucan complexes is probably variable depending on tissues types; for instance in seedling roots XXT5 is largely dispensable, whereas in hypocotyl XXT5 plays a major role in determining and/or maintaining the composition of xyloglucan biosynthetic complex(es).
Conclusions
The results presented in the current study demonstrate that Rluc-PCA adapted for use in transient expression in N. benthamiana allows easy, rapid, and mid-throughput screening of PPIs among Golgi-resident membrane proteins. Integration of Gateway technology enables versatile choice of additional PPI systems (BiFC and split-ubiquitin assay in yeast) to be applied. The system can be readily used for studies of PPIs in the lumen of the endomembrane system as well as in the cytosol.
Supplementary data
Supplementary data are available at JXB online
Figure S1. Maps of destination vectors produced in this study
Figure S2. Refinement of Rluc-PCA parameters
Figure S3. Prediction of IRX9 protein topology
Figure S4. Immunoblot of competition assay
Figure S5. Immunoblot of XyG proteins
Figure S6. Application of Rluc-PCA to test xylan related PPIs
Figure S7. Random interaction of MUR3-bait in split-ubiquitin assay
Table S1. Primers sequences used in this study
Table S2. OD dependency assay
Acknowledgements
This work was supported by the Danish Advanced Technology Foundation (Biomass for the 21st century, grant number 001-2011-4); The Danish Council for Strategic Research (Plant Power, grant number 12-131834); Nordic Research Energy (AquaFEED, grant number 24); European Union′s Seventh Framework Programme FP7 (ENERGY-2010–1 DirectFuel, grant number 256808); The People Programme Marie Curie Actions (PHOTO.COMM, grant number 317184), and The U.S. Department of Energy Office of Science and Office of Biological and Environmental Research (contract no. DE–AC02–05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy).
We thank Stephen W. Michnick (Université de Montréal, Succursale Center-Ville, Montréal, QC, Canada) for providing the hRluc containing vectors, PKACat.hRluc-F[1] and PKACat.hRluc-F[2] and Jacob K. Jensen (Michigan State University, USA) for providing the 35S–ARAD1–cMyc construct. We also thank Sara Fasmer Hansen (Copenhagen University, Denmark) for critical review and discussion and Yuta Hihara, Johannes Evald Buus, Daniel Godske Eriksen, Ditte Bøgeskov Hansen, and Nanna Brøns Jungersen for experimental assistance. No conflict of interest is declared.
Glossary
Abbreviations:
- BiFC
bimolecular fluorescence complementation
- Cub
carboxy terminal fragment of ubiquitin
- FRET
Förster resonance energy transfer
- GOI
gene of interest
- GT
glycosyltransferase
- hRluc
human-codon optimized Renilla reniformis luciferase
- Nub
amino terminal fragment of ubiquitin
- POI
protein of interest
- PPI
protein–protein interaction
- RLU
relative luminescence unit
- Rluc
Renilla reniformis luciferase
- Rluc-PCA
Renilla luciferase complementation assay
- ST
Rat sialyltransferase
- TF
transcription factor
- XyG
xyloglucan.
References
- Arabidopsis Interactome Mapping Consortium. 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo MA, Sakuragi Y, Zhu X, Burrell AJ, Mohanty SS, Atwood JA, Orlando R, Scheller HV, Mohnen D. 2011. Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proceedings of the National Academy of Sciences, USA 108, 20225–20230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellincampi D, Cervone F, Lionetti V. 2014. Plant cell wall dynamics and wall-related susceptibility in plant–pathogen interactions. Frontiers in Plant Science 10.3389/fpls.2014.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blancaflor EB. 2002. The cytoskeleton and gravitropism in higher plants. Journal of Plant Growth Regulation 21, 120–136. [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Cruz SS, Martin B, Betteridge A, Hawes C. 1998. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. The Plant Journal 15, 441–447. [DOI] [PubMed] [Google Scholar]
- Brown DM, Goubet F, Wong VW, Goodacre R, Stephens E, Dupree P, Turner SR. 2007. Comparison of five xylan synthesis mutants reveals new insight into the mechanisms of xylan synthesis. The Plant Journal 52, 1154–1168. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zhang Z, Stephens E, Dupree P, Turner SR. 2009. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis . The Plant Journal 57, 732–746. [DOI] [PubMed] [Google Scholar]
- Carpita NC. 2012. Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Current Opinion in Biotechnology 23, 330–337. [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Keegstra K. 2006. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. Journal of Biological Chemistry 281, 34197–34207. [DOI] [PubMed] [Google Scholar]
- Cavalier DM, Lerouxel O, Neumetzler L, et al. 2008. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. The Plant Cell 20, 1519–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazzonelli C, Velten J. 2006. An in vivo, luciferase-based, Agrobacterium-infiltration assay system: implications for post-transcriptional gene silencing. Planta 224, 582–597. [DOI] [PubMed] [Google Scholar]
- Chou Y-H, Pogorelko G, Zabotina OA. 2012. Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiology 159, 1355–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocuron J-C, Lerouxel O, Drakakaki G, Alonso AP, Liepman AH, Keegstra K, Raikhel N, Wilkerson CG. 2007. A gene from the cellulose synthase-like C family encodes a β-1,4 glucan synthase. Proceedings of the National Academy of Sciences, USA 104, 8550–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J, Brandizzi F, Liepman AH, Keegstra K. 2010. Arabidopsis mannan synthase CSLA9 and glucan synthase CSLC4 have opposite orientations in the Golgi membrane. The Plant Journal 64, 1028–1037. [DOI] [PubMed] [Google Scholar]
- de Graffenried CL, Bertozzi CR. 2004. The roles of enzyme localisation and complex formation in glycan assembly within the Golgi apparatus. Current Opinion in Cell Biology 16, 356–363. [DOI] [PubMed] [Google Scholar]
- Driouich A, Follet-Gueye M-L, Bernard S, Kousar S, Chevalier L, Vicré-Gibouin M, Lerouxel O. 2012. Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Frontiers in Plant Science 3, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupree P, Sherrier DJ. 1998. The plant Golgi apparatus. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1404, 259–270. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Faik A, Bar-Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K. 2000. Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in Pea. Journal of Biological Chemistry 275, 15082–15089. [DOI] [PubMed] [Google Scholar]
- Faik A, Jiang N, Held M. 2014. Xylan biosynthesis in plants, simply complex. In: McCann MC, Buckeridge MS, Carpita NC, eds. Plants and BioEnergy. New York: Springer, 153–181. [Google Scholar]
- Fujikawa Y, Kato N. 2007. Technical advance: Split luciferase complementation assay to study protein–protein interactions in Arabidopsis protoplasts. The Plant Journal 52, 185–195. [DOI] [PubMed] [Google Scholar]
- Gehl C, Kaufholdt D, Hamisch D, Bikker R, Kudla J, Mendel RR, Hänsch R. 2011. Quantitative analysis of dynamic protein–protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. The Plant Journal 67, 542–553. [DOI] [PubMed] [Google Scholar]
- Gehl C, Waadt R, Kudla J, Mendel R-R, Hänsch R. 2009. New GATEWAY vectors for high throughput analyses of protein–protein interactions by bimolecular fluorescence complementation. Molecular Plant 2, 1051–1058. [DOI] [PubMed] [Google Scholar]
- Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA. 2007. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Research 35, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. 2002. Transformation of yeast by the Liac/SS carrier DNA/PEG method. Methods in Enzymology 350, 87–96. [DOI] [PubMed] [Google Scholar]
- Harholt J, Jensen J, Verhertbruggen Y, et al. 2012. ARAD proteins associated with pectic arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta , 1–14. [DOI] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV. 2006. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis . Plant Physiology 140, 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. 2010. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. Journal of Biological Chemistry 285, 17771–17777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhong RQ, Morrison WH, Ye ZH. 2003. The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta 217, 912–921. [DOI] [PubMed] [Google Scholar]
- Jensen JK, Schultink A, Keegstra K, Wilkerson CG, Pauly M. 2012. RNA-seq analysis of developing nasturtium seeds (Tropaeolum majus): identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Molecular Plant 5, 984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. 1994. Split ubiquitin as a sensor of protein interactions in-vivo . Proceedings of the National Academy of Sciences, USA 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Xuan Y, Xu M, et al. 2014. Border control—A membrane-linked interactome of Arabidopsis . Science 344, 711–716. [DOI] [PubMed] [Google Scholar]
- Jungmann J, Munro S. 1998. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with [alpha]-1,6-mannosyltransferase activity. The EMBO Journal 17, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Jones J. 2010. The split luciferase complementation assay. Methods in Molecular Biology 655, 359–376. [DOI] [PubMed] [Google Scholar]
- Konishi T, Takeda T, Miyazaki Y, Ohnishi-Kameyama M, Hayashi T, O’Neill MA, Ishii T. 2007. A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 17, 345–354. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Lao J, Oikawa A, Bromley JR, et al. 2014. The plant glycosyltransferase clone collection for functional genomics. The Plant Journal 79, 517–529. [DOI] [PubMed] [Google Scholar]
- Madson M, Dunand C, Li X, Verma R, Vanzin GF, Caplan J, Shoue DA, Carpita NC, Reiter W-D. 2003. The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. The Plant Cell 15, 1662–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magliery TJ, Wilson CGM, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. 2005. Detecting protein−protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. Journal of the American Chemical Society 127, 146–157. [DOI] [PubMed] [Google Scholar]
- McCormick C, Duncan G, Goutsos KT, Tufaro F. 2000. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proceedings of the National Academy of Sciences, USA 97, 668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A, Tsien RY. 2000. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods in Enzymology 327, 472–500. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G. 1993. Kin recognition: A model for the retention of Golgi enzymes. FEBS Letters 330, 1–4. [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Hansen BG, Nørholm MHH, Jensen JK, Halkier BA. 2006. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Research 34, e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, et al. 2004. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proceedings of the National Academy of Sciences, USA 101, 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa A, Lund CH, Sakuragi Y, Scheller HV. 2013. Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends in Plant Science 18, 49–58. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. 2008. Cell-wall carbohydrates and their modification as a resource for biofuels. The Plant Journal 54, 559–568. [DOI] [PubMed] [Google Scholar]
- Peña MJ, Zhong R, Zhou G-K, Richardson EA, O’Neill MA, Darvill AG, York WS, Ye Z-H. 2007. Arabidopsis irregular xylem8 and irregular xylem9: Implications for the complexity of glucuronoxylan biosynthesis. The Plant Cell 19, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. 1999. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284, 1976–1979. [DOI] [PubMed] [Google Scholar]
- Perrin RM, Jia Z, Wagner TA, O’Neill MA, Sarria R, York WS, Raikhel NV, Keegstra K. 2003. Analysis of xyloglucan fucosylation in Arabidopsis . Plant Physiology 132, 768–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehler J. 2005. New methodologies for measuring protein interactions in vivo and in vitro . Current Opinion in Structural Biology 15, 4–14. [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Herter T, Petzold CJ, Ishii T, Mukhopadhyay A, Usadel B, Scheller HV. 2011. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis . The Plant Cell 23, 1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuragi Y, Nørholm MH, Scheller H. 2011. Visual mapping of cell wall biosynthesis. Methods in Molecular Biology 715, 153–167. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. 2010. Hemicelluloses. In: Merchant S, Briggs WR, Ort D, eds. Annual Review of Plant Biology. Palo Alto: Annual Reviews, 263–289. [DOI] [PubMed] [Google Scholar]
- Schoberer J, Liebminger E, Botchway SW, Strasser R, Hawes C. 2013. Time-resolved fluorescence imaging reveals differential interactions of N-glycan processing enzymes across the Golgi Stack in planta . Plant Physiology 161, 1737–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge U-I, Kunze R. 2003. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiology 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RM, Chan FK-M, Zacharias DA, Swofford R, Holmes KL, Tsien RY, Lenardo MJ. 2000. Measurement of molecular interactions in living cells by fluorescence resonance energy transfer between variants of the green fluorescent protein. Science Signaling 2000, pl1. [DOI] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S. 1998. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo . Proceedings of the National Academy of Sciences, USA 95, 5187–5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan E, Aquin S, Berger N, Landry CR, Nyfeler B, Bouvier M, Michnick SW. 2007. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo . Proceedings of the National Academy of Sciences, USA 104, 16916–16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C, Woo J, Cai X, Xu X, Servick S, Johnson CH, Nebenführ A, Von Arnim AG. 2006. A suite of tools and application notes for in vivo protein interaction assays using bioluminescence resonance energy transfer (BRET). The Plant Journal 48, 138–152. [DOI] [PubMed] [Google Scholar]
- Søgaard C, Stenbæk A, Bernard S, Hadi M, Driouich A, Scheller HV, Sakuragi Y. 2012. GO-PROMTO illuminates protein membrane topologies of glycan biosynthetic enzymes in the Golgi apparatus of living tissues. PLoS ONE 7, e31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I. 2005. KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis . The Plant Cell 17, 1764–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers J, Vernhettes S, Desprez T, Vincken J-P, Visser RGF, Trindade LM. 2009. Interactions between membrane-bound cellulose synthases involved in the synthesis of the secondary cell wall. FEBS Letters 583, 978–982. [DOI] [PubMed] [Google Scholar]
- Tu L, Banfield D. 2010. Localization of Golgi-resident glycosyltransferases. Cellular and Molecular Life Sciences 67, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. The Plant Journal 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wolf S, Mouille G, Pelloux J. 2009. Homogalacturonan methyl-esterification and plant development. Molecular Plant 2, 851–860. [DOI] [PubMed] [Google Scholar]
- Zabotina OA. 2012. Xyloglucan and its biosynthesis. Frontiers in Plant Science 3, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Avci U, Cavalier D, Pattathil S, Chou Y-H, Eberhard S, Danhof L, Keegstra K, Hahn MG. 2012. Mutations in multiple XXT genes of Arabidopsis reveal the complexity of xyloglucan biosynthesis. Plant Physiology 159, 1367–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabotina OA, Van De Ven WTG, Freshour G, Drakakaki G, Cavalier D, Mouille G, Hahn MG, Keegstra K, Raikhel NV. 2008. Arabidopsis XXT5 gene encodes a putative α-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. The Plant Journal 56, 101–115. [DOI] [PubMed] [Google Scholar]
- Zamyatnin AA, Solovyev AG, Bozhkov PV, Valkonen JPT, Morozov SY, Savenkov EI. 2006. Assessment of the integral membrane protein topology in living cells. The Plant Journal 46, 145–154. [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A. 2010. A glucurono(arabino)xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Physiology 154, 78–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.