Highlight
Expression of the diterpene synthase gene for gibberellin biosynthesis occurs in tissues different from those in which its isoform for phytoalexin biosynthesis is expressed, reflecting their distinct biological roles in rice.
Key words: Biosynthetic enzyme, diterpene, gene expression, gibberellin, growth, rice.
Abstract
Gibberellins (GAs) are diterpenoid phytohormones that regulate various aspects of plant growth. Tetracyclic hydrocarbon ent-kaurene is a biosynthetic intermediate of GAs, and is converted from geranylgeranyl diphosphate, a common precursor of diterpenoids, via ent-copalyl diphosphate (ent-CDP) through successive cyclization reactions catalysed by two distinct diterpene synthases, ent-CDP synthase and ent-kaurene synthase. Rice (Oryza sativa L.) has two ent-CDP synthase genes, OsCPS1 and OsCPS2. It has been thought that OsCPS1 participates in GA biosynthesis, while OsCPS2 participates in the biosynthesis of phytoalexins, phytocassanes A–E, and oryzalexins A–F. It has been shown previously that loss-of-function OsCPS1 mutants display a severe dwarf phenotype caused by GA deficiency despite possessing another ent-CDP synthase gene, OsCPS2. Here, experiments were performed to account for the non-redundant biological function of OsCPS1 and OsCPS2. Quantitative reverse transcription–PCR (qRT–PCR) analysis showed that OsCPS2 transcript levels were drastically lower than those of OsCPS1 in the basal parts, including the meristem of the second-leaf sheaths of rice seedlings. qRT–PCR results using tissue samples prepared by laser microdissection suggested that OsCPS1 transcripts mainly localized in vascular bundle tissues, similar to Arabidopsis CPS, which is responsible for GA biosynthesis, whereas OsCPS2 transcripts mainly localized in epidermal cells that address environmental stressors such as pathogen attack. Furthermore, the OsCPS2 transgene under regulation of the OsCPS1 promoter complemented the dwarf phenotype of an OsCPS1 mutant, oscps1-1. The results indicate that transcripts of the two ent-CDP synthase genes differentially localize in rice plants according to their distinct biological roles, OsCPS1 for growth and OsCPS2 for defence.
Introduction
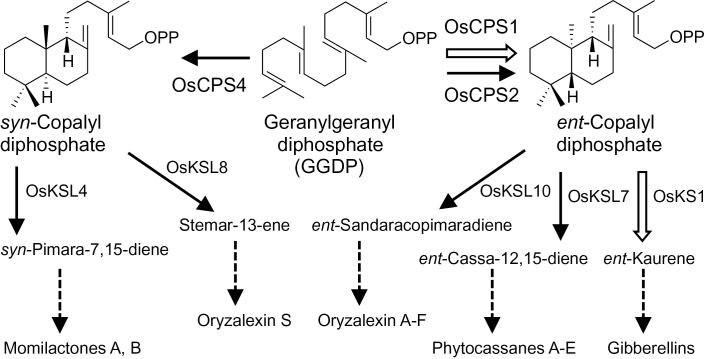
Diterpenoids are a class of terpenoids mainly derived from the C20 prenyl substrate geranylgeranyl diphosphate (GGDP). The linear substrate GGDP is converted into various cyclic hydrocarbons by specific diterpene cyclases. The carbon skeletons are successively chemically modified by enzymes including P450 monooxygenases, dehydrogenases, methyltransferases, glucosyl transferases, and others. Gibberellins (GAs) are labdane-related diterpene phytohormones that regulate various aspects of plant growth, such as germination, stem elongation, and flowering (Yamaguchi, 2008; Hedden and Thomas, 2012). GAs are biosynthesized from the intermediate tetracyclic hydrocarbon ent-kaurene, which is converted from GGDP by two-step cyclization (Fig. 1). The two steps are catalysed by two distinct diterpene cyclases, ent-copalyl diphosphate (ent-CDP) synthase and ent-kaurene synthase. Other GA biosynthetic genes have been identified and characterized in detail (Yamaguchi, 2008; Hedden and Thomas, 2012).
Fig. 1.
Early steps in the biosynthetic pathway of labdane-related diterpenoids in rice. The steps in the formation of intermediate cyclic hydrocarbons of gibberellins (open arrows) and diterpene phytoalexins (solid arrows) are shown with the names of diterpene cyclases next to the arrows.
Rice (Oryza sativa L.) produces not only GAs but also labdane-related diterpene phytoalexins, including phytocasssanes (Koga et al., 1995, 1997; Yajima and Mori, 2000), oryzalexins (Akatsuka et al., 1985; Kato et al., 1993, 1994; Tamogami and Mitani, 1993), and momilactones (Kato et al., 1973; Cartwright et al., 1981). Phytoalexins are low molecular weight compounds produced for defence against pathogens (Ahuja et al., 2012). The biosynthetic hydrocarbon intermediates of phytocassanes A–E, oryzalexins A–F, oryzalexin S, and momilactones A and B are ent-cassa-12, 15-diene, ent-sandaracopimaradiene, stemar-13-ene, and syn-pimara-7, 15-diene, respectively (Fig. 1). The first and last two are converted from GGDP through ent-CDP and syn-CDP, a diastereomer of ent-CDP, respectively (Fig. 1). The diterpene cyclase genes responsible for these steps have been identified in the rice genome (Peters, 2006; Toyomasu, 2008): OsCPS2, OsCPS4, OsKSL4, OsKSL7, OsKSL8, and OsKSL10 (Fig. 1). Diterpene cyclases responsible for ent-kaurene biosynthesis in rice are OsCPS1 and OsKS1 (Fig. 1) because OsCPS1 and OsKS1 loss-of-function mutants display a severe dwarf phenotype caused by GA deficiency (Sakamoto et al., 2004). Thus, rice has two ent-CDP synthase genes, OsCPS1 for GA biosynthesis and OsCPS2 for phytoalexin biosynthesis. Although rice possesses another ent-CDP synthase gene, OsCPS2, loss-of-function OsCPS1 mutants display a severe dwarf phenotype (Sakamoto et al., 2004). This finding indicates that OsCPS2 cannot prevent the OsCPS1 mutant dwarf phenotype and has no redundant biological function with OsCPS1. OsCPS2 expression is up-regulated, unlike OsCPS1, in response to biotic or abiotic stress, including UV irradiation, chitin elicitor treatment, jasmonic acid treatment, and pathogen attack (Otomo et al., 2004; Prisic et al., 2004; Sakamoto et al., 2004; Okada et al., 2007; Hasegawa et al., 2010). Thus, the positive correlation of OsCPS2 expression and phytoalexin accumulation strongly suggested that OsCPS2 is responsible for phytoalexin biosynthesis.
A difference in enzymatic properties between recombinant OsCPS1 and OsCPS2 was previously shown (Hayashi et al., 2008). However, these studies did not clearly explain the non-redundant function of OsCPS1 and OsCPS2. Therefore, herein the localization of these transcripts in rice plants and the subcellular localization of their translated products were compared in order to account for their non-redundancy. Consequently, it was found that transcripts of OsCPS1 and OsCPS2 are differentially localized in rice plants according to their biological roles, and a complementation experiment using an OsCPS1 mutant by ectopic expression of OsCPS2 was performed to verify the conclusion genetically.
Materials and methods
Plant materials
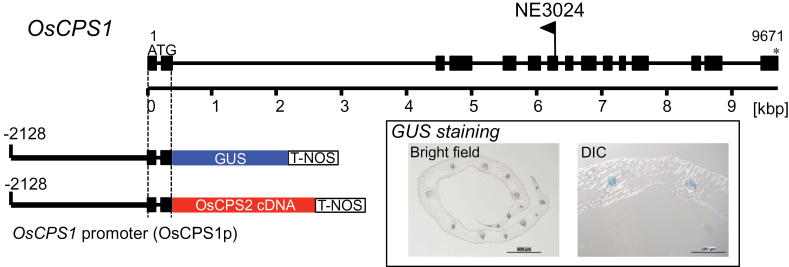
Rice (Oryza sativa L. cv. Nipponbare) seedlings were grown at 25 °C under continuous light conditions in a growth chamber until the third-leaf stage, and the upper and basal 2cm regions were excised from the second-leaf sheath and used for quantitative analyses of transcripts. The oscps1-1 mutant (Sakamoto et al., 2004) is a Tos17-inserted mutant NE3024 (Nipponbare background; Supplementary Fig. S1 available at JXB online), and its M1 seeds were purchased from the Rice Genome Resource Center, National Institute of Agrobiological Sciences. A heterozygous M1 plant was used for transformation.
Laser microdissection
The upper parts of the second-leaf sheaths of third-leaf stage rice seedlings were fixed in ethanol:acetic acid (3:1, v/v). Paraffin embedding and laser microdissection were performed as previously described (Takahashi et al., 2010). In brief, 16 μm thin sections were cut from paraffin blocks and mounted on PEN membrane class slides (Life Technologies Corporation, CA, USA) for laser microdissection. The vascular bundle-rich and epidermis-rich tissues were collected from the leaf sheath sections using a Veritas Laser Microdissection System LCC1704 (Life Technologies Corporation). In addition, mesophyll-rich tissues were collected.
RNA extraction and quantitative reverse transcription–PCR (qRT–PCR)
Total RNA was extracted and purified from the leaf sheath samples using an RNAqueous kit (Invitrogen, Carlsbad, CA, USA), and cDNA templates were synthesized from total RNA using a QuantiTect Reverse Transcription kit (Qiagen KK, Tokyo, Japan). Total RNA was extracted from tissue samples prepared by laser microdissection, using a PicoPure RNA Isolation kit (Life Technologies Corporation) according to the manufacturer’s instructions. The quantity of RNA was determined by the Quant-iT RiboGreen RNA Assay Kit (Life Technologies Corporation). RNA integrity was assessed using an Agilent 2100 Bioanalyzer with an Agilent RNA 6000 Nano kit (Agilent Technologies, CA, USA). cDNA templates were synthesized and amplified using a WT-Ovation RNA Amplification System version 1.0 (NuGEN Technologies, CA, USA). qRT–PCR was performed using a Thermal Cycler Dice Real Time System TP800 (Takara Bio, Otsu, Japan) and SYBR Premix Ex Taq Perfect Real Time version 2 (Takara Bio), as previously described (Toyomasu et al., 2008). The concentration of each transcript was normalized to 18S rRNA. Nucleotide sequences of the primers used are listed in Supplementary Table S1 at JXB online. The primer set for OsCPS1 can amplify a fragment derived from the wild-type OsCPS1 mRNA but not a fragment from Tos17-inserted OsCPS1 mRNA (Supplementary Fig. S1).
Green fluorescent protein (GFP) assay
The cDNA fragments encoding the N-terminal 153 and 108 amino acids of OsCPS1 and OsCPS2, respectively (OsCPS1-N153 and OsCPS2-N108, Supplementary Fig. S2A at JXB online), were amplified by RT–PCR using the Advantage 2 PCR System (Takara Bio) and subcloned into the pGEM-T Easy vector (Promega, WI, USA). Primer sets (sense/antisense) for amplification of OsCPS1-N153 and OsCPS2-N108 fragments are XbaI-CPS1-F/BamHI-CPS1-N153-R and XbaI-CPS2-F/BamHI-CPS2-N108-R, respectively (Supplementary Table S2). A fragment encoding the chimeric peptide OsCPS1-N91:OsCPS2-N108 (Supplementary Fig. S2B) was prepared by fusion of two corresponding cDNA fragments. Fragments encoding the N-terminal 91 amino acids of OsCPS1 (OsCPS1-N91) and OsCPS2-N108 were amplified using KOD Plus version 2 (Toyobo, Osaka, Japan) and the primer sets XbaI-CPS1-F/CPS1N91CPS2-R and CPS1N91CPS2-F/BamHI-CPS2-N108-R, respectively (Supplementary Table S2). The OsCPS1-N91 fragment was fused to the 5’ terminus of the OsCPS2-N108 fragment using an In-Fusion HD cloning kit (Takara Bio) and subcloned into the pGEM-T Easy vector. Each insert was ligated into a pTH121 vector (Zhu et al., 1997) using the XbaI and BamHI sites. Each constructed plasmid was introduced into second-leaf sheaths of 1-week-old rice seedlings using the PDS-1000 He Biolistic Particle Delivery System (Bio-Rad, CA, USA). Following bombardment, the cells were incubated on solid Murashige and Skoog (MS) medium for 24h at 28 °C in the dark before observation. Microscopic observation was performed with an Olympus AX80T microscope (Olympus, Tokyo, Japan).
β-Glucuronidase (GUS) assay
The GUS cDNA and NOS terminator originating in the pBI221 vector were inserted into the binary vector pZH2B (Kuroda et al., 2010) using BamHI and EcoRI sites (pZH2B-GUS-Nos; Supplementary Fig. S3 at JXB online). The genomic DNA fragment of 2.1kb 5’ upstream of the OsCPS1 ATG start site plus the coding sequence in the second exon of OsCPS1 (OsCPS1p) was amplified by PCR using KOD Plus version 2 and primer set IF-AscI-CPS1p-F/IF-BamHI-CPS1p-R (Supplementary Table S3), and directly subcloned into pZH2B-GUS-Nos, which was digested with AscI and BamHI, using an In-Fusion HD cloning kit (Supplementary Fig. S3). The plasmid pZH2B-OsCPS1p::GUS was introduced into Nipponbare rice cells through Agrobacterium transfection following the method previously described (Kuroda et al., 2010). Transgenic rice was grown and used for GUS assay. GUS assay was performed as previously described (Yamaguchi et al., 2001), using an Axioplan 2 microscope system (Zeiss Japan, Tokyo, Japan).
Complementation experiments
The NOS terminator originating in the pBI221 vector was inserted into pZH2B using SacI and EcoRI sites (pZH2B-Nos; Supplementary Fig. S3 at JXB online). The full-length open reading frame (ORF) cDNA of OsCPS2 was amplified by RT–PCR using the primer set KpnI-CPS2-F and KpnI-CPS2-R (Supplementary Table S) and subcloned into the pZH2B-Nos vector using the KpnI site (Supplementary Fig. S3). The OsCPS1p fragment was cleaved from the pZH2B-OsCPS1p::GUS plasmid and ligated into pZH2B-OsCPS2 using AscI and SmaI sites (pZH2B-OsCPS1p::OsCPS2; Supplementary Fig. S3). A heterozygous M1 plant of oscps1-1 was selected as a transformation host by PCR genotyping using MightyAmp DNA polymerase (Takara Bio) and the primer sets CPS1-WT-F/CPS1-WT-R (~230bp) for the wild-type gene and Tos17-F/CPS1-WT-R (~500bp) for the mutant gene (Supplementary Fig. S1). pZH2B-OsCPS1p::OsCPS2 was introduced into the heterozygous rice plant by Agrobacterium infection (Kuroda et al., 2010), and T1 seeds of 12 transgenic lines were obtained. The T1 seedlings were grown and subjected to genotyping. The host OsCPS1 genotype was confirmed by PCR using the above primer sets, and the transgene OsCPS2 cDNA was detected by PCR using the primer set CPS2-QRT-F/CPS2-QRT-R (Supplementary Table S1), ~250bp from the endogenous gene (with intron) and ~170bp from the transgene cDNA (no intron). The targeted-genotype plants (homozygous oscps1-1 mutants with OsCPS2 cDNA and segregated wild-type plants without OsCPS2 cDNA) were grown and T2 seeds were obtained for further analyses. The upper and basal parts of T2 seedlings were collected and used for gene expression analysis, after genotyping of each seedling by PCR, as described above.
Results
Subcellular localization of OsCPS1 and OsCPS2
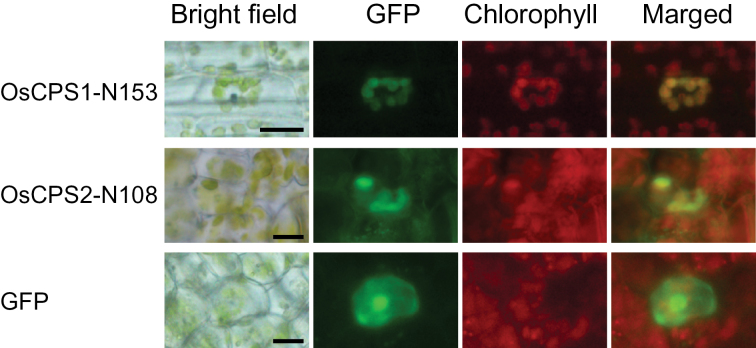
Diterpene cyclases, including ent-CDP synthase and ent-kaurene synthase, in higher plants have been considered to localize in the plastid (Toyomasu and Sassa, 2010). In general, transit peptides for plastid targeting are present at the N-termini of the plant diterpene cyclases (Toyomasu and Sassa, 2010), and Arabidopsis CPS, which is responsible for GA biosynthesis, has been shown to be transported into the plastid (Sun and Kamiya, 1994). OsCPS1 (867 amino acids, LOC_Os02g17780) and OsCPS2 (800 amino acids, LOC_Os02g36210) also have transit peptide-like sequences at their N-termini (Otomo et al., 2004; Prisic et al., 2004). To verify the plastid localization of OsCPS1 and OsCPS2, the N-terminal transit peptide-like sequences OsCPS1-N153 and OsCPS2-N108 (Supplementary Fig. S2A at JXB online) were fused to GFP and expressed in rice mesophyll cells. GFP fluorescence in plastids indicated that both OsCPS1-N153–GFP and OsCPS2-N108–GFP were transported into the plastids (Fig. 2). These results suggest that OsCPS2 as well as GA biosynthetic OsCPS1 localize in the rice cell plastid.
Fig. 2.
Subcellular localization of OsCPS1 and OsCPS2. The N-terminal region including the transit peptide of each CPS fused to GFP at its N-terminus was expressed in rice mesophyll cells, and GFP fluorescence was observed by fluorescence microscopy, as described in the Materials and methods. OsCPS1-N153 and OsCPS2-N108 indicate the N-terminal 153 and 108 amino acids of OsCPS1 and OsCPS2, respectively. Scale bars indicate 10 μm.
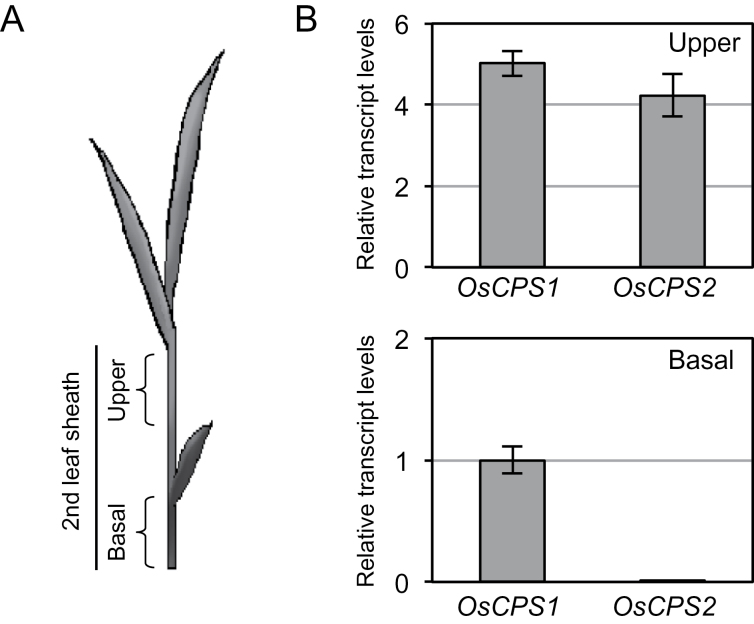
Expression patterns of OsCPS1 and OsCPS2 in the second-leaf sheath of rice seedlings
The data suggested the same subcellular localization of OsCPS1 and OsCPS2 in the rice cell. Next, expression analysis of OsCPS1 and OsCPS2 was performed to compare the localization of these transcripts in rice plants. The second-leaf sheath grows sensitively in response to exogenously applied GA; therefore, it has been used in bioassays to assess GA activity (Murakami, 1968; Nishijima and Katsura, 1989). The second-leaf sheath was used here as material for qRT–PCR analysis. The upper and basal 2cm were excised from second-leaf sheaths of third-leaf stage rice seedlings (Fig. 3A) and used for RNA extraction. The qRT–PCR analysis showed that the OsCPS2 transcript level was much lower than that of OsCPS1 in the basal part including the meristem tissues, whereas OsCPS2 expression was almost similar to OsCPS1 expression in the upper part (Fig. 3B).
Fig. 3.
Localization of transcripts of OsCPS1 and OsCPS2 in the second-leaf sheath of rice. (A) The upper and basal parts of the rice second-leaf sheath were used for expression analyses by qRT–PCR. (B) Relative transcript levels of two CPS genes. The concentration of each transcript was normalized to that of 18S rRNA, and adjusted to a value of 1 for OsCPS1 in the basal parts with SEs (n=3).
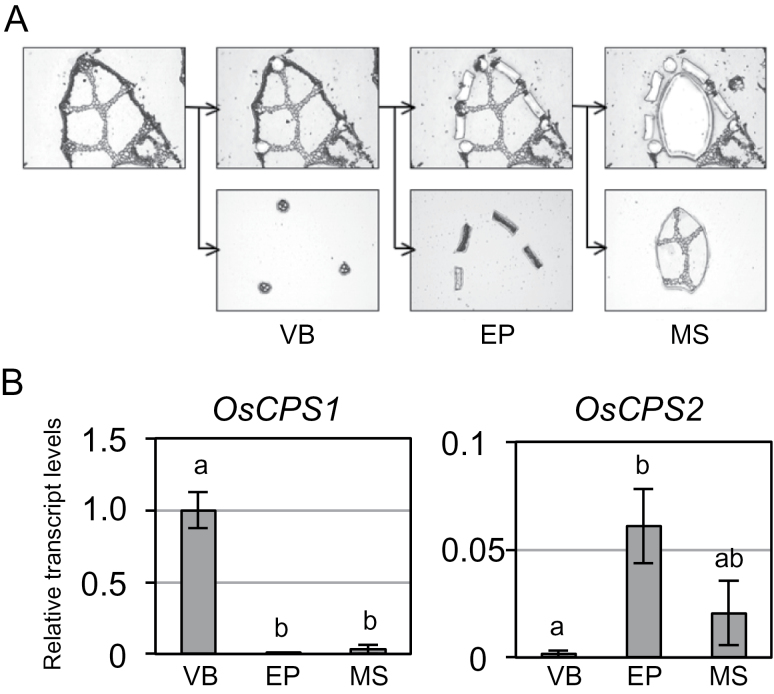
Furthermore, the transcript contents were quantified in separate tissues from the upper part of the second-leaf sheath in which the transcript contents of the two OsCPS genes were almost similar. Vascular bundle-rich, epidermis-rich, and residual tissues (mesophyll-rich tissues) were excised from leaf sheath slices, collected by laser microdissection (Fig. 4A), and used for RNA extraction. qRT–PCR was performed using amplified cDNAs derived from RNA obtained as a template. OsCPS1 transcripts were significantly more abundant in vascular bundle-rich tissues than in other tissues, whereas OsCPS2 transcript levels were more abundant in epidermis-rich and mesophyll-rich tissues than in vascular bundle-rich tissues (Fig. 4B). Expression of the GA biosynthetic gene AtCPS, which encodes ent-CDP synthase in Arabidopsis, is observed around vascular tissues (Silverstone et al., 1997; Yamaguchi et al., 2001), similar to OsCPS1, but unlike OsCPS2.
Fig. 4.
Localization of transcripts of OsCPS1 and OsCPS2 to different tissues in the upper part of the second-leaf sheath of rice. (A) Tissue samples collected by laser microdissection. Vascular bundle-rich (VB), epidermal-rich (EP), and mesophyll-rich (MS) tissues were collected from segments of the upper parts of the second-leaf sheaths of rice. (B) Relative transcript levels of two rice CPS genes in VB, EP, and MS tissues. Expression analysis was performed by qRT–PCR. The concentration of each transcript was normalized to that of 18S rRNA and adjusted to a value of 1 for OsCPS1 in VB with SEs (n=3). Data were subjected to analysis of variance (ANOVA) in each graph.
Complementation of the severe dwarf phenotype of the OsCPS1 mutant by OsCPS2 expression under the OsCPS1 promoter
A complementation experiment with a loss-of-function OsCPS1 mutant by OsCPS2 under the regulation of the OsCPS1 promoter was performed to confirm genetically that the OsCPS1 and OsCPS2 expression sites are different in rice. First a suitable DNA region of the OsCPS1 promoter wase identified using a GUS reporter gene. A genomic DNA fragment of 2.1kb 5′ upstream of the OsCPS1 ATG start site plus the coding sequence in the second exon of OsCPS1 (OsCPS1p) was amplified using PCR and fused to GUS (Fig. 5) in the pZH2B binary vector (Kuroda et al., 2010). It has been shown previously that the first 1–2 introns of AtCPS are required for proper expression in Arabidopsis (Silverstone et al., 1997). GUS staining was observed in the vascular bundle of the second-leaf sheath of rice seedlings (Fig. 5; Supplementary Fig. S4 at JXB online). The close correspondence of GUS staining to the accumulation pattern of OsCPS1 transcripts by tissue-specific qRT–PCR (Fig. 4B) suggested that OsCPS1p promoter activity was effective. When the OsCPS1p fragment is fused to the full-length OsCPS2 ORF cDNA in-frame, the resulting translated product is OsCPS2, which has the OsCPS1-N91 at its N-terminus (Supplementary Fig. S2B). Both OsCPS1-N91 and a chimeric peptide OsCPS1-N91:OsCPS2-N108 led GFP to the plastid (Supplementary Fig. S5), similar to OsCPS2-N108 (Fig. 2). These results suggest that a pseudo-mature form of OsCPS2 is transported into plastids when driven by OsCPS1p.
Fig. 5.
Gene structure of OsCPS1 and its promoter region used in the complementation experiment. Filled boxes represent exons and lines represent introns of OsCPS1 from the translation start codon to the stop codon. The flag shows the insertion site of Tos17 in the oscps1-1 mutant (NE3024). The 2.1kb region of the OsCPS1 promoter (OsCPS1p) used for GUS assay and ectopic expression of OsCPS2 is shown. The inset shows images of the sliced second-leaf sheath of GUS-stained rice T1 seedlings into which OsCPS1p::GUS was introduced. Bars in the left panel (bright field) and right panel (differential interference contrast microscopy, DIC) indicate 500 μm and 200 μm, respectively. (This figure is available in colour at JXB online.)
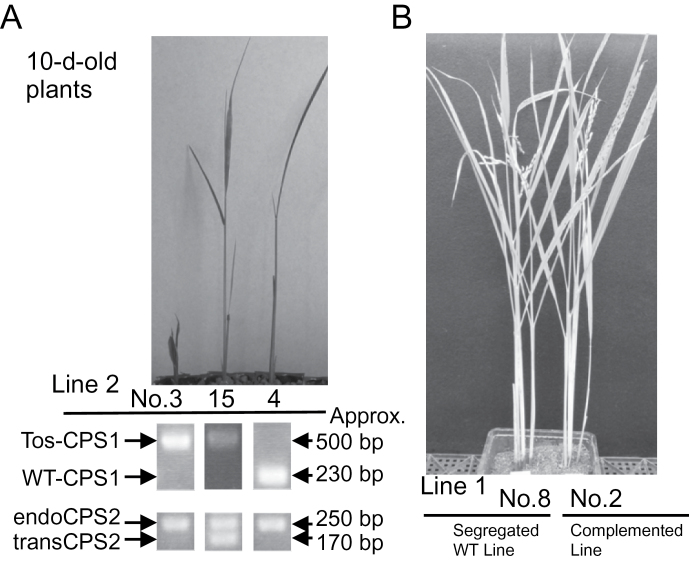
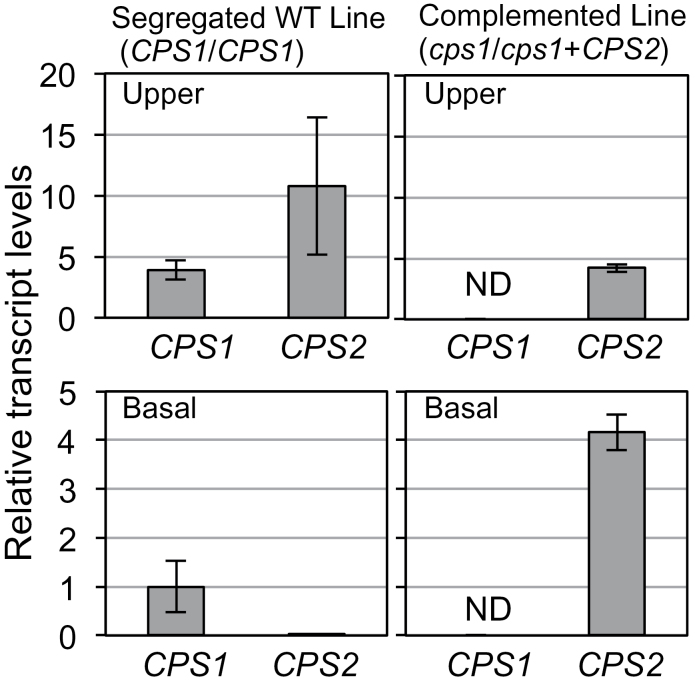
The OsCPS1p fragment was accordingly fused to OsCPS2 ORF cDNA (Fig. 5) in the pZH2B vector and introduced into a heterozygous oscps1-1 mutant by Agrobacterium infection. oscps1-1 is a Tos17-inserted OsCPS1 mutant that displays a severe dwarf phenotype caused by GA deficiency (Sakamoto et al., 2004). Because the homozygous oscps1-1 mutant was incapable of dedifferentiation, a heterozygous mutant was transformed by Agrobacterium infection, and self-pollination of transgenic plants produced T1 seeds. Transgenic homozygous oscps1-1 (complemented line), non-transgenic homozygous oscps1-1 (knockout line), and non-transgenic wild-type (segregated wild-type line) plants were observed by genotyping the T1 seedlings (Fig. 6A). Four complemented line T1 plants were identified, and displayed almost the same height as that of the segregated wild-type lines (Fig. 6B). OsCPS1 and OsCPS2 expression patterns were analysed in the second-leaf sheath to check successful ectopic expression of OsCPS2. qRT–PCR showed that OsCPS2 transcripts accumulated in basal parts of the second-leaf sheath of T2 complemented line seedlings, similar to OsCPS1 transcripts in wild-type Nipponbare (Fig. 3B) and in segregated wild-type line seedlings, whereas wild-type OsCPS1 transcripts were not detected in complemented line plants (Fig. 7). These results indicate that OsCPS2 expression under the OsCPS1 promoter rescued the oscps1-1 mutant phenotype.
Fig. 6.
Complementation experiment of a loss-of-function OsCPS1 mutant by ectopic expression of OsCPS2. T1 seeds were obtained from several lines of T0 plants, transgenic heterozygous oscps1-1 (NE3024) plants into which OsCPS1p::OsCPS2 cDNA (Fig. 5) was introduced. (A) Results of genotyping and images of T1 seedlings of line 2, the knockout line (no. 3), complemented line (no. 15), and the segregated wild-type line (no. 4). Tos-CPS1, bands from Tos17-inserted OsCPS1 genome DNA (Tos17-F and CPS1-WT-R in Supplementary Fig. S1 available at JXB online); WT-CPS1, bands from wild-type OsCPS1 genomic DNA (CPS1-WT-F and CPS1-WT-R in Supplementary Fig. S1); endoCPS2, bands from native OsCPS2 genomic DNA (with intron); transCPS2, bands from transgene OsCPS2 cDNA (no intron). The sense primer (CPS2-QPCR-F; Supplementary Table S) and antisense primer (CPS2-QPCR-R; Supplementary Table S1) for OsCPS2 genotyping were designed from the nucleotide sequences of exons 10 and 11, respectively. (B) Images of adult T1 rice plants of line 1, the segregated wild-type line (no. 8), and the complemented line (no. 2).
Fig. 7.
Expression patterns of OsCPS1 and OsCPS2 in second-leaf sheaths of the complemented line rice seedlings. The upper and basal parts of the second-leaf sheath (Fig. 3A) of T2 seedlings, obtained from the segregated wild-type T1 line (line 1, no. 8) and the complemented T1 line (line 1, no. 2) rice plants, were used for expression analyses by qRT–PCR. The concentration of each transcript was normalized to that of 18S rRNA, and adjusted to a value of 1 for OsCPS1 in the basal parts of the segregated wild-type line plants with SEs (n=3). ND, not detected.
Discussion
Phytohormone GAs are biosynthesized from GGDP, a common precursor of diterpenoids, through several steps catalysed by various biosynthetic enzymes including diterpene cyclases in plastids, P450 monooxygenases in the endoplasmic reticulum, and 2-oxoglutarate-dependent dioxygenases in the cytoplasm (Yamaguchi, 2008; Hedden and Thomas, 2012). The sites of expression of genes encoding the soluble dioxygenases GA 20-oxidase and GA 3-oxidase, which are responsible for direct synthesis of physiologically active GAs, have been characterized in rice previously (Kaneko et al., 2003), whereas the sites of expression of diterpene cyclase genes, including OsCPS1 and OsKS1, involved in the initial step of bioactive GA biosynthesis have not been elucidated. The diterpene cyclases responsible for biosynthesis of ent-kaurene, a GA biosynthetic intermediate, have been identified not only in plants but also in bacteria (Morrone et al., 2009) and fungi (Kawaide et al., 1997; Toyomasu et al., 2000), both of which produce GAs as specialized metabolites. Although two distinct cyclases successively convert GGDP into ent-kaurene via ent-CDP in higher plants, one peptide bi-functional diterpene cyclase produces ent-kaurene from GGDP in bryophytes (Hayashi et al., 2006; Kawaide et al., 2011). Coniferous gymnosperms also have ent-CDP synthase and ent-kaurene synthase for GA biosynthesis, while they have bi-functional labdane-related diterpene cyclases involved in specialized metabolites, resin acids produced via (+)-CDP (Keeling et al., 2010; Zerbe et al., 2012). Synthesis of ent-CDP is the first branch point of GA biosynthesis from GGDP, but ent-CDP and ent-kaurene may not be specific intermediates of only GAs. For example, Stevia rebaudiana produces steviol glycosides via ent-kaurene. In S. rebaudiana, ent-kaurene is synthesized by highly accumulated ent-CDP synthase and ent-kaurene synthase, both of which are responsible for GA biosynthesis, in leaf parenchyma (Richman et al., 1999). In addition, rice produces labdane-related phytoalexins converted from ent-CDP; phytocassanes A–E and oryzalexins A–F. Rice has an ent-CDP synthase, OsCPS2, specific for phytoalexin biosynthesis as well as OsCPS1 specific for GA biosynthesis (Otomo et al., 2004; Prisic et al., 2004), whereas S. rebaudiana uses ent-CDP synthase for GA biosynthesis to synthesize steviol glycosides.
Here it is shown that transcript levels of OsCPS2 were drastically lower than those of OsCPS1 in the basal parts of rice seedling second-leaf sheath. The basal parts include the meristematic tissues necessary for rice growth. It has been shown that growth signals mediated by GAs engage in antagonistic cross-talk with defence signals mediated by jasmonic acid via the DELLA–JAZ interaction (Hou et al., 2013). DELLA and JAZ are key repressors in GA and jasmonic acid signalling, respectively. Therefore, expression of the defence gene OsCPS2 might be suppressed in the basal parts with high growth activity, although it has been unclear whether cross-talk of these hormones is involved in the OsCPS2 regulation in the basal parts of rice seedlings. Furthermore, qRT–PCR analysis using separate tissues prepared by laser microdissection indicated that OsCPS1 transcripts mainly accumulated in vascular bundle-rich tissues, whereas OsCPS2 transcripts were mainly observed in epidermis-rich tissues. Thus, the different localization of OsCPS1 and OsCPS2 transcripts in rice plants was found.On the other hand, it was suggested that both OsCPS2 and OsCPS1 function in the plastid. It was previously shown that GAs are biosynthesized mainly from GGDP that is derived through the methylerythritol 4-phosphate (MEP) pathway in the plastid (Kasahara et al., 2002). Transcript levels of several genes in rice responsible for the MEP pathway are drastically up-regulated after elicitor treatment, suggesting that the MEP pathway also participates in diterpene phytoalexin biosynthesis in rice (Okada et al., 2007). Therefore, it is reasonable that OsCPS2 functions in the plastid like GA biosynthetic OsCPS1.
Although recombinant OsCPS2 converts GGDP to ent-CDP in vitro, OsCPS2 cannot rescue loss-of-function OsCPS1 mutants under control of the native promoter (Sakamoto et al., 2004). In the present study, it was shown that OsCPS2 under regulation by the OsCPS1 promoter complemented the OsCPS1 mutant phenotype. The qRT–PCR results showed differences in tissue-specific expression of the two CPS genes. These results strongly suggest that proper tissue-specific expression of ent-CDP synthase genes is critical for GA biosynthesis. Results of a GUS reporter gene assay in germinating Arabidopsis seeds suggest that AtCPS transcripts for GA biosynthesis localize in vascular tissues, although no signal was detected by in situ hybridization (Silverstone et al., 1997). Another study showed that transcripts of dioxygenases, responsible for later steps of GA biosynthesis, localize mainly in the cortex and endodermis, and promoter-swapping experiments indicate that intercellular transport of the biosynthetic intermediate ent-kaurene is required to produce bioactive GAs (Yamaguchi et al., 2001). An SrCPS signal involved in stevioside biosynthesis in S. rebaudiana was detected in mesophyll by in situ hybridization, whereas its signal for GA biosynthesis in vascular tissues was not detected using this method (Richman et al., 1999), similar to AtCPS (Silverstone et al., 1997). It has been suggested that a spatially different localization of CPS transcripts separates stevioside biosynthesis from GA biosynthesis in S. rebaudiana. In any case, CPS expression patterns associated with GA biosynthesis are considered well conserved among higher plant species. However, it is reasonable that transcription of stress-inducible OsCPS2 mainly occurs near the epidermis, producing phytoalexins in response to environmental stress. The present results confirm that the first step in GA biosynthesis occurs in the vascular tissues for effective production of bioactive GAs. The monocots wheat and maize not only have the ent-CDP synthase genes TaCPS3 and An1, which are responsible for GA biosynthesis, but also have stress-inducible ent-CDP synthase genes TaCPS1 and An2 (Harris et al., 2007; Wu et al., 2012). Similar to rice, TaCPS1 and An2 may be expressed in different tissues from those of the GA biosynthetic genes TaCPS3 and An1.
It was shown herein that OsCPS1 and OsCPS2 transcripts were differentially localized in rice plants according to their distinct biological roles, one for growth and the other for defence, although these translated products have the same enzymatic activity in vitro. It is concluded that OsCPS2 contributes little to GA biosynthesis and cannot prevent the GA-deficient dwarf phenotype of loss-of-function OsCPS1 mutants because of different localization of OsCPS2 transcripts from OsCPS1 transcripts.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Primer design for transcript analyses and genotyping
Figure S2. Transit peptide-like sequences of OsCPSs
Figure S3. Construction of plasmids for introducing the transgene
Figure S4. GUS staining of rice seedlings.
Figure S5. Subcellular localization of GFP fused to OsCPS1-N91 and OsCPS1-N91:OsCPS2-N108 at its N-terminus
Table S1. Sequences of primers used for qRT–PCR.
Table S2. Sequences of primers used for GFP experiments.
Table S3. Sequences of primers used for complementation experiments.
Acknowledgements
This work was supported in part by JSPS KAKENHI grant nos 17580093 and 24580155. We thank Dr Eiji Nambara (Toronto University) for insightful discussions and comments on the manuscript.
Glossary
Abbreviations:
- CDP
copalyl diphosphate
- GA
gibberellin
- GFP
green fluorescent protein
- GGDP
geranylgeranyl diphosphate
- GUS
β-glucuronidase
- MEP
methylerythritol 4-phosphate
- qRT–PCR
quantitative reverse transcription–PCR.
References
- Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends in Plant Science 17, 73–90. [DOI] [PubMed] [Google Scholar]
- Akatsuka T, Takahashi N, Kodama O, Sekido H, Kono Y, Takeuchi S. 1985. Novel phytoalexins (Oryzalexins A, B and C) isolated from rice blast leaves infected with Pricularia oryzae. Part 1: isolation, characterization and biological activities of oryzalexins. Biosciences, Biotechnology, and Biochemistry 49, 1689–1694. [Google Scholar]
- Cartwright DW, Langcake PW, Pryce RJ, Leworthy DP, Ride JP. 1981. Isolation and characterization of two phytoalexins from rice as momilactones A and B. Phytochemistry 20, 535–537. [Google Scholar]
- Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ. 2007. The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Molecular Biology 59, 881–894. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Mitsuhara I, Seo S, Imai T, Koga J, Okada K, Yamane H, Ohashi Y. 2010. Phytoalexin accumulation in the interaction between rice and the blast fungus. Molecular Plant-Microbe Interactions 23, 1000–1011. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Kawaide H, Notomi M, Sakigi Y, Matsui A, Nozaki H. 2006. Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens . FEBS Letters 580, 6175–6181. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Toyomasu T, Hirose Y, Onodera Y, Mitsuhashi W, Yamane H, Sassa T, Dairi T. 2008. Comparison of the enzymatic properties of ent-copalyl diphosphate synthases in the biosynthesis of phytoalexins and gibberellins in rice. Biosciences, Biotechnology, and Biochemistry 72, 523–530. [DOI] [PubMed] [Google Scholar]
- Hedden P, Thomas SG. 2012. Gibberellin biosynthesis and its regulation. Biochemical Journal 444, 11–25. [DOI] [PubMed] [Google Scholar]
- Hou X, Ding L, Yu H. 2013. Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Reports 32, 1067–1074. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Itoh H, Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Ashikari M, Matsuoka M. 2003. Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? The Plant Journal 35, 104–115. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Hanada A, Kuzuyama T, Takagi M, Kamiya Y, Yamaguchi S. 2002. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of gibberellins in Arabidopsis . Journal of Biological Chemistry 277, 45188–45194. [DOI] [PubMed] [Google Scholar]
- Kato T, Kabuto C, Sasaki N, Tsunagawa M, Aizawa H, Fujita K, Kato Y, Kitahara Y, Takahashi N. 1973. Momilactones, growth inhibitors from rice, Oryza sativa L. Tetrahedron Letters 14, 3861–3864. [Google Scholar]
- Kato H, Kodama O, Akatsuka T. 1993. Oryzalexin E, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry 33, 79–81. [Google Scholar]
- Kato H, Kodama O, Akatsuka T. 1994. Oryzalexin F, a diterpene phytoalexin from UV-irradiated rice leaves. Phytochemistry 36, 299–301. [Google Scholar]
- Kawaide H, Hayashi K, Kawanabe R, Sakigi Y, Matsuo A, Natsume M, Nozaki H. 2011. Identification of the single amino acid involved in quenching the ent-kauranyl cation by a water molecule in ent-kaurene synthase of Physcomitrella patens . FEBS Journal 278, 123–133. [DOI] [PubMed] [Google Scholar]
- Kawaide H, Imai R, Sassa T, Kamiya Y. 1997. ent-Kaurene synthase from the fungus Phaeosphaeria sp. L487: cDNA isolation, characterization, and bacterial expression of a bifunctional diterpene cyclase in fungal gibberellin biosynthesis. Journal of Biological Chemistry 272, 21706–21712. [DOI] [PubMed] [Google Scholar]
- Keeling CI, Dullat HK, Yuen M, Ralph SG, Jancsik S, Bohlmann J. 2010. Identification and functional characterization of monofunctional ent-copalyl diphosphate and ent-kaurene synthases in white spruce reveal different patterns for diterpene synthase evolution for primary and secondary metabolism in gymnosperms. Plant Physiology 152, 1197–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga J, Ogawa N, Yamauchi T, Kikuchi M, Ogasawara N, Shimura M. 1997. Functional moiety for the antifungal activity of phytocassane E, a diterpene phytoalexin from rice. Phytochemistry 44, 249–253. [Google Scholar]
- Koga J, Shimura M, Oshima K, Ogawa N, Yamauchi T, Ogasawara N. 1995. Phytocassanes A, B, C and D, novel diterpene phytoalexins from rice, Oryza sativa L. Tetrahedron 51, 7907–7918. [Google Scholar]
- Kuroda M, Kimizu M, Mikami C. 2010. A simple set of plasmids for the production of transgenic plants. Biosciences, Biotechnology, and Biochemistry 74, 2348–2351. [DOI] [PubMed] [Google Scholar]
- Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ. 2009. Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum . FEBS Letters 583, 475–480. [DOI] [PubMed] [Google Scholar]
- Murakami Y. 1968. The microdrop method, a new rice seedling test for gibberellins and its use for testing extracts of rice and morning glory. Botanical Magazine, Tokyo 79, 33–43. [Google Scholar]
- Nishijima T, Katsura N. 1989. A modified micro-drop bioassay using dwarf rice for detection of femtomol quantities of gibberellins. Plant and Cell Physiology 30, 623–627. [Google Scholar]
- Okada A, Shimizu T, Okada K, Kuzuyama T, Koga J, Shibuya N, Nojiri H, Yamane H. 2007. Elicitor induced activation of the methylerythritol phosphate pathway toward phytoalexins biosynthesis in rice. Plant Molecular Biology 65, 117–187. [DOI] [PubMed] [Google Scholar]
- Otomo K, Kenmoku H, Oikawa H, König WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T. 2004. Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. The Plant Journal 39, 886–893. [DOI] [PubMed] [Google Scholar]
- Peters RJ. 2006. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry 67, 2307–2317. [DOI] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ. 2004. Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiology 136, 4227–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman AS, Gijzen M, Starratt AN, Yang Z, Brandle JE. 1999. Diterpene synthesis in Stevia rebaudiana: recruitment and up-regulation of key enzymes from the gibberellin biosynthetic pathway. The Plant Journal 19, 411–421. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, et al. 2004. An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiology 134, 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Chang C-W, Krol E, Sun T-P. 1997. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana . The Plant Journal 12, 9–19. [DOI] [PubMed] [Google Scholar]
- Sun T-P, Kamiya Y. 1994. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthase A of gibberellin biosynthesis. The Plant Cell 6, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kamakura H, Sato Y, Shiono K, Abiko T, Tsutsumi N, Nagamura Y, Nishizawa NK, Nakazono M. 2010. A method for obtaining high quality RNA from paraffin sections of plant tissues by laser microdissection. Journal of Plant Research 123, 807–813. [DOI] [PubMed] [Google Scholar]
- Tamogami S, Mitani M. 1993. Oryzalexin S structure: a new stemarane-type rice plant phytoalexin and its biogenesis. Tetrahedron 49, 2025–2032. [Google Scholar]
- Toyomasu T. 2008. Recent advances regarding diterpene cyclase genes in higher plants and fungi. Biosciences, Biotechnology, and Biochemistry 72, 1168–1175. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kagahara T, Okada K, Koga J, Hasegawa M, Mitsuhashi W, Sassa T, Yamane H. 2008. Diterpene phytoalexins are biosynthesized in and exuded from the roots of rice seedlings. Biosciences, Biotechnology, and Biochemistry 72, 562–567. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Kawaide H, Ishizaki A, Shinoda S, Otsuka M, Mitsuhashi W, Sassa T. 2000. Cloning of a full-length cDNA encoding ent-kaurene synthase from Gibberella fujikuroi: functional analysis of a bifunctional deterpene cyclase. Biosciences, Biotechnology, and Biochemistry 64, 660–664. [DOI] [PubMed] [Google Scholar]
- Toyomasu T, Sassa T. 2010. Diterpenes. In: Mander L, Lui H-W, eds. Comprehensive natural products II. Chemistry and biology , Vol. 1 Oxford: Elsevier, 643–672. [Google Scholar]
- Wu Y, Zhou K, Toyomasu T, et al. 2012. Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane-related diterpenoid metabolism in the cereals. Phytochemistry 84, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima A, Mori K. 2000. Synthesis and absolute configuration of (–)-phytocassane D, a diterpene phytoalexin isolated from the rice plant, Oryza sativa . European Journal of Organic Chemistry 2000, 4079–4091. [Google Scholar]
- Yamaguchi S. 2008. Gibberellin metabolism and its regulation. Annual Review of Plant Biology 59, 225–251. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kamiya Y, Sun T-P. 2001. Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. The Plant Journal 28, 443–453. [DOI] [PubMed] [Google Scholar]
- Zerbe P, Chiang A, Bohlmann J. 2012. Mutational analysis of white spruce (Picea glauca) ent-kaurene synthase (PgKS) reveals common and distinct mechanisms of conifer diterpene synthases of general and specialized metabolism. Phytochemistry 74, 30–39. [DOI] [PubMed] [Google Scholar]
- Zhu XF, Suzuki K, Saito T, Okada K, Tanaka K, Nakagawa T, Matsuda H, Kawamukai M. 1997. Geranylgeranyl pyrophosphate synthase encoded by the newly isolated gene GGPS6 from Arabidopsis thaliana is localized in mitochondria. Plant Molecular Biology 35, 331–341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.