Summary
AUXIN BINDING PROTEIN1 (ABP1) mutants have properties of auxin- and red light-signalling mutants. A novel concept for growth control by ABP1 and phytochromes is indicated by this functional link.
Key words: AUXIN-BINDING PROTEIN1 (ABP1), abp1 mutants, early auxin-regulated genes, early light-regulated genes, gravitropism, phototropism, phytochrome, hypocotyl elongation, shade avoidance, Arabidopsis thaliana.
Abstract
The function of the extracytoplasmic AUXIN-BINDING-PROTEIN1 (ABP1) is largely enigmatic. We complemented a homozygous T-DNA insertion null mutant of ABP1 in Arabidopsis thaliana Wassilewskia with three mutated and one wild-type (wt) ABP1 cDNA, all tagged C-terminally with a strepII–FLAG tag upstream the KDEL signal. Based on in silico modelling, the abp1 mutants were predicted to have altered geometries of the auxin binding pocket and calculated auxin binding energies lower than the wt. Phenotypes linked to auxin transport were compromised in these three complemented abp1 mutants. Red light effects, such as elongation of hypocotyls in constant red (R) and far-red (FR) light, in white light supplemented by FR light simulating shade, and inhibition of gravitropism by R or FR, were all compromised in the complemented lines. Using auxin- or light-induced expression of marker genes, we showed that auxin-induced expression was delayed already after 10min, and light-induced expression within 60min, even though TIR1/AFB or phyB are thought to act as receptors relevant for gene expression regulation. The expression of marker genes in seedlings responding to both auxin and shade showed that for both stimuli regulation of marker gene expression was altered after 10–20min in the wild type and phyB mutant. The rapidity of expression responses provides a framework for the mechanics of functional interaction of ABP1 and phyB to trigger interwoven signalling pathways.
Introduction
For many years the function(s) of AUXIN BINDING PROTEIN1 (ABP1) remained enigmatic. In earlier work, ABP1 functions were associated with the plasma membrane (Napier, 1995). Besides regulation of K+ channel activity and membrane potential, protein kinase activity, phospholipase A activity, calcium influx, and other very rapid responses were described, which all are too rapid to be initiated by transcription and protein biosynthesis. Instead, post-translational mechanisms are suggested to initiate these rapid responses. For these ABP1 is thought to function as an auxin receptor (Scherer, 2011).
A conditional ABP1 mutant was created by expressing an antibody against ABP1 in the apoplast which suppressed ABP1 functions like leaf expansion, endomitosis, cell division, and cell expansion (David et al., 2007; Braun et al., 2008; Paque et al., 2014), results verified with an inducible mutant (Jones et al., 1998; Chen et al., 2001a ). The only known T-DNA insertion mutant of this gene proved to be embryo-lethal (Chen et al., 2001b). The point mutation abp1-5, obtained by TILLING, was useful to uncover the interaction of ABP1, PIN proteins, and ROP/RIC signalling in protein trafficking (Robert et al., 2010; Xu et al., 2010). More detailed investigations using the heterozygous ABP1/abp1 T-DNA insertion line revealed that functions like auxin-induced gene expression, phototropism and gravitropism, and auxin transport are defective in this mutant (Effendi et al., 2011; Effendi and Scherer, 2011). Recently ABP1 has been linked to red light physiology, using ABP1/abp1 and abp1-5 (Effendi et al., 2013), and to control of TIR1 activity (Effendi et al., 2011; Tromas et al., 2013).
Both ABP1/abp1 and abp1-5 have weak phenotypes so that progress in ABP1 research based on these mutants is still limited. On the other hand, the embryo lethality of a homozygous T-DNA insertion plant (Chen et al., 2001b) opened up the possibility to complement this plant not only with wild-type but also with point-mutated cDNAs. We describe here such a series of mutants based on complementation of the knock-out plant that show more severe auxin-related phenotypes than previous abp1 mutants. These results reveal that not only auxin but also phytochrome signalling is compromised in these lines.
Material and methods
Quantum chemical modelling
A theoretical examination of the geometry, electronic structure, and electronic binding energies (∆E) of the auxin binding pocket of ABP1 were performed. The structural data was obtained from the crystal structure of ABP1 (Protein Data Bank with accession codes 1LRH). The pocket containing the 1-NAA molecule and the surrounding amino acids at 6 Angstroms (~400 atoms) was isolated (amino acids: I22, L24, W44, Q46, I48, T54, P55, H57, H59, E63, F65, H106, V108, V121, I130, L132, F149, W151). The geometric structure of the wild-type pocket was optimized taking into account previous analysis of auxin molecules (Ferro et al., 2006) and protein cavities (Rolo-Naranjo, et al., 2010). The optimization was carried out using Density Function Theory (DFT) using the b3-lyp function (Becke, 1988, Lee et al., 1988, Stephens et al., 1994) including the Van der Waals correction D3 (Grimme et al., 2010) and atomic basis sets at triple zeta level (def-TZVP) (Eichkorn et al., 1997). The input geometry constrained 17 atoms in order to conserve the pocket structure and the start charge of the pocket was 2+ owing to the influence of Zn2+. All calculations have been performed with the program package TURBOMOLE (http://www.turbomole.com).
Different computational chemistry experiments were conducted to analyse the influence of mutations of the amino acids at positions 25, 54, 106, and 151 and the substitution of IAA in the position of 1-NAA. The substitution (mutants) H106 to N106, L25 to Y25, T54 to I54 were modelled and their geometries re-optimized at DFT level with b-lyp and the D3-correction. The re-optimizations included both pocket–auxin pairs and pockets alone to investigate ∆E. The ∆E energies were calculated by a single point calculation with b3-lyp and TZVP basis set following the equation: ∆Ebind=∆Epocket – aux – (∆Epocket + ∆Eaux) comparing each mutant with the wild type. The calculations solve the electronic problem accurately and, neglecting changes of pressure and volume in the cell, we hold that the electronic energy and the enthalpy are approximately equal (∆E=∆H). Our calculation will not allow for entropic processes according to the ∆G. For further analysis of the potential surface and electric field of the pocket we used the theory of deformed atoms in molecules (DAM; Rico et al., 2004) as well as the comparison of electronic features using quantum similarity measures (Ferro et al., 2006), applied now at pocket level using the auto values ZAA(Ω)=∫ρA(r)Ω(r)ρA(r)dr, where the operator at Ω were Coulomb and Overlap. This analysis offers details about the differences of the electronic features of each pocket.
Plant material and growth conditions
Arabidopsis thaliana Wassilevskija (Ws) heterozygous wt plants containing a T-DNA insertion and kanamycin resistance were used for transformation. ABP1 cDNA containing FLAG-tag and strep-tag II directly before the C-terminal KDEL under control of the 35S promoter was provided by T. Reinard (University of Hannover).This construct was then cloned into pENTR D-TOPO (Invitrogen) where site-directed mutation was performed using QuikChange™ Site-Directed Mutagenesis Kit (Stratagen). Entry vectors were cloned into destination vector pB2GW7 (basta resistance: Karimi et al., 2002). The complete ABP1 cDNA sequences in the vectors were sequenced after transformation into Agrobacterium and the designed mutations verified (MWG-Biotech AG Eurofins Genomics, Ebersberg, info-eu@eurofins.com). Confirmed vectors were used to transform Arabidopsis thaliana heterozygous ABP1/abp1 plants (Chen et al., 2001b). Progenies of the transformed plants were selected on agar plates containing kanamycin (50 µg/ml) and BASTA (30 µg/ml). Surviving seedlings were PCR genotyped to identify homozygous null ABP1 wt plants (primer list: see Supplementary Table S2). Double homozygous lines were selected from these.
Seedling experiments were performed on sterile 1% (w/v) agar (growth experiments), 0.5% (w/v) gelrite to stabilize tropism experiments (Santner and Watson, 2006), or liquid (seedlings for RNA extraction) half-strength Murashige and Skoog (MS) medium containing 1% (w/v) sucrose at 22 °C for 10 d or as otherwise indicated (Figs 2 and 3). Experiments were repeated two to three times independently (n=75–90).
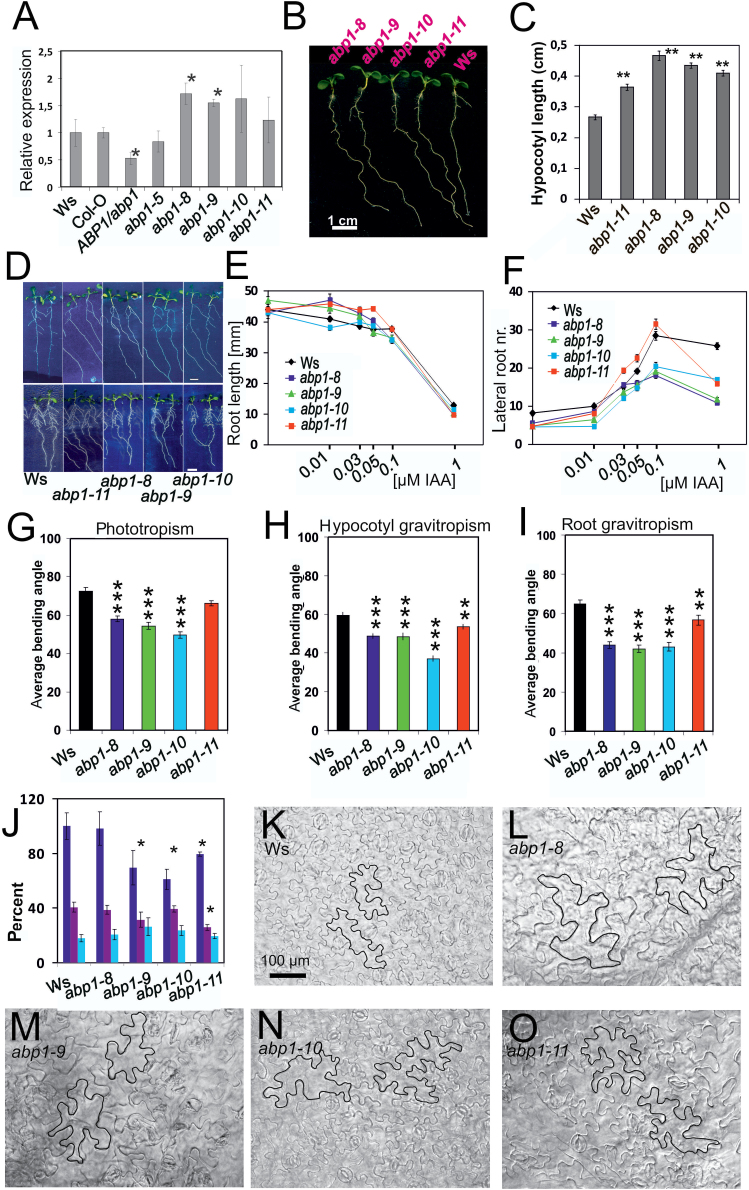
Fig. 2.
Developmental physiology of abp1 mutant seedlings. (A) Level of ABP1 transcript in abp1 mutants in comparison to the respective wild types ABP1 (abp1-5 is in Col-O, all others in Ws). Value for wt ABP1 is set as 1 (asterisk: different by P<0.05). (B) Representative light-grown seedlings (7 d; Ws, abp1 mutants). Bar=5mm. (C) Hypocotyl length of light-grown seedlings (14 d) (n=23–30; SEM, P<0.01). (D) Root development without auxin (upper row) and in the presence of 0.1 µM IAA (lower row) (n=30–39; SEM). (E) Auxin sensitivity of primary root length and (F) and lateral root number (n=30–39; SEM). Error bars are either visible or smaller than symbols. Non-overlapping symbols or error bars in E and F were significantly different from each other (P<0.01 or lower). (G) Delayed phototropic responses of hypocotyls of dark-grown (3 d) seedlings. Phototropism was induced by lateral blue light (10 µmol m–1 s–2) for 8h (SEM, n=70–130). (H) Delayed gravitropic responses in hypocotyls of 3-day-old dark-grown seedlings after 24h tilting by 90° (n=63–140). (I) Delayed gravitropic responses of roots of dark-grown seedlings (3 d) after 24h tilting by 90° (n=52–90). (G–I; average bending angles±SEM * P<0.05; ** P<0.01; *** P<0.001). (J) Acropetal auxin transport in roots of 4-day-old light-grown seedlings. Dark blue bars: 5–10mm from tip; purple bars: 10–15mm from tip; light blue bars: 15–20mm from tip. (n=40; *: P<0.05). (K–O) Epidermal pattern of primary leaves. (K) Wassilewskia wt; (L) abp1-8; (M) abp1-9; (N) abp1-10; (O) abp1-11. In each photo two cells are outlined for comparison. Bars=100µm. Cell areas and lobe numbers of epidermis cells are presented in Supplementary Fig. S2.
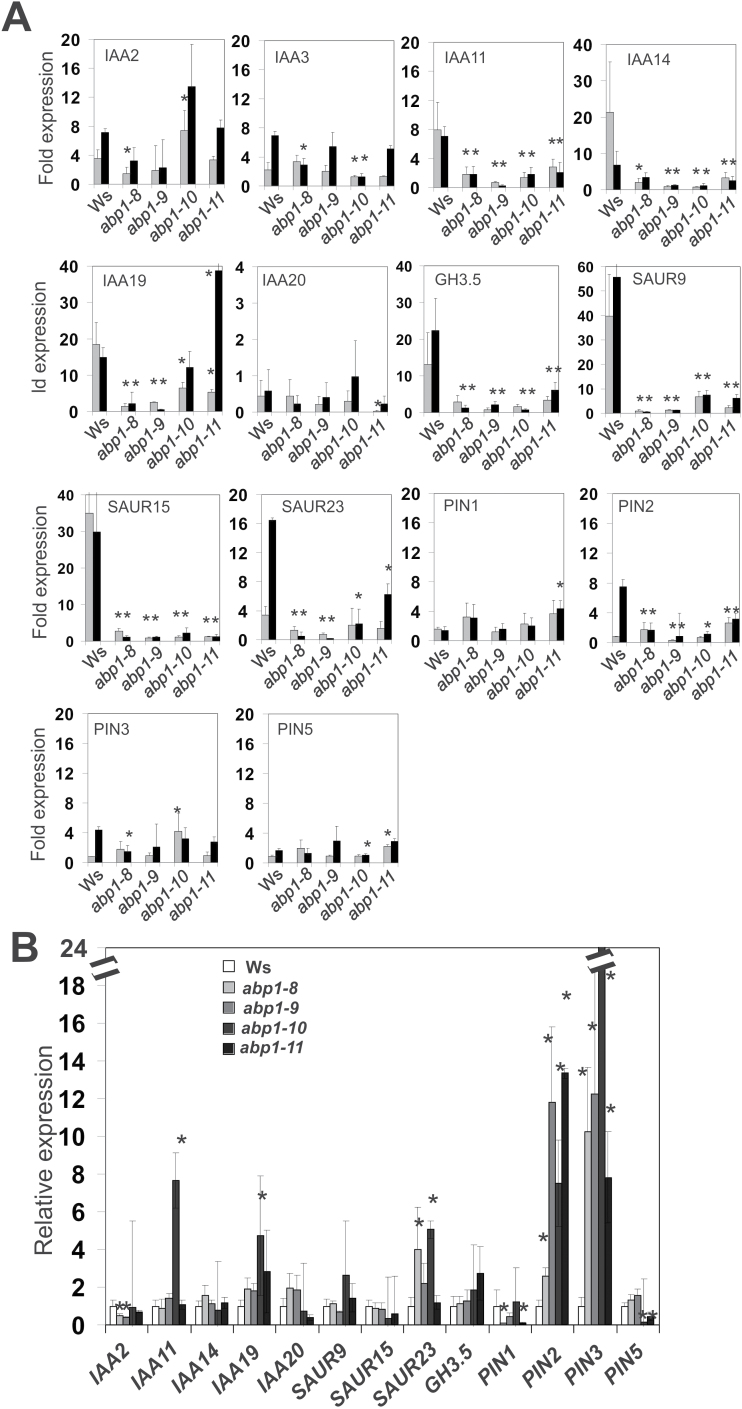
Fig. 3.
Expression of early auxin genes and several PIN genes in 14-day-old light-grown seedlings. Seedlings were either treated with 10 µM IAA in (A) (grey bars: 10min, black bars: 30min) or mock in (B). qRT-PCRs were from three biological replicates with three technical replicates for each gene. Statistical analysis was performed as described by Livak and Schmittgen (2001) and verified using the method of Pfaffl et al. (2002). At t=0min fold expression was set as 1 for the wt in (B). Asterisks indicate significant difference to the wt (* P<0.05; ** P<0.01).
Auxin sensitivity was repeated twice by transferring light-grown seedlings at day 4 to media containing increasing IAA concentrations (0–10 µM) or mock for further growth for 6 days (Figs 2D–F; 5L, M). Hypocotyl lengths in light-grown or auxin-treated seedlings were calculated by subtracting the lengths obtained without auxin (Fig. 5L, M). Dark-grown seedlings were pre-grown in liquid half-strength MS for two days without auxin (Fig. 5L). Auxin was added and the increments of hypocotyl lengths after 12h were determined. Basipetal auxin transport was measured according to Lewis and Muday (2009). Radioactive auxin was applied to the root tip and segments cut after 8h (5–10mm, 10–15mm, and 15–20mm from tip) and counted after 18h. The 5–10mm segment in the wt was set as 100% and others calculated accordingly (Fig. 2J).
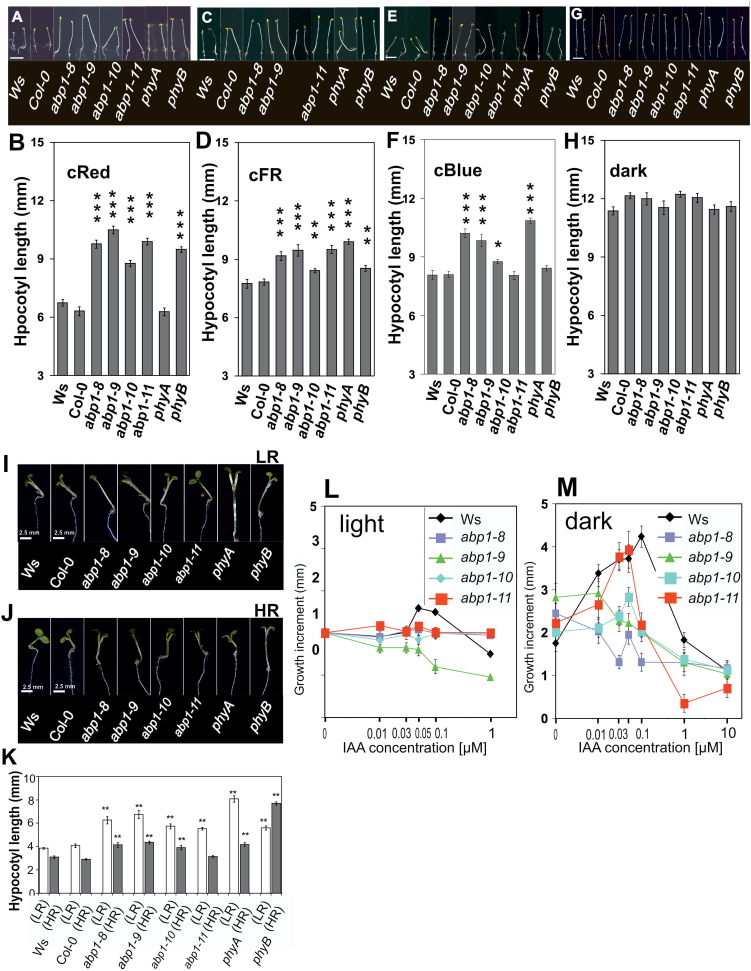
Fig. 5.
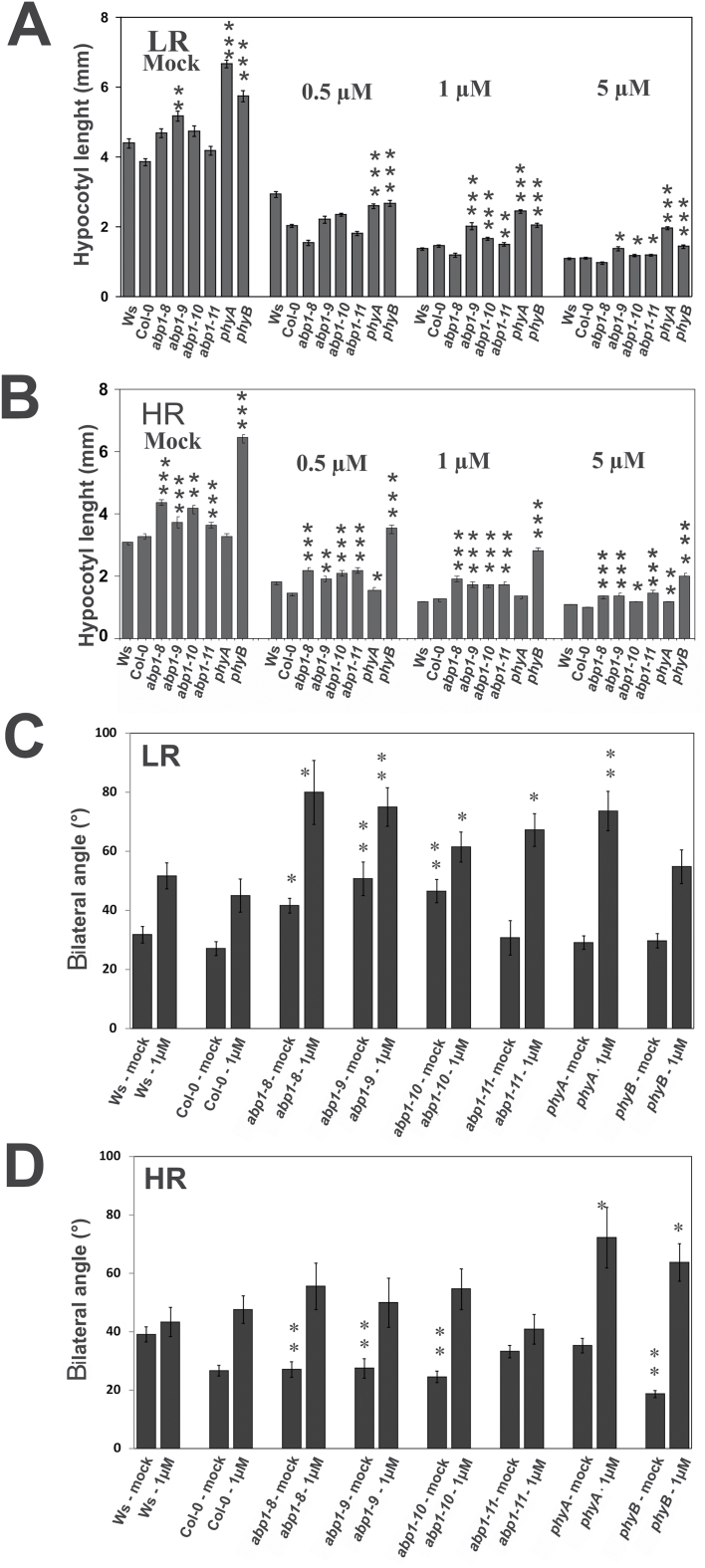
Elongation in monochromatic continuous light (R, FR, B) of 4 d-old seedlings in Ws and abp1 mutants, phyA and phyB. (A, C, E, G) Representative images of seedlings grown in FR, R, B (0.1 µmol m–1 s–2 each) or dark, respectively. Bar=5mm. (B, D, F, H) Hypocotyl lengths (n>80; SEM). (I–K) Responses of hypocotyls of 3-d-old seedlings grown in W and then transferred for three more days to W with added low ratio R:FR (FR-enriched light) and high ratio R:FR (R-enriched light) in Ws wt, abp1 mutants, and phyA and phyB. (I, J) Representative images of seedlings. (K) Hypocotyl lengths. (n>120; SEM). (B, D, F, H, K) Shown by asterisks is significance between the wt and mutants (*P<0.05; ** P<0.01; *** P<0.001). When error bars do not overlap values of different bars in one graph are significantly different from each other (P<0.05)). (L, M) Hypocotyl elongation in light (L) and dark (M) in the presence of increasing auxin concentrations. (L) Hypocotyl length increment induced by auxin of light-grown seedlings (see also Fig. 2A). (n=20–30, SEM). (M) Seedlings were grown in dark for 2 d and the length increment during subsequent 12h was recorded (n=20–30, SEM). Error bars are either visible or smaller than symbols. Data in L and M are significantly different between control and auxin-treated seedlings for each line when symbols or error bars do not overlap.
Plants were cultured in soil on a growth chamber at 22 °C constant 8h/16h (light/dark; SD) on peat-based compost soil (Einheitserde, http://www.einheitserde.de/) containing 30% silica sand. Leaves were measured from the three largest leaves from each of 60 adult plants per genotype. Rosette leaf number at 59 d and flowering time was obtained in two independent replications (30 plants each). Flowering time (first flower with white petals) for each genotype was recorded. Apical dominance at 90–92 days was measured as the number of branches at the bottom of fully grown plants with 100 plants each grown in SD (Fig. 4H).
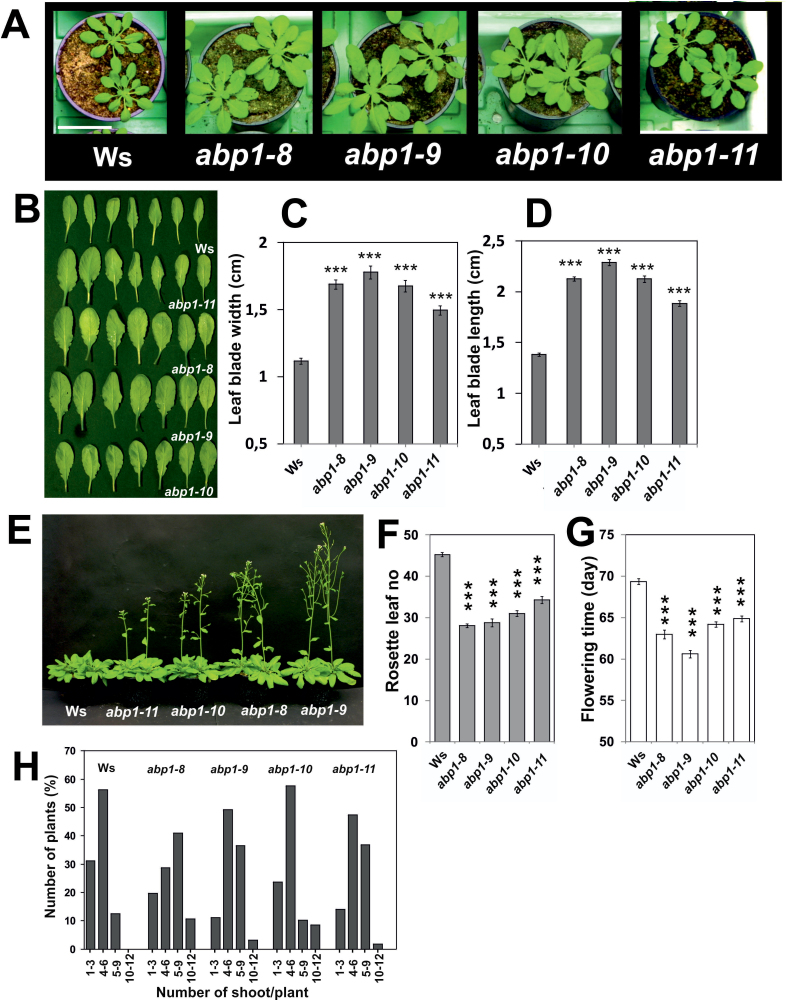
Fig. 4.
Phenotypes of abp1 mutants grown in short days (8h/16h light/dark). (A) Representative images of 37-day-old rosettes of Ws and abp1 mutants (abp1-8, abp1-9, abp1-10, abp1-11). Bar=5cm. (B) Representative images of leaves of plants shown in (A). (C) Leaf blade width and (D) blade length measured from 59-d-old plants (n=132–190; SEM; P<0.01). (E) Flowering plants at day 59 (n=30). (F) Rosette leaf number at flowering date (n=30). (G) Flowering time. Values (F, G) are means with SEM (P<0.001). Shown by asterisks is significance between the wt and mutants. (H) Apical dominance. Branches at the bottom were counted from 100 plants each at 90–92 d. [C, D, F, G: when error bars do not overlap values are significantly different from each other (P<0.05 or lower)].
For seedling light experiments, seeds were stratified for 4 d, plates were placed in horizontal position at 22 °C under white light (W) for 2h before transfer for 1 d into darkness. Then they were kept for 3 d either in constant R, FR, or B (0.1 µmol m–1 s–2 or 1 µmol m–1 s–2) or dark (Fig. 5A–K). For shade avoidance experiments, seeds on plates prepared like as were exposed to 24.5 µmol m–1 s–2 constant white LED light for 3 d. Then to W either low R/FR ratio (0.098) or high R/FR ratio (2.1) was added for 3 d (spectra: see Effendi et al., 2013). For RNA extraction, seedlings received the low red (LR) or high red (HR) treatment for 1h after 3 d in W (Fig. 7; Fig. 8). In experiments with NPA (naphthylpthalamic acid) this was added to the plates from the start of the experiment (Fig. 6). Data were obtained from three independent replications and each replication was consisted of more than 40 seedlings. Light experiments were done without sucrose in the medium in an LED chamber (CLF, Plant Climatics) (Effendi et al., 2013) (Fig. 5). All quantifications were done by scanning the plates with CanonScan 8800F (resolution of 600 dots per inch; Canon, http://www.canon-europe.com) and evaluating lengths or angles with AXIOVISIOLE version 4.6 software (Zeiss, http://www.zeiss.com/) and analysed using the t-test in Excel.
Fig. 7.
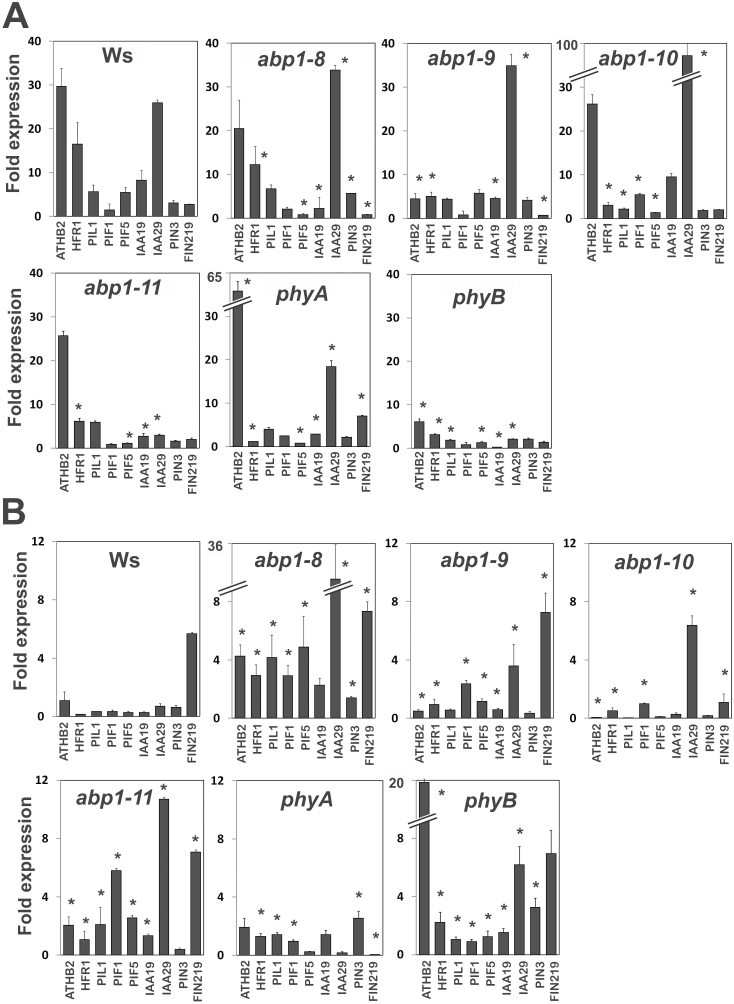
Expression response of shade-induced genes to 1h (A) low ratio R:FR (FR-enriched light) or (B) high ratio R:FR (R-enriched light). Seedlings were grown for 3 d in white light. qPCR data were obtained from at least three biological replications with three technical replications for each gene target. Statistical analysis was as described (Livak and Schmittgen, 2001; Pfaffl et al., 2002). At t=0min fold expression was set as 1 for each genotype (not on the graph). Values are means with SEM (* P<0.05).
Fig. 8.
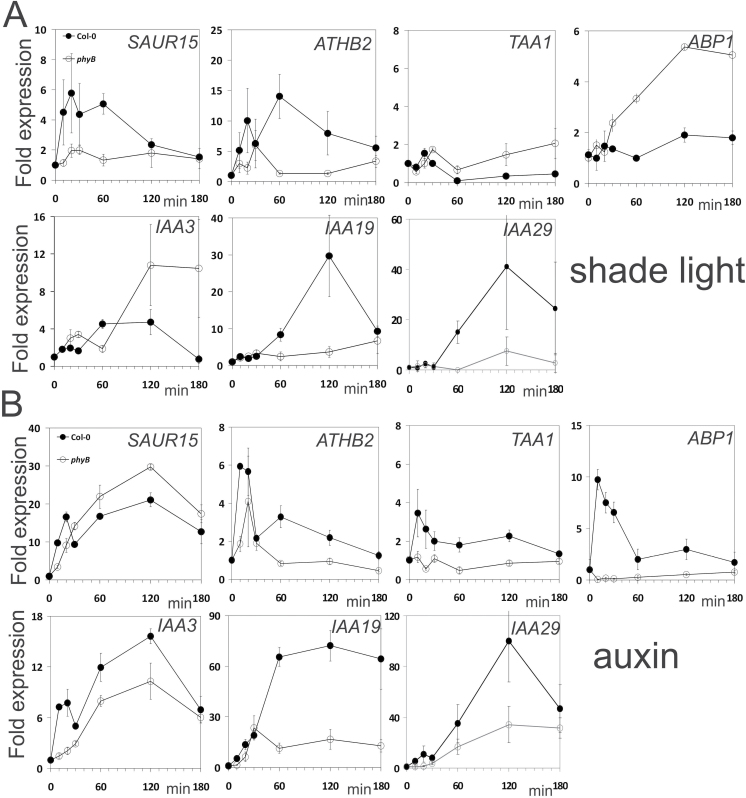
Differential co-regulation of expression of auxin- and light-induced genes in phyB and Col wt seedlings. Seedlings were grown for 3 d in w light and then treated in (A) with additional FR in LR light or (B) treated with 10 µM IAA for the times indicated in the graphs. Black symbols: Col; white symbols: phyB. qPCR data were obtained from two biological replications with three technical replications for each gene target. Statistical analysis was as described (Livak and Schmittgen, 2001; Pfaffl et al., 2002). At t=0min fold expression was set as 1 for each genotype. Values within one graph are significantly different when error bars or symbols do not overlap (P<0.05).
Fig. 6.
Effect of NPA on elongation and gravitropism in LR or HR. Seedlings were grown for 3 d in W and then for 3 d in low ratio R:FR + W (FR-enriched light) and high ratio R:FR (R-enriched light) on agar without and with indicated NPA concentrations. (A, B) Hypocotyl lengths. (C, D) Absolute values of hypocotyl angles as deviating from the plumb line. Asterisks indicate significances between the wt and mutants: * P<0.05; ** P<0.01; *** P<0.001 mutants (n=35 for each genotype; SEM). When error bars of different bars within one experiment do not overlap values are significantly different from each other within one panel.
Nucleic acid analysis
Seedlings were grown on half-strength MS liquid media in W for 14 d for auxin treatment (for light treatments see above). Seedlings were then equilibrated for 2h in fresh half-strength MS liquid media and then 10 µM IAA or mock was added. qPCR and statistics were performed as described (Livak and Schmittgen, 2001; Pfaffl et al., 2002; Effendi et al. 2013). Test gene and reference gene primers are listed in Supplementary Table S2.
Results
Modified auxin binding binding box in ABP1
We designed and developed new Arabidopsis abp1 mutants by transforming the kanamycin resistant ABP1/abp1 mutant (Chen et al., 2001b) with wild type (wt) ABP1 cDNA or ABP1 cDNA containing point mutations in the auxin-binding site of ABP1 (Woo et al., 2002) (Fig. 1) using Basta selection. A strep II tag and a FLAG tag were inserted immediately upstream of the C-terminal ER retention motif KDEL. We were able to isolate four stable abp1 mutants, abp1-8 (T54>I54), abp1-9 (L25>Y25), abp1-10 (H106>N106), and abp1-11 (no point mutation but tagged) in the background of the homozygous T-DNA insertion null mutant. The isolation showed that doubly resistant transformed T1 seedlings could be obtained and selected. Other lines did not propagate or produced very few doubly resistant plant progeny. From progeny of the mutant lines we theoretically expected one in four plants to have no wt ABP1 owing to homozygosity of the T-DNA insertion, but we needed to genotype 500–700 individuals until we found the desired mutant, still heterozygous for the basta marker. Selfing then gave lines homozygous for the basta marker. Owing to the difficulty in producing suitable lines we chose to consider those four orthologous lines as a set rather than isolating several lines of each mutant.
Fig. 1.
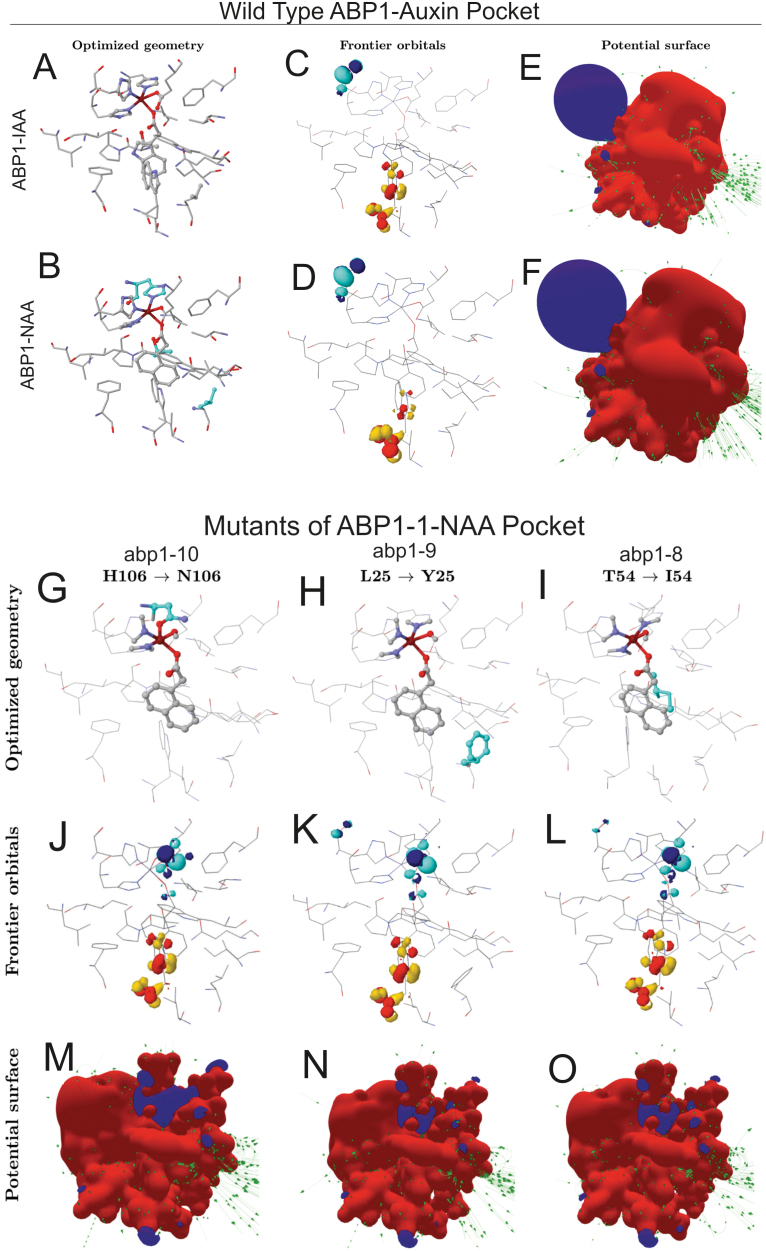
Modelling of changes in the geometry and electronic structure of the ABP1 pocket resulting from substitutions in the amino acid sequence. (A–F) The panels of the wild type ABP1-auxin binding pocket show optimized geometry (A, B), frontier orbitals (C, D), and potential surfaces (E, F). Mutated amino acids are highlighted in blue in panel B and individually in panels G–I. Note that in mutants not only geometries are changed (G–I) but also the geometries of the highest occupied molecular orbital [HOMO (red-yellow)] on W151 and the lowest unoccupied molecular orbital [LUMO (blue-cyan)] on H59 of the Zn2+ complex (K–N).
Quantum chemical modelling
We concentrated the analysis on an accurate quantum chemical model using density functional theory (DFT) for describing geometry, chemical bonds, electronic properties, and electronic binding energies of the wild-type auxin-binding pocket of ABP1 and the three site-specific mutations. The optimized structures of all binding pockets (Fig. 1A, B, G–I) demonstrated that the coordination number of the Zn2+ atom was 5 with square-based pyramidal geometry (Alberts et al., 1998), one of which coordinates the carboxyl group of the auxin ligand.
The electron donor-acceptor regions or frontier molecular orbitals (Fig. 1D, J–L) of the wt auxin pocket (Fig. 1C, D) play an important role to determine the activity of auxin and auxin-like molecules (Ferro et al., 2006). The calculations of binding pocket geometry were complemented by visualizing the pocket surface (Fig. 1E, F). We determined the interaction to ligands by observing the frontier orbitals (HOMO, highest occupied molecular orbital; LUMO, lowest unoccupied molecular orbital; Fig. 1C, D). The electron donor W151 (wt) is plotted in yellow-red (HOMO) and the acceptor, concentrated around H59, is plotted in blue-cyan (LUMO) surrounding the Zn2+, and both are independent of the presence of 1-NAA or IAA. In the presence of either 1-NAA or IAA the localization of the frontier molecular orbitals is nearly identical and the surface pattern potentially exposed to ligand is strongly polarized (Fig. 1E, F). The region formed by the Zn2+ complex presents the negative potential (blue), and the remainder of the pocket is dominated by the positive potential (red) of other amino acids. Both results, the position of the frontier orbitals in the pocket and the polarized potential, are consistent. The green lines represent the electrostatic field lines or force produced by the atoms.
Calculations were done for the substitutions of the amino acids L25>Y25 (abp1-9), T54>I54 (abp1-8), and H106>N106 (abp1-10) in the wt structure of the auxin pocket of ABP1 (Fig. 1G–O). The polarization of the surface potential observed in the wt pocket was lost in the mutations (Fig. 1J–L). In addition, the HOMO–LUMO localization depicted (Fig. 1G–I) showed that every mutation changes the localization of the electron acceptor (LUMO) from the H59 to the E63 (Fig. 1J–L).
The changes in the Coulomb matrix have previously been connected with the biological activity of the auxin molecules and in binding specificity of auxin molecules (Ferro et al., 2006). To correlate the physiological properties of the mutants with modelling, we focussed on further quantum chemical calculations of dE, Overlap, and Coulomb matrices. The electronic binding energies suggest that the mutants H106>N106 [abp1-10: –37.51 dE (Kcal/mol)], and T54>I54 [abp1-8: –35.90 dE (Kcal/mol)] offer less stability for binding the auxin molecule. Two mutants, abp1-10 and abp1-8, also showed similar trends in the changes of the Overlap and Coulomb auto values (Supplementary Table S1) indicating similar binding properties. Though the calculated binding energies are similar in both the wt [–41.10 dE (Kcal/mol)] and abp1-9 [–42.14 dE (Kcal/mol)], differences in Overlap and Coulomb matrices and geometry will influence binding because the smaller L25 is replaced by the bulky Y25, increasing electronic interaction with auxin similar to W151 (Woo et al., 2002) but, at the same time, also restricting pocket space. Accordingly, the Coulomb auto value, representing the charge surface of the pocket for electrostatic interactions with the ligand, and the Overlap auto value, representing the electron density of the pocket surface, in abp1-9 showed the strongest differences to the wt of these two parameters (Supplementary Table S1). Combined with the effects of dE this indicates a decrease of function in abp1-9. The strep tag is already outside of the main part of the ABP1 and thus most probably does not interfere with auxin binding (see Supplementary Fig. S1).
Auxin-related functions are compromised and auxin sensitivity is lower in abp1 mutants
The mutants transcribed the mutated ABP1 genes at about 1–1.7-fold the level of the wt Ws (Fig. 2A). The ABP1/abp1 mutant (Ws) was transcribed only at about 50% wt level and the abp1-5 mutant (Col-0) at about 80% of the corresponding wt.
All four abp1 mutants had longer hypocotyls (Fig. 2B, C) than wt, although in abp1-11 this phenotype was modest. When abp1 mutants were grown on auxin, abp1-8, abp1-9, and abp1-11 roots were longer than wt roots. All other genotypes had lengths similar to the wt (Fig. 2D, E). In response to 0.03 µM or higher auxin, a clear decrease in lateral root number was found in abp1-8, abp1-9, and abp1-10, but not in abp1-11 (Fig. 2D, F). These data indicated lower auxin sensitivities for the three abp1 mutants.
Phototropic and gravitropic bending of hypocotyls and gravitropic bending of roots of abp1 mutants was slower than for wt and abp1-11 (Fig. 2G– I). Gravitropic bending of hypocotyls and roots of dark-grown abp1 mutants was delayed (Fig. 2H, I) and bending angles of hypocotyls and roots were clearly smaller (Fig. 2H, I). Acropetal auxin transport from shoot base to root tip was delayed in all mutants except abp1-8 (Fig. 2J).
Leaf cell growth and epidermal cell lobe numbers are ABP1-dependent (Xu et al., 2010). Epidermal cells were larger in abp1-8, abp1-9, and abp1-10 (Fig. 2K–N), but only weakly so in abp1-11 (Fig. 2O; Supplementary Fig. S2D). Suppression of epidermal cell lobes per cell area was most pronounced in abp1-8 and in abp1-9 (Fig. 2L, M; Supplementary Fig. 2S). From these data we conclude that the three abp1 site-directed mutants generally were less sensitive in their responses to auxin, or to responses involving auxin transport than wt. Such auxin-related properties were less prominent in abp1-11, which resembled more the wt, with the small phenotype possibly associated with the presence of the tag.
Early expression of auxin-induced genes in abp1 mutants is insensitive to auxin
We chose rapidly responding genes IAA2, IAA3, IAA11, IAA14, IAA19, IAA20, GH3-5, SAUR9, SAUR15, and SAUR23 and PIN genes PIN1, PIN2, PIN3, and PIN5 to test for the role of ABP1 on the control of gene expression (Effendi and Scherer, 2011; Effendi et al., 2011 2013 2014; Labusch et al. 2013). In the wt most marker genes were up-regulated after 10min of auxin application, whereas IAA3, IAA20, and the PIN genes were unchanged. After 30min wt expression of PIN2 and PIN3, but not PIN5 and IAA20 were also up-regulated (Fig. 3A).
Auxin-induced expression of the marker genes was delayed in all four abp1 mutants compared with the wt, with the exception of IAA2, IAA3, PIN1, PIN3, and PIN5. The greatest differences were found for the three SAUR genes, GH3-5, IAA14, and IAA19. Taken together, the delayed expression of auxin-induced marker genes clearly indicated insensitivity to auxin in abp1-8, abp1-9, and abp1-10 compared with wt. Delayed expression was generally small in abp1-11.
Morphology and flowering of adult plants
The ABP1/abp1 plants were in the Ws background (Chen et al., 2001b) where a deletion in phyD renders this gene non-functional (Aukermann et al., 1997). The lack of a phyD gene influences early flowering (Effendi et al., 2014). We observed that abp1 mutants had longer and wider leaf blades (Fig. 4A–D), which is reminiscent of phyA mutants when grown under identical conditions (Supplementary Fig. S3). Flowering was earliest in abp1-9 followed by abp1-8 and abp1-10 and finally the wt (Fig. 4E–G). In comparison to the wt abp1-11 flowered early, but later than the other abp1 mutants. In addition, we found decreased apical dominance in short days in abp1-8 and abp1-9 lines, relatively weak decreases in abp1-10 and abp1-11.
abp1 mutants have altered responses to continuous light and shade
We investigated the growth of abp1 mutants in continuous monochromatic FR, R, or blue (B) light and in darkness to test for the involvement of a photoreceptor (Fig. 5). In continuous R, all abp1 seedlings had significantly longer hypocotyls than wt seedlings (Fig. 5A, B), similar to phyB but not to phyA seedlings. In continuous FR, all abp1 mutants also displayed longer hypocotyls in comparison to wt seedlings, but were shorter than phyA seedlings (Fig. 5C, D). As hypocotyl elongation is inhibited by continuous FR in a fluence- and PHYA-dependent manner (Whitelam et al., 1993) the data indicate that abp1 mutants interfere with phyA-mediated responses. However, not all phyA deficiency responses in de-etiolated seedlings were observed in abp1 mutants because they had opened and expanded cotyledons and displayed no apical hook, both responses not found in phyA seedlings (Fig. 5C). Hypocotyl elongation of abp1 mutants displayed small differences under continuous B like phyA seedlings (Fig. 5E, F). Because of the small magnitude of B insensitivity of abp1 seedlings, similar to phyA seedlings, assigning B insensitivity to either compromised phyA function or to insensitivity of a B receptor was not possible (Fankhauser and Casal, 2004). A dark phenotype was not obvious (Fig. 5G, H). Qualitatively similar but quantitatively smaller results were obtained when plants were grown in 1 µm–1 s–2 monochromatic light (see Supplementary Fig. S4).
In white light (W) a decrease in the R/FR ratio is the main cue for plants to perceive the presence of neighbours as physiological shade. W supplemented by a low ratio R:FR (LR) leads to strong elongation. W with added high ratio R:FR (HR) represses elongation. Responses to shade depend mainly on a low phyB signalling input (Fankhauser and Casal, 2004). Hypocotyl lengths in LR and HR were analysed (Fig. 5I–K). Seedlings of abp1 mutants displayed significantly longer hypocotyls than wt under LR, whereas abp1-11 seedlings were only slightly longer (Fig. 5I, K). Surprisingly, abp1 mutants in HR also had hypocotyls longer than the wt, except abp1-11 (Fig. 5J, K). This indicated that the strong abp1 mutant alleles might be defective in phyB-mediated responses to physiological shade.
We tested the effect of auxin on light- and dark-grown seedlings. IAA applied in the light increased hypocotyl elongation slightly in the wt. In abp1-9 auxin inhibited slightly and in abp1-8, abp1-10, and abp1-11 it had no effect (Fig. 5L). However, exogenous IAA in 2-day-old dark-grown seedlings did stimulate elongation in wt and abp1-11, with an optimum at 0.05 µM IAA. This was not the case in abp1-8, abp1-9, or abp1-10 (Fig. 5M) suggesting that ABP1 was a receptor for growth in dark-grown tissue. Reduced growth repression in R (Fig. 5A, B) in the abp1 mutants was consistent with the observation that a fully functional ABP1 supports repression of growth in the light.
NPA as an indicator for interaction of ABP1 and phytochromes in shade-induced elongation and inhibition of hypocotyl gravitropism
The elongation response to shade includes regulation of polar auxin transport (Nagashima et al., 2008a /b). Therefore, we tested the influence of the polar transport inhibitor naphthylpthalamic acid (NPA) on elongation in LR and HR light (Fig. 6A, B). NPA inhibited elongation strongly at 0.5 µM. In LR, phyB and more so phyA plants were more resistant to NPA than the wt. This property was observed only in abp1-9 to some extent at 0.5 µM and 1 µM NPA so that an NPA insensitivity was not clearly indicated in abp1 mutants (Fig. 6A, B). In HR, phyB seedlings were clearly more resistant to NPA than all other genotypes (Fig. 6B).
We noticed that inhibition of hypocotyl gravitropism was increased by the combination of either LR or HR with NPA (Nagashima et al., 2008a /b). In LR abp1-8, abp1-9, abp1-10, but not abp1-11, grew less vertical compared with wt (Fig. 6C). This gravitropism inhibition also was the case in the presence of 1 µM NPA for all four abp1 mutants. In LR alone, phyA and phyB seedlings responded as the wt, but phyA seedlings in LR in the presence of NPA clearly displayed inhibited hypocotyl gravitropism so that the response of the abp1 mutants was more similar to phyA than to phyB seedlings.
In HR alone abp1 seedlings grew more upright than wt seedlings, similar to phyB seedlings. This is consistent with a compromised phyB signalling as found in low continuous R (Fig. 5A, B). In HR and added NPA the abp1 mutants showed a tendency towards inhibited gravitropism but this was not statistically significant as in phyA and phyB seedlings. LR and HR clearly inhibited gravitropism in abp1 mutants as in phyA and phyB seedlings (Fig. 6C, D) (Liscum and Hangarter, 1993; Robson and Smith, 1996). In abp1-11, R or FR inhibition of gravitropism was absent, in LR and in the presence of NPA inhibition was apparent. Taking all four sets of data together, inhibition of gravitropism was weakest in abp1-11 compared with the three abp1 mutants.
Expression of light-induced genes in abp1 mutants
We investigated expression of ten shade-induced genes (ATHB2, HFR1, PIL1, PIF1, PIF5, IAA19, IAA29, PIN3, FIN219; Fig. 7). References for primers are in Supplementary Table S2. We restricted FR or R light to a short induction period of 1h in W (Wang et al. 2011).
In LR, phytochrome mutants and all abp1 mutants clearly showed a different expression of shade-induced genes than wt. In phyA, being wt with respect to phyB, high induction of IAA29 similar to wt was found, and this was also found in abp1-8, abp1-9 and abp1-10 but not in phyB and abp1-11. HFR1 was repressed both in phyA, phyB and in the abp1 mutants. ATHB2 was de-repressed in phyA and abp1-8, abp1-10, and abp1-11, but not in phyB and abp1-9. In phyB all test genes were repressed in comparison to the wt. Overall, expression of shade marker genes in abp1 mutants was, in general, more similar to phyA than to phyB in LR.
In HR and with phyB eight out of nine genes tested were more strongly de-repressed than in the wt, whereas in phyA only five genes were de-repressed but generally less than in phyB. FIN219, a phyA-dependent gene (Wang et al., 2011), was repressed in phyA. In abp1 mutants several genes (4–8) were de-repressed, and therefore in HR they were clearly more similar to phyB than to phyA.
Expression of shade-induced and auxin-induced genes in phyB seedlings
The expression of a number of genes (IAA3, IAA19, IAA19, SAUR15, ATHB2, FIN219) is co-regulated by auxin and shade (Steindler et al., 1999; Devlin et al., 2003; Kunihiro et al., 2011; Leivar et al., 2012) and we included the auxin biosynthesis and shade-induced gene, TAA1 (Tao et al., 2008) and ABP1 (Effendi et al., 2011 2013) into the analysis. We showed that abp1 mutants misregulate expression of auxin-regulated genes after 10–30min (Fig. 3) and of light-regulated genes after 1h (Fig. 7). However, little is known whether phyB seedlings misregulate the expression of auxin-induced genes. Therefore, we investigated the kinetics of induction of these genes by auxin or shade in phyB seedlings (Fig. 8).
In the wt (Col), shade induced a rise in expression of SAUR15, ATHB2, IAA3, and IAA19 and weakly in IAA29 at 10–20min. The responses peaked at 1–2h with a tendency to decline at 3h (Fig. 8A). Expression of ABP1 and TAA1 stayed low in wt and phyB seedlings. In phyB seedlings, shade-induced expression of ABP1, TAA1, and IAA3 was higher than in wt seedlings suggesting that phyB represses these genes. All time courses were fast which indicates a rather direct signal pathway from phyB to its target genes.
We quantified auxin-induced expression in wt seedlings (Fig. 8B). As expected, all genes were induced, although TAA1 only at a low level. Again, expression started to rise at 10–20min, peaked and declined towards 3h. In phyB seedlings, expression of most markers was lower than in wt seedlings; only SAUR15 was higher in phyB than in wt and, surprisingly, expression of ABP1 was not elevated by auxin in phyB. Hence, the phyB mutant had a partially aberrant auxin physiology with respect to the expression of marker the genes used here.
Discussion
Genetic engineering of stable mutant alleles of ABP1 by complementation
There is only one ABP1 gene in the Arabidopsis genome (Chen et al., 2001b). The embryo lethality of the T-insertion mutant and the still largely unsuccessful attempts to search for other types of mutants (Braun et al., 2008; Robert et al., 2010; Xu et al., 2010; Effendi et al., 2011) prompted us to engineer strong point mutation alleles by complementation of the knockout to aid investigation of ABP1 functions.
The novel abp1 mutants are loss-of-function
Modelling of the mutated binding sites showed that the protein surface contacting 1-NAA is distorted in all mutants (Fig. 1 and Supplementary Table S1). The thermodynamic surface description of the binding pockets and calculated binding energies for the wt and the abp1 mutants provided an explanation for why we could obtain only a few mutant alleles and why all were loss-of-function mutants.
The mutated and the wt protein in all four complementation lines is tagged, but upstream of the ER retention signal KDEL. Alterations to KDEL and additions like GFP have substantial effects on the exocytosis/endocytosis balance (Robert et al., 2010; Wang et al., 2013). Thus, an important aspect of our lines is that they are characterized in a wide variety of experiments and abp1-11, expressing the tagged wt ABP1, is phenotypically similar to wt. Owing to the immense difficulties associated with isolation of these mutants we isolated only one line per genotype so we cannot fully exclude positional effects of the new cDNAs on the respective phenotypes of the four mutants.
The C-terminus and thus its tag protrudes from the protein (Woo et al., 2002) and therefore can be expected to negatively interfere in abp1-11 with the interaction to essential partners, such as with the four recently identified receptor kinases (Dai et al., 2013; Xu et al., 2014). This notion is supported by the finding that the C-terminus may be mobile and participates in its signalling function (Thiel et al., 1993). Effects originating from overexpression of the mutated ABP1 cDNA are less likely because the overexpression level reached only1.5–1.7-fold in the mutants and 1.3-fold in abp1-11 as compared with the wt (Fig. 2A). Moreover, expression of 50% ABP1 in ABP1/abp1 (Fig. 2A) can cause a phenotype very similar to the one observed here in the point mutants (Effendi et al., 2011) so that overexpression is an unlikely cause for the mild phenotype in abp1-11.
A near-wild-type phenotype was recorded for abp1-11 in induction of lateral roots by IAA, phototropism, hypocotyl gravitropism, root gravitropism and lobe formation (Fig. 2). Auxin-induced elongation growth in the dark was dependent on the presence of wt ABP1 in abp1-11 but was not found in the other three mutants (Fig. 5M). Delay of marker gene expression was apparent but quantitatively lowest in abp1-11 as compared with abp1-8, abp1-9, or abp1-10 (Fig. 3). The permanently high expression of PIN2 and PIN3 in all lines (Fig. 3B) may relate to disturbances in tropisms (Petrášek and Friml, 2009) and the response to shade (Keuskamp et al., 2010). In abp1-8 this increased expression of PIN2 was not found and PIN3 expression was lowest as compared with the wt. This could provide an explanation for the similarity of auxin transport in abp1-8 and the wt (Fig. 2I). In response to various light conditions, abp1-11 had a light-related phenotype weaker than the other three mutants: in hypocotyl length in W light (Fig. 2B); leaf blade shape (Fig. 4A–D), flowering time (Fig. 4E–G), and hypocotyl elongation in response to high ratio R:FR+W (Fig. 5K) and constant B (Fig. 5F); and increase of the bilateral angle in low ratio R:FR (Fig. 6C) and high ratio R:FR (Fig. 6D) in the presence of 1 µM NPA.
Auxin phenotypes in abp1 mutants are linked to auxin transport
Most auxin actions are interwoven with changes in polar auxin transport (Petrášek and Friml, 2009). Here (Fig. 2) and in ABP1/abp1 (Effendi et al., 2011) we showed that basipetal auxin transport in root tips of abp1 mutants was delayed. Functions such as lateral root formation, tropisms (Petrášek and Friml, 2009), and emergence and growth of epidermal cell lobes (Xu et al., 2010) were all affected in abp1 mutants and are all dependent on polar auxin transport, supporting the suggestion that ABP1 function is linked to auxin transport-dependent functions.
ABP1 at the apoplastic side of the plasma membrane and the ER lumen cannot directly interact with TIR1 in the nucleus, yet ABP1 is necessary for efficient stimulus–response coupling between the two receptors within 10min (Figs 3 and 8). Four transmembrane receptor kinases were recently found to bind to ABP1 and provide a mechanism for the long sought transmembrane signalling (Dai et al., 2013; Xu et al., 2014). Auxin-induced expression of ABP1 is detected after 10min (compare Fig. 8B and Effendi et al., 2011; Effendi and Scherer, 2011), but a secreted protein needs roughly 1h to reach the plasma membrane (Scherer, 2011) so that TIR1 cannot regulate the presence or activity of ABP1 in less than 1h (Fig. 9). Short-term effects of NPA inhibition showed down-regulation of PIN protein activity, which consequently would lead to an increased auxin concentration in the cytosol (Covanová et al., 2013) with the logical consequence of re-quantifying TIR1/AFB-dependent transcriptional abundance of auxin marker genes (Scherer et al., 2012).
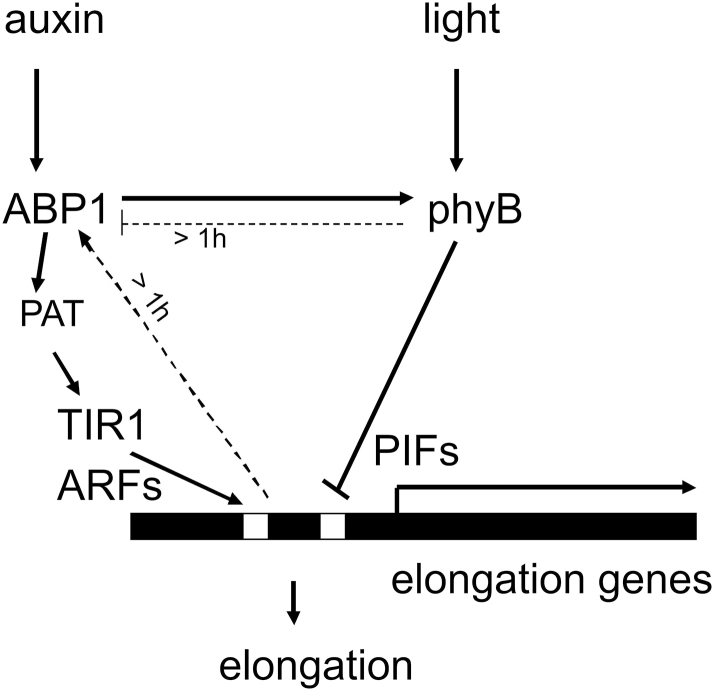
Fig. 9.
Model of suggested linkages between the receptors ABP1 and phyB and early downstream responses. Only responses during the first hour of stimulus are depicted. Solid arrows indicate functional links without implying a detailed mechanism. Dotted arrows indicate slow transcriptional regulation. Auxin short term responses include the regulation of polar auxin transport (PAT) by ABP1 (Robert et al., 2010) and the influence on TIR1 by the cytosolic auxin concentration.
ABP1 and phytochrome regulate growth in a tight functional interaction
The most significant result of this study was the compromised R light signalling in abp1 mutants, although this was indicated in a weaker fashion in abp1-5 and ABP1/abp1 (Effendi et al., 2013). In brief, abp1 mutants are compromised in a number of phyB functions (elongation in R, apical dominance, early flowering, inhibition of gravitropism, misregulation of shade marker genes in LR similar to a phyB mutant), and some phyA functions (broad leaves, elongation in FR, inhibition of gravitropism, misregulation of shade marker genes in HR similar to a phyA mutant).
Interaction between auxin and light in plant growth regulation has been intensively investigated, particularly in responses to shade light (Ruberti et al., 2012). Hypocotyl elongation of the abp1 mutants was partially insensitive to both continuous FR and R (Fig. 5). Insensitivity to B was also observed and could be a consequence of compromised phyA signalling (Fankhauser and Casal, 2004). ABP1 seems to have a dual role, repression of elongation in the light in conjunction with phytochromes, but supporting elongation in the dark (Fig. 5L, M), which offers an explanation for use of etiolated tissue in the classical auxin growth test. In line with the observations on gravitropism by others (Nagashima et al., 2008a/b), compromised phytochrome signalling is indicated by our observations on effects of R and FR light in conjunction with NPA on gravitropism (Fig. 6).
Our data indicate that ABP1 can crosstalk with phyB and phyA, even though these are located in the cytosol and nucleus. Our short-term marker gene expression experiments provide a basis to understand how to link the responses to their receptors (Figs 3, 7, 8, model in Fig. 9). The advantage of short-term kinetics is that negative or positive back-coupling responses can be minimized. In phenotypic assays the final outcome is the sum of many events over time. Thus, even though regulation of auxin-induced expression of auxin marker genes is executed by TIR1 (Mockaitis and Estelle, 2008), their delayed expression in abp1 mutants is observed after only 10min (Fig. 3). Therefore, ABP1 for this response acts functionally upstream of TIR1 and exerts a strong influence.
That changes in the status of phyB have such early consequences for auxin signalling was unexpected, and seems to integrate phyB into auxin signalling (see also Reddy and Finlayson, 2014). Expression of marker genes under R or FR control using phytochromes as sensors was altered in abp1 mutants after 1h (Fig. 7). Therefore mutations in the ABP1 auxin receptor change the light-induced expression of shade marker genes. This argues either for a (i) parallel co-regulation of marker gene expression by auxin-dependent and light-dependent transcription factors or (ii) for a change of the phyB activity status induced by the mutated ABP1, or both (Fig. 9).
For the co-regulation of expression of elongation genes, phyB acts as a repressor activated by R (Steindler et al., 1999; Devlin et al., 2003; Tao et al., 2008; Kunihiro et al., 2011; Leivar et al., 2012). Potential mechanisms of one receptor to regulate the activity of the other are not clear. Because shade does not up-regulate ABP1 expression in the wt (Fig. 8A), the possibility of positive modulation of ABP1 activity by phyB can be excluded. Rather in the phyB mutant ABP1 transcription was slowly increased in shade light, not in the wt. This indicates that a long-term inhibition of ABP1 protein activity by phyB is a possibility (Fig. 9).
Could ABP1 regulate phyB activity? Signalling from ABP1 to phytochromes could start with ABP1 interaction with a transmembrane co-receptor that has the capacity to modulate the phyB phosphorylation status (Effendi et al., 2013; Medzihradszky et al., 2013; Nito et al., 2013; Xu et al., 2014). This speculative mechanism could explain how an auxin transmembrane signal to a network of cytosolic proteins could be transmitted.
The physiology of hypocotyl elongation in our abp1 mutants is fully compatible with this model. Elongation in the dark is ABP1-modulated (Fig. 5M) and repressed by phyB in the light. IAA cannot overcome this light repression (Fig. 5L). Shade releases the repression (Fig. 5I–K) by inactivation of phyB. Partial R and FR insensitivity of elongation in abp1 mutants (Fig. 5A–H) is also consistent with this model in that ABP1 supports the action of phyB in light and repression of phyB action is weakened in an abp1 mutant.
Auxin transport and auxin biosynthesis were suggested to play a role in shade-induced elongation (Nagashima et al., 2008a/b; Tao et al., 2008), which is not mutually exclusive for the functions discussed above. The effects of NPA on elongation and on gravitropism in LR and HR (Fig. 6) are consistent with the concept of weakened phyB action. Regulation of auxin transport in shade was suggested to depend on PIN efflux facilitators (Keuskamp et al., 2010) and ABCB efflux transporters (Nagashima et al., 2008a/b), the latter being shown to be directly inhibited by NPA (Bailly et al., 2012).
Ordering early responses into a timeline as a first strategy (Figs 3, 7, 8) is a way to mechanistically explain the functional interaction of ABP1 and phytochromes (Fig. 9) and provides a fresh starting point to investigate auxin and R signalling and growth control.
Author contributions
YE performed and designed experiments, NF did quantum modelling, MG did transport experiments, CL performed transcription experiments, GFES designed the research and GFES, YE, NF, and MG wrote the paper.
Supplementary Material
Acknowledgements
We are grateful for financial support from the Deutsches Zentrum für Luft- und Raumfahrt (contract number 50WB0627, 50BW0933 and 50BW1333 to GS). We thank Thomas Reinhard (Hannover) for making the initial vector containing the ABP1 cDNA available to us and Laurence Charrier for performing transport measurements. We thank T. Buckhout for critically reading the manuscript. Mutants phyA-211 (Col) and phyB-9 (Col-0) were obtained from Christian Luschnig (BOKU, Vienna) and Mathias Zeidler (University of Giessen).
Supplementary data
Supplementary data are available at JXB online
Figure S1. 3-dimensional structure of the strep tag attached at the C-terminus of ABP1.
Figure S2. Quantification of lobe formation in abp1 mutants.
Figure S3. Rosettes of phyA and phyB mutants grown side by side with respective wt plants.
Figure S4. Elongation in monochromatic continuous light (R, FR, B) of 4 d old seedlings (1 µmol m–1 s–2 each).
Table S1. Quantum chemical modelling of the auxin binding box in abp1 mutants.
Table S2. Primers for PCR and accession numbers.
References
- Alberts IL, Nadassy K, Wodak SJ. 1998. Analysis of zinc binding sites in protein crystal structures. Protein Science 7, 1700–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. 1997. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. The Plant Cell 9, 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A, Yang H, Martinoia E, Geisler M, Murphy AS. 2012. Plant lessons: exploring ABCB functionality through structural modeling. Frontiers in Plant Science 2, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becke AD. 1988. Density-functional exchange-energy approximation with correct asymptotic-behavior. Physical Reviews A 38, 3098–3100. [DOI] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ. 2008. Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic development in Arabidopsis and tobacco. The Plant Cell 20, 2746–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Shimomura S, Sitbon F, Sandberg G, Jones AM. 2001a. The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. The Plant Journal 28, 607–617. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. 2001b. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes and Development 15, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covanová M, Sauer M, Rychtář J, Friml J, Petrášek J, Zažímalová E. 2013. Overexpression of the AUXIN BINDING PROTEIN1 modulates PIN-dependent auxin transport in tobacco cells. PLoS One 8, e70050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai N, Wang W, Patterson SE, Bleecker AB. 2013. The TMK subfamily of receptor-like kinases in Arabidopsis display an essential role in growth and a reduced sensitivity to auxin. PLoS One 8, e60990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David KM, Couch D, Braun N, Brown S, Grosclaude J, Perrot-Rechenmann C. 2007. The auxin-binding protein 1 is essential for the control of cell cycle. The Plant Journal 50, 197–206. [DOI] [PubMed] [Google Scholar]
- Devlin P, Yanovsky MJ, Kay SA. 2003. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiology 133, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi Y, Jones AM, Scherer GFE. 2013. AUXIN-BINDING-PROTEIN1 (ABP1) in phytochrome-B-controlled responses. Journal of Experimental Botany 64, 5065–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi Y, Radatz K, Rietz S, Labusch C, Rietz S, Wimalasekera R, Helizon H, Zeidler M, Scherer GFE. 2014. Mutants of phospholipase A (pPLA-I) have a red light and auxin phenotype. Plant, Cell and Environment 37, 1626–1640. [DOI] [PubMed] [Google Scholar]
- Effendi Y, Rietz S, Fischer U, Scherer GFE. 2011. The heterozygous abp1/ABP1 insertional mutant has defects in functions requiring polar auxin transport and in regulation of early auxin-regulated genes. The Plant Journal 65: 282–294. [DOI] [PubMed] [Google Scholar]
- Effendi Y, Scherer GFE. 2011. AUXIN BINDING-PROTEIN1 (ABP1), a receptor to regulate auxin transport and early auxin genes in an interlocking system with PIN proteins and the receptor TIR1. Plant Signaling and Behavior 6, 1101–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichkorn K, Weigen F, Treutler O, Ahlrichs R. 1997. Auxiliary basis sets for main row atoms and transition metals and their use to approximate Coulomb potentials. Theoretical Chimica Acta 97, 119 –124. [Google Scholar]
- Fankhauser C, Casal JJ. 2004. Phenotypic characterization of a photomorphogenic mutant. The Plant Journal 39, 747–760. [DOI] [PubMed] [Google Scholar]
- Ferro N, Gallegos A, Bultinck P, Jacobsen HJ, Carbó-Dorca R, Reinard T. 2006. Coulomb and overlap self-similarities: a comparative selectivity analysis of structure-function relationships for auxin-like molecules. Journal of Chemical Informatics Modeling 46, 1751–1762. [DOI] [PubMed] [Google Scholar]
- Grimme S, Antony J, Ehrlich S, Krieg H. 2010. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. Journal of Chemical Physics 132, 154104–154109. [DOI] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns AN. 1998. Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282, 1114–1117. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LACJ, Peeters AJM, Pierik R. 2010. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proceedings of the National Academy of Sciences, USA 107, 22740–22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro A, Yamashino T, Nakamichi N, Niwa Y, Nakanishi H, Mizuno T. 2011. Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana . Plant and Cell Physiology 52, 1315–1329. [DOI] [PubMed] [Google Scholar]
- Labusch C, Shishova M, Effendi Y, Li M, Wang X, Scherer GFE. 2013. Patterns and timing in expression of early auxin-induced genes in phospholipase A (pPLA) T-DNA insertion mutants reveal a function in auxin signaling. Molecular Plant 6, 1473–1486. [DOI] [PubMed] [Google Scholar]
- Lee C, Yang W, Parr RG. 1988. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Physical Reviews B 37, 785–789. [DOI] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. 2012. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis . The Plant Cell 24, 1398–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Muday GK. 2009. Measurement of auxin transport in Arabidopsis thaliana . Nature Protocols 4, 437–451. [DOI] [PubMed] [Google Scholar]
- Liscum E, Hangarter RP. 1993. Genetic evidence that the red-absorbing form of phytochrome B modulates gravitropism in Arabidopsis thaliana . Plant Physiology 103, 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real time quantitative PCR and the 2–∆∆CT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Medzihradszky M, Bindics J, Ádám É, et al. 2013. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis . The Plant Cell 25, 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M. 2008. Auxin receptors and plant development: a new signaling paradigm. Annual Reviews of Cell and Developmental Biology 24, 55–80. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Suzuki G, Uehara Y, et al. 2008a. Phytochromes and cryptochromes regulate the differential growth of Arabidopsis hypocotyls in both a PGP19-dependent and a PGP19-independent manner. The Plant Journal 53, 516–529. [DOI] [PubMed] [Google Scholar]
- Nagashima A, Uehara Y, Sakai T. 2008b. The ABC subfamily B auxin transporter AtABCB19 is involved in the inhibitory effects of N-1-naphthyphthalamic acid on the phototropic and gravitropic responses of Arabidopsis hypocotyls. Plant and Cell Physiology 49, 1250–1255. [DOI] [PubMed] [Google Scholar]
- Napier RM. 1995. Towards an understanding of ABP1. Journal of Experimental Botany 46, 1787–1795. [Google Scholar]
- Nito K, Wong CC, Yates JR, 3rd, Chory J. 2013. Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Reports 27, 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Mouille G, Grandont L, et al. 2014. AUXIN BINDING PROTEIN1 links cell wall remodeling, auxin signaling, and cell expansion in Arabidopsis . The Plant Cell 26, 280–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Friml J. 2009. Auxin transport routes in plant development. Development 136, 2675–2688. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan G, Dempfle L. 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Finlayson SA. 2014. Phytochrome B promotes branching in Arabidopsis by suppressing auxin signaling. Plant Physiology 164, 1542–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico JF, López R, Ema I, Ramírez G, Ludeña EV. 2004. Analytical method for the representation of atoms-in-molecules densities. Journal of Comparative Chemistry 25, 1355–1363. [DOI] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, et al. 2010. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis . Cell 143, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, Smith H. 1996. Genetic and transgenic evidence that phytochrome A and B act to modulate the gravitropic orientation of Arabidopsis thaliana hypocotyls. Plant Physiology 110, 211–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolo-Naranjo A, Codorniu-Hernández E, Ferro N. 2010. Quantum chemical associations ligand-residue: their role to predict flavonoid binding sites in proteins. Journal of Chemical Informatics Modeling 50, 924–933. [DOI] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Ciolfi A, Possenti M, Carabelli M, Morelli G. 2012. Plant adaptation to dynamically changing environment: The shade avoidance response. Biotechnology Advances 30, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Santner AA, Watson JC. 2006. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis . The Plant Journal 45, 752–764. [DOI] [PubMed] [Google Scholar]
- Scherer GFE. 2011. AUXIN-BINDING-PROTEIN1, the second auxin receptor: what is the significance of a two-receptor concept in plant signal transduction? Journal of Experimental Botany 62, 3339–3357. [DOI] [PubMed] [Google Scholar]
- Scherer GFE, Labusch C, Effendi Y. 2012. Phospholipases and the network of auxin signal transduction with ABP1 and TIR1 as two receptors: a comprehensive and provocative model. Frontiers in Plant Science 3, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steindler C, Mateucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. 1999. Shade avoidance responses are mediated by the ATHB-2 HD-Zip protein, a negative regulator of gene expression. Development 126, 4235–4245. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ. 1994. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. Physical Chemistry 98, 11623–11627. [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, Blatt MR, Fricker MD, White IR, Millner P. 1993. Modulation of K+ channels in Vicia stomatal guard cells by peptide homologues to the auxin-binding proteins C-terminus. Proceedings of the National Academy of Sciences, USA 90, 11493–11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Paque S, Stierlé V, Quettier AL, Muller P, Lechner E, Genschik P, Perrot-Rechenmann C. 2013. Auxin-binding protein 1 is a negative regulator of the SCF(TIR1/AFB) pathway. Nature Communications 4, 2496. [DOI] [PubMed] [Google Scholar]
- Wang C, Yan X, Chen Q, Jiang N, Fu W, Ma B, Liu J, Li C, Bednarek SY, Pan J. 2013. Clathrin light chains regulate clathrin-mediated trafficking, auxin signaling, and development in Arabidopsis . The Plant Cell 25, 499–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JG, Chen CH, Chien CT, Hsieh HL. 2011. FAR-RED INSENSITIVE219 modulates CONSTITUTIVE PHOTOMORPHOGENIC1 activity via physical interaction to regulate hypocotyl elongation in Arabidopsis . Plant Physiology 156, 631–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. 1993. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. The Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo EJ, Marshall J, Bauly J, Chen JG, Venis M, Napier RM, Pickersgill RW. 2002. Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO Journal 21, 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, et al. 2014. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343, 1025–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. 2010. Cell surface- and Rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis . Cell 143, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.