Abstract
Inactivation of intact influenza viruses using formaldehyde or β-propiolactone (BPL) is essential for vaccine production and safety. The extent of chemical modifications of such reagents on viral proteins needs to be extensively investigated to better control the reactions and quality of vaccines. We have evaluated the effect of BPL inactivation on two candidate re-assortant vaccines (NIBRG-121xp and NYMC-X181A) derived from A/California/07/2009 pandemic influenza viruses using high-resolution FT-ICR MS-based proteomic approaches. We report here an ultra performance LC MS/MS method for determining full-length protein sequences of hemagglutinin and neuraminidase through protein delipidation, various enzymatic digestions, and subsequent mass spectrometric analyses of the proteolytic peptides. We also demonstrate the ability to reliably identify hundreds of unique sites modified by propiolactone on the surface of glycoprotein antigens. The location of these modifications correlated with changes to protein folding, conformation, and stability, but demonstrated no effect on protein disulfide linkages. In some cases, these modifications resulted in suppression of protein function, an effect that correlated with the degree of change of the modified amino acids’ side chain length and polarity.
Keywords: β-Propiolactone, Biomedicine, Influenza vaccine, MS, PTM, Virus inactivation
1 Introduction
Influenza viruses cause widespread human respiratory infections, impacting public health in the form of both seasonal epidemics and less frequent pandemic outbreaks 1. Vaccination is the most cost-effective and efficient public health measure for disease prevention 2,3. Currently available vaccines are usually live attenuated or inactivated types of influenza viruses, which are derived from embryonated eggs and cell culture-based propagations. Frequent mutations of viral proteins (“antigentic drift”) and the occasional presence of novel subtypes (“antigenic shift”) of influenza A viruses result in significant losses of communal immunity. Therefore, vaccines must be strategically designed and administered on an annual basis to combat currently circulating strains. The timely development of safe and effective vaccines is a year-round monitoring and manufacturing effort with limited capacity to meet emergency needs at the time of a pandemic outbreak 4,5. The safety and efficacy of vaccines are high public health priorities. Therefore quality control measures for influenza vaccine production include strict control of virus formulation, purification, growth rate, antigen yield, stability, immunogenicity, and enzymatic activity. Viral particles harvested from a growth substrate, such as embryonated chicken eggs or mammalian cells, are typically inactivated by trace amounts of formaldehyde or β-propiolactone (BPL) during the vaccine production process 6. Inactivation occurs via chemical reactions with viral constituents 7, ideally without disrupting the integrity of whole virus particles or the immunogenicity of the antigenic epitopes of key surface proteins (e.g., hemagglutinin (HA) and neuraminidase (NA)). However, excessive inactivation reagents may cause unanticipated modifications to the vaccine antigens that result in diminished potency. Evidence from previous studies has shown that the hemagglutination titer, antigenicity, and NA activity of the human influenza virus were significantly reduced by incorporation of BPL 6–10. A suggested cause was the alteration of protein conformation (specifically at antigenic epitopes) caused by exposure to excessive reagents used in the viral inactivation process. In this study, we sought to determine the location and extent of exogenous modifications on the outer surface membrane protein antigens of influenza virus, HA and NA, in order to better understand how this inactivation reagent may affect vaccine efficacy and quality control throughout the large-scale vaccine production.
To understand the potential effects of inactivation reagents on viral vaccine strains in general, Uittenbogaard et al. investigated the chemically induced modifications of BPL by in vitro reactions with some nucleobase analogues, nucleosides, and synthetic peptides 7. Despite the fact that the chosen analytical targets were from a restricted group of small peptides, BPL was found to ubiquitously react with several electron-pair-donor amino acid residues with reactivity decreasing in the order of Cys > Met > His > Asp/Glu > Tyr > Lys > Ser > Thr. Such reactions generated novel (mono- and bis-) alkylated and acylated molecules formed with dramatic changes in both amino acid polarity and length of amino acid side chains. The impact of these modifications on intact influenza virus particles during an inactivation process remains to be further understood. Here, the proteomic investigation of two monovalent inactivated vaccines derived from influenza virus strains (NIBRG-121xp and NYMC-X181A from A/California/7/2009) has been carried out using high-resolution ultra performance LC (UPLC) and linear ion-trap FT ICR (LTQ FT-ICR) MS. We have determined the complete sequences of HA and NA through membrane delipidation, various enzymatic digestions, and extensive characterization of glycopeptides. Comprehensive analyses of thousands of modified peptides revealed unique sites on the surface regions of the protein structures that reacted with BPL, as well as subtle differences between the candidate reassortants. Our findings thus provide structural insights into the protein modifications and surface topology, and increase our understanding how the protein immunogenicity and enzymatic activity are affected by BPL inactivation of the monovalent influenza viruses.
2 Materials and methods
2.1 Proteins, chemical reagents, and materials
DTT, hydroxylamine, iodoacetamide, ammonium bicarbonate (NH4HCO3), 2,5-DHB, formic acid (FA), TFA, ACN, chymotrypsin, proteinase K, and pepsin were purchased from Sigma-Aldrich (Oakville, ON, USA). Sequence-grade bovine trypsin and endoproteinase Asp-N was obtained from Roche Diagnostics Corporation (Indianapolis, IN, USA). Porous graphitic carbon (PGC) cartridges were obtained from Mandel Scientific (Guelph, ON, USA).
2.2 Purification of influenza proteins
Egg-derived monovalent influenza vaccines derived (inactivated, split virion) from the virus strains of NIBRG-121xp (HA: K136N, D239G; NA: N88G) and NYMC-X181A (HA: K236T, Q240R) were acquired as described previously 11. The process and effect of BPL inactivation of influenza viruses has been studied by several groups 8,12,13 Samples (1.5 mL) of the candidate vaccines were prefiltered through a membrane centrifugal filtration unit (Satorius Stedim Biotech, Germany) with 10 000 MW cut off and washed with 25 mM NH4HCO3. The purified proteins were then treated with 10 mM DTT at 60°C for 1 h. After cooling down, an equal volume of hydroxylamine solution (Sigma, 2 M, pH 7.4) was added, and the sample was allowed to shake for 2 h at room temperature in order to completely remove the conjugated lipids from HA 14,15. To block free cysteines, iodoacetamide was added to a final concentration of 55 mM and the mixture was incubated at room temperature in dark for 1 h. The alkylated sample was finally dialyzed against 5 mM NH4HCO3 and dried in a Savant vacuum centrifuge (Thermo Fisher Scientific, ON, Canada).
2.3 Enzymatic digestion and mass spectrometric analysis
Aliquots (∼20 μg) of the purified influenza proteins were reconstituted in 20 μL of 25 mM NH4HCO3 (pH 7.6) or 10 mM Tris buffer (pH 8.0) and digested using trypsin, endoproteinase Asp-N, chymotrypsin, proteinase K, and pepsin at 37°C for 4 h. Each of the digests were subsequently diluted 50 times in 0.2% FA and analyzed using a nanoAcquity UPLC (Waters, Milford, MA) coupled with an LTQ FT-ICR (Thermo Fisher, San Jose, CA, USA) mass spectrometer. UPLC separation was achieved using a binary RP gradient (solvent A—0.1% FA in water, solvent B—0.1% FA in ACN). Peptides were first trapped by a RP Symmetry C18 column (180 μm id × 20 mm length, 5 μm diameter particles) at 5 μL/min (solvent A/B—99/1) for 3 min, and subsequently separated on a C18 analytical column (100 μm id × 100 mm, 1.7 μm particle diameter, BEH 130) at 400 nL/min. Peptide elution was achieved using a linear gradient from 5 to 30% solvent B over 40 min, followed by a linear gradient to 85% solvent B over 10 min. FT-MS scans were acquired with high resolution (100 000) from m/z 300 to 2000, and low-resolution MS/MS measurements in LTQ mode were obtained by data-dependent scans of the top eight most intense precursor ions at multiply charged states of 2+, 3+, and 4+. Dynamic exclusion was enabled for a period of 180 s. Off-line UPLC MS and MS/MS analyses of the tryptic digests were performed on the AB Sciex QStar XL MALDI quadrupole TOF (MALDI QqTOF) mass spectrometer equipped with an orthogonal (oMALDI) source operating with a nitrogen laser (337 nm). UPLC fractions were collected at 2 min intervals and were spotted onto a MALDI target plate with 0.5 μL matrix (2,5-DHB, 160 mg/mL in ACN/0.1% FA) predeposited on each spot.
2.4 Glycopeptide purification using PGC
The vaccine sample of 1 mL (∼500 μg/mL) was treated by delipidation and alkylation as indicated earlier, then the purified proteins were digested by 5 μg of trypsin in 25 mM NH4HCO3 for 4 h followed by overnight digestion with chymotrypsin or proteinase K at a ratio of 1:50 (enzyme/substrate ratio) at 37°C. PGC cartridges were washed with 3 mL of 80% v/v ACN followed by 3 mL of water. The protein digest was fully loaded on PGC cartridges, and then washed with 500 μL of water for three times. The glycopeptides were sequentially eluted by 25% ACN in 0.1% TFA, 50% ACN in 0.1% TFA, and 75% ACN in 0.1% TFA. Each fraction was freeze-dried by SpeedVac, and finally dissolved in 50 μL of 0.2% FA for mass spectrometric analysis.
2.5 Database search and peptide identification
Peptide identification was performed using MASCOT Server (version 2.3.0, Matrix Science, London, UK), and LC MS/MS raw data were searched against the NCBI nonredundant database and an in-house influenza vaccine protein database 16. The search parameters for data from samples digested with trypsin were restricted to fully tryptic peptides with a maximum of two missed cleavages. Data from Asp-N, chymotrypsin, and proteinase K digestions were searched allowing for nonspecific enzyme cleavage. Cysteine carbamidomethylation (+57.02146 Da) was designated as a fixed modification, and deamidation of asparagine and glutamine (+0.98402 Da), methionine oxidation (+15.99492 Da), single modification by BPL of amino acids (Cys, Asp, Glu, His, Lys, Met, Ser, Thr, Tyr; +72.02113 Da), double modification by BPL of amino acids (Cys, Asp, Glu, His, Met; +144.04226 Da), and pyro-Glu of Gln conversion (−17.02655 Da) at the N-terminus were considered as variable modifications. Mass tolerances were set up to 10 ppm for the FT-MS ions and 1 Da for ion-trap MS/MS fragment ions. Peptide assignments were filtered by an ion score cut off of 20, and the significance threshold was adjusted to 0.001 to achieve a false discovery rate (FDR) of less than 1%. The identified spectra were also verified manually.
3 Results and discussion
3.1 Complete sequence determination of influenza virus antigens
Accurate identification of influenza virus strains requires complete sequence determination of membrane surface glycoproteins, which can be difficult to achieve by conventional LC-MS methods. Detection of peptides is often limited by too large or small size of proteolytic products derived from a single enzyme digestion (e.g., trypsin), insolubility of hydrophobic fragments, as well as negatively charged PTMs such as N-linked glycosylation and S-acylation of HA and NA. Our previous UPLC MS/MS analyses demonstrated that the sequence coverage could be considerably improved up to a maximum of 86% (HA) or 83% (NA) by deglycosylation with PNGase F and combined enzymatic digestions of trypsin and chymotrypsin 16. The missing portions of HA sequences were located near regions of uncleaved N-linked glycosylation sites, where the attached fucosyl glycan conjugates to the proteins were resistant to proteolysis by PNGase F; and the cytoplasmic tail in the C-terminal transmembrane domains caused by palmitoylation and stearoylation of cysteines 16–18. These modified peptides were either negatively charged or water-insoluble, resulting in ion suppression and low-sensitivity detection using positive ionization mode of ESI and/or MALDI when mixing with nonmodified peptides. To solve this problem, we delipidated intact, membrane bound proteins with 1 M hydroxylamine, and isolated the glycopeptides by graphite affinity purification of nonspecific enzyme digests. Through such analyses, we were able to achieve 100% sequence coverage of HA and NA by high-resolution UPLC and MS. As summarized in Supporting Information Table 1 and Fig. 1, identification of the N- and C-termini of the proteins was achieved by highly confident peptide identification from various enzymatic digests followed by MASCOT database search. In agreement with the previous observation 19, the N-terminal signal peptide at residues 1–17 was deleted in the mature form of HA, and the C-terminus of the protein was identified by peptides containing three alkylated cysteines. Since the asparagine residue in other C-terminal peptide sequences (residues 554–566, 555–566, 553–560) of HA was found without change (Supporting Information Table 1), deamidation of Asn557 at NG motif of peptide 557–566 can be reasonably inferred to be a result of a chemical conversion event during the sample preparation 20. The complete sequence of NA was also confirmed by LC-MS/MS, and the N-terminal methionine as a starting amino acid of the protein encoded by the nucleotide sequence AUG is retained in the protein sequence; its deletion likely inhibited by the presence of proline at the third residue 21.
This method, utilizing protein delipidation and multiple enzymatic digestions together with glycopeptide isolation, thus provided a complete set of overlapping peptides for reliable identification of influenza vaccine antigens and additional PTMs of the proteins, such as those modified by BPL in the inactivated influenza virus antigens as discussed later.
3.2 Protein modifications by BPL
We initially observed modifications of proteins by BPL by off-line UPLC MALDI QqTOF MS analyses of the tryptic digests of monovalent influenza vaccines, in which a set of unknown peptides were found to have a mass difference of +72 Da compared to their nonmodified counterparts (Fig.1). Relative intensities of these modified peptides varied with the sequence regions of the proteins. MALDI MS/MS sequencing was used to identify the peptides and modification sites, which were found to occur at a variety of amino acid residues, suggesting widespread chemical reactions of BPL with the influenza proteins. The analyses confirmed the covalent additions of BPL to the reactive sites of peptides which occur via two unique pathways giving rise to either alkylated or acylated products as summarized in Fig.2 7. To gain a complete view of the chemical modifications on intact proteins, proteomic analysis was performed on various enzymatic digests (with trypsin, chymotrypsin, endoproteinase Asp-N, protease K, and pepsin) of the influenza vaccines derived from the strains NIBRG-121xp and NYMC-X181A, respectively. Using nanoflow UPLC LTQ-FT MS/MS and subsequent database searches we retrieved a total of 1765 modified peptides from HA and NA. Following removal of false positive results, manual inspection of MS/MS spectra confirmed a number of modified residues: 83 sites on HA and 43 sites on NA in NIBRG-121xp derived vaccine, 99 sites on HA and 39 sites on NA in NYMC-X181A derived vaccine (Supporting Information Fig. 2). Sequence alignment of these vaccines showed conserved site identities of 75% (74/99) for HA and 56% (24/43) for NA at the modified residues, whereas other distinct residues appeared at unique locations accounting for the exposed structural surface regions caused by different protein conformations.
Figure 1.
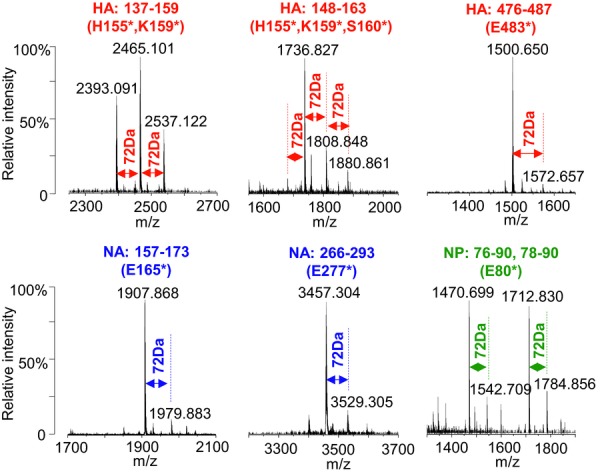
Relative abundances of the modified tryptic peptides (+72 Da) of HA, NA, and NP in the influenza vaccines from the strains of NIBRG-121xp and NYMC-X181A. The data were obtained by off-line UPLC fraction collections followed by MALDI Q-Star XL QqTOF MS analyses on a trypsin digest, and an asterisk indicates the residue of the peptide fragments was modified by BPL.
Figure 2.
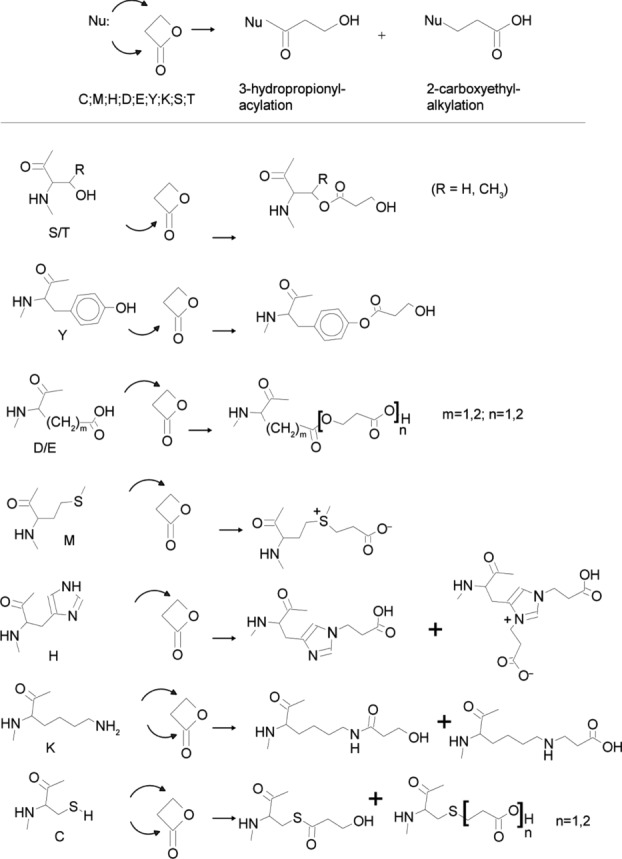
Summary of the chemical reactions of BPL with polar amino acids, adapted from Uittenbogaard et al. 7.
Detailed investigation of the protein sequences in the NIBRG-121xp sample revealed many BPL modifications of HA and NA at polar amino acids (Met, His, Asp/Glu, Tyr, Lys, Ser/Thr (+72.02113 Da)), though not cysteine residues as discussed later. The N- and C-terminal regions of HA were not modified by BPL, probably due to the steric hindrance provided by long membrane lipid chains near the C-terminus of the protein as well as the large glycan side chains near the N-terminus. Sixteen more modification sites were identified on the HA from vaccines derived from NYMC-X181A versus that of NIBRG-121xp (Supporting Information Fig. 2). Extension of the exposed sequences at the protein structure implied more solvent-accessible sites of HA (NYMC-X181A) reacted with BPL than that of the HA (NIBRG-121xp). The phenomenon is consistent with the nearly identical sequences of two protein mutants but slight difference in protein conformation, due to a unique glycosylation site introduced by a point mutation of K136N in HA (NIBRG-121xp), which is supposed to be favorable to protein folding 5,11. As expected, BPL modifications of Glu132, Lys136, and Lys147 were present in the surface area (segment 132–147) of HA (NYMC-X181A), but absent in the HA (NIBRG-121xp). In addition, a group of six residues in close proximity to each other (Met493, Glu494, Asp502, Tyr503, Lys505, and Tyr506) reacted with BPL in HA (NYMC-X181A), but not in HA (NIBRG-121xp).
In the case of NA, both the N- and C-termini of the protein (Met1 and Lys469) were partially modified by BPL (Supporting Information Table 1 and Fig. 2). The protein sequence of NA (NYMC-X181A) has only four more modified sites than that of NA (NIBRG-121xp), in which two small distinct regions were seen. In addition to the C-terminus (segment 462–469), the second region is located close to the mutation site of Asn88Gly, where a glycan motif has been lost from the influenza A/California strain, leading to BPL modifications of Ser79, Ser82, and Lys84. Overall, our analyses here identified both ubiquitous and unique sites from HA and NA where BPL reactions occurred. The unique locations offered valuable information to identify subtle differences between the protein structures of vaccine isolates.
3.3 Glycan chain protection from the chemical modifications of BPL
Glycosylation of HA and NA helps maintain the proper folding of protein structures, the integrity of influenza viruses, and the immunogenicity of antigens. The gain or loss of glycosylation sites can have dramatic effects on the biosynthesis, stability, receptor binding, immune response, and other functions of viral glycoproteins as reported previously 5,22. The monomers of the HA and NA investigated in this study contain 11 (121xp-HA), 10 (X181A-HA), 7 (121xp-NA), and 8 (X181A-NA) putative N-linked glycosylation sites based on the consensus NxS/T/C motifs. MS/MS sequencing of the isolated glycopeptides has simultaneously been used to determine the heterogeneous glycan structures attached to the side chain of asparagines, which were completely or partially modified on HA and NA (details not shown). In terms of structural diversity at particular sites, steric hindrance of large glycan chains prevents the chemical reactions with BPL from occurring on nearby amino acid residues. Specifically, Asn40, Asn104, Asn136, Asn293, Asn304, and Asn498 of HA were fully glycosylated, and the residues near these sites were free of BPL modifications. We did detect a set of glycopeptides at residues 301–322 with additional masses of 72 Da (Supporting Information Table 2), however MS/MS sequencing revealed that the glycosylated and BPL-modified residues (Asn304 and Lys319, respectively) were 15 residues apart. There were also cases of partial N-linked glycosylation (e.g., Asn479, Asn490, Asn498) near the C-terminal region of HA which allowed partial BPL modifications to occur in this region.
From these observations, we can conclude that the N-linked glycosylation of proteins largely prevents neighboring amino acid residues from reacting with BPL, and the presence of nearby BPL modifications indicates partial or nonglycosylation status at the NxS/T/C motif.
3.4 Cysteine residues of influenza virus antigens were not affected by BPL inactivation
Disulfide linkages in influenza vaccine proteins are also important contributors to protein stability and conformational integrity. Since BPL reacts with free cysteine residues, but not cystine 7, our workflow can be used to distinguish between the two, with free cysteines modified by BPL and disulfide bound cysteines modified by carbamidomethylation. Our results showed that all cysteines in HA and NA (15 and 19, respectively) were carbamidomethylated. However, to lesser degree, some cysteine-containing peptides were also found to be modified by +72 Da or +144 Da at Cys59, Cys72, Cys84, Cys320, Cys481, and Cys562 of HA, Cys184 and Cys335 of NA (Supporting Information Table 3). Assignment of those “BPL-modified peptides” revealed very high MASCOT scores, indicating unambiguous identification of the peptide sequences. However, the mass error of more than 5 ppm in individual peptides is normally unacceptable for high-resolution FT-ICR MS measurements, suggesting the need for a careful examination of the putative modifications. Indeed, manual inspection of the MS/MS spectra localized the only mass shift of +73 Da at the cysteine residues (Fig.3), excluding the possibility of combined two point modifications at both cysteine (+72 Da) and deamidated Asn or Gln (+1 Da). Thus, the peptides can be unambiguously identified as oxidized carbamidomethylation on cysteines alone. Supporting evidence was obtained through analyses of the isotope distribution of peptides, and accurate masses of oxidized carbamidomethyl cysteine in the peptides resulted in an increased mass of 73.0164 Da differing by 0.0113 Da from the predicted value of a putative BPL modification plus deamidation (73.0051 Da) of Asn or Gln, which allow all peptides to achieve a high mass accuracy of better than 2 ppm (Supporting Information Table 3). A second mis-assignment of a 144 Da mass increment was tentatively defined as a double BPL attachment to some peptides in the database search, for example, the peptide 281–293, 281CYPDSSEITCVCR293, of NA at m/z 867.3511(2+; Supporting Information Table S3). The MS/MS spectrum displayed a complete set of y ions, and showed that the 144 Da increase was localized at the first amino acid residue, Cys281 (Supporting Information Fig. 3A). Although the peptide sequence was identified by high MASCOT scores from both trypsin and Asp-N digestions, again, the mass error of 7 or 8 ppm failed to reach the expected mass accuracy of the FT-ICR MS instrument. A retrospective examination of the NA sequence revealed that the assigned N-terminal residue (Cys281) is preceded by a serine residue (Ser280), allowing for the inclusion of Ser280 (+87.0320 Da) together with a change of the Cys281 modification from a double BPL attachment (+144.0423 Da) to carbamidomethylation (+57.0215 Da) results in a mass shift of 0.0112 Da, which reduces mass error from 7 to 1 ppm. Both peptide sequences match the set of high-intensity C-terminal y-ion fragments generated by CID (Supporting Information Fig. 3A). Likewise, the peptide of m/z 747.3317 derived from a trypsin digestion followed by pepsin was incorrectly assigned as a peptide fragment 334–347 (334SCGPVSSNGANGYK347) from NA with BPL modifications at Ser334 (+72 Da) and Cys335 (+144 Da). Based on accurate mass measurement and reinterpretation of the MS/MS spectrum (Supporting Information Table 3 and Fig. 3B), the N-terminus was confirmed to have two more residues Thr332 (101.0477 Da) and Gly333 (57.0215 Da) together with Ser334 (unmodified, 87.0320 Da) and alkylated Cys335 (+57.0215 Da), which yield a total mass increase of 215.0907 Da plus a deamidation of asparagine to aspartic acid (+0.98402 Da) at NG motifs of the peptide sequence, rather than that with two BPL modifications of Ser334 (+72.02113 Da) and Cys335 (+144.04226 Da). Not surprisingly, the mistakes were produced by automated database search on the datasets generated from nonspecific enzymatic digests, fortunately, high mass accuracy analysis could distinguish these differences in the peptide sequences and related modifications. Protein sulfenic acids are transient intermediates in the formation of disulfide bonds, which are formed by oxidation, and play a significant role in enzyme catalysis, protein turnover, regulation, and signaling transduction 23–25. Although the precise location of disulfide bonds in influenza virus proteins remains to be fully understood, some have been partially resolved by protein crystal structure 26,27, as well as chemical and enzymatic studies 28,29. 3D structure modeling has shown that the intramolecular disulfides of Cys72-Cys84 and Cys107-Cys153 localize closely to the antigenic sites of the HA globular head domain, whereas disulfide pairs Cys59-Cys292, Cys296-Cys320, and Cys21-Cys481 are on the exposed surface region of HA stem domains (Supporting Information Fig. 4). In the case of NA, the disulfide pairs of Cys129-Cys124, Cys231-Cys238, Cys279-Cys292, Cys290-Cys281, and Cys421–446 are adjacent to the catalytic cavity of NA, which are critical for maintaining enzymatic stability and activity 30, and the surface-exposed Cys184 and Cys335 were oxidized against the modification by BPL (Supporting Information Fig. 4). Our results clearly showed that all cysteine residues of HA and NA are involved in either intra- or intersubunit disulfide crosslink, or S-palmitoylation, which do not react with BPL, and accordingly, no BPL-modified cysteine was found in the proteins. Inactivation of influenza viruses using BPL therefore likely has little influence on the integrated protein frame structure of HA and NA.
Figure 3.
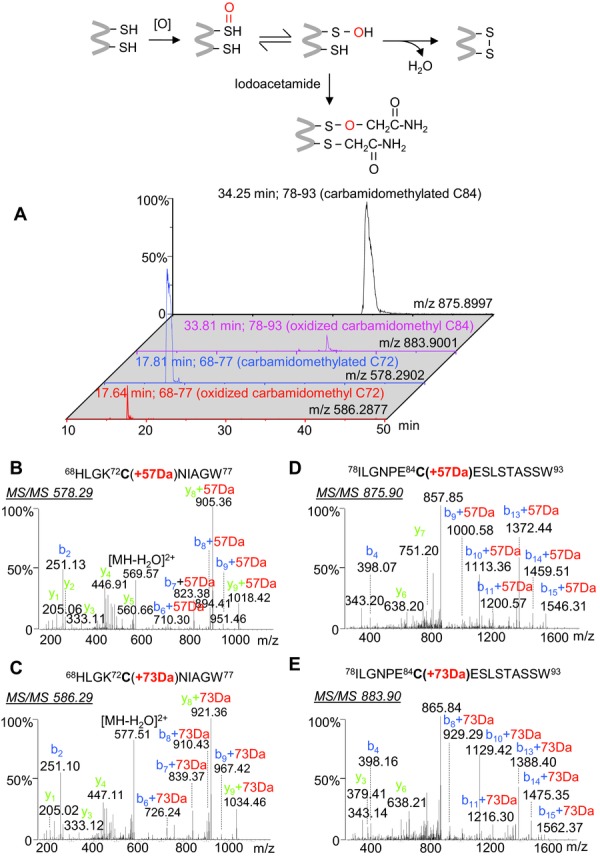
Identification of modified peptides containing oxidized carbamidomethyl Cys residue (73 Da) instead of a single BPL-modified Cys plus a deamidation of Asn/Gln (73 Da). (A) Extracted ion chromatogram of the peptide ions. (B–E) MS/MS spectra of the peptide ions at m/z 578.29, m/z 586.29, m/z 875.90, and m/z 883.90, respectively.
To rule out the possibility of missing detection of BPL-cysteine modifications due to an unexpected removal of side groups by protein delipidation during sample processing, we reinterrogated the database search results for nucleocapsid protein sequences (gi| 256259586) of the influenza vaccine and successfully identified BPL-modified cysteine-containing peptides. As an example, Supporting Information Fig. 5 shows that the mass increase of 72 Da was localized at Cys44 of the tryptic peptide 39–48 (R.FYIQMCTELK.L) by MS/MS. Here the free cysteine reacted with BPL in which the accurate mass of m/z 674.3205 (calculated molecular weight, 1346.6254 Da) was a close match to the predicted value of 1346.6250 Da. The use of hydroxylamine for protein delipidation is not expected to influence on the high-sensitivity determination of BPL modifications of proteins in terms of the structural stability and mass spectrometric ionization efficiency of the derivatives.
3.5 Structural and functional effects of BPL inactivation on influenza virus antigens
As mentioned, chemical reaction of viral proteins with BPL on active residues is essential for the full inactivation of influenza viruses used in vaccine preparations, and our analyses have confirmed the presence of extensive modifications on polar amino acids located at the surfaces of the proteins. The extent of the reaction, however, needs to be tightly controlled to retain the structural integrity of the viral protein complexes and immunogenicity of influenza vaccines. Reagents selected to inactivate the virus have to meet several criteria including small molecule size and high reactivity to viral proteins, such as formaldehyde or BPL. Formaldehyde is a well-known cross-linking agent that is often used to inactivate and stabilize proteins and reacts with the side-chains of Arg, Cys, His, and Lys residues to form methylol groups, Schiff bases, and methylene bridges 31,32. In a similar manner, BPL is capable of interacting with various amino acids, including Cys, Met, His, Asp, Glu, Tyr, Lys, Ser, and Thr, yielding alkylated and acylated products 7,33. However, unlike formaldehyde, BPL modification can cause extensive formation of bis-alkylated groups; undesired long chains that can significantly affect the function of protein antigens.
Figure4A shows the extracted ion chromatograms of an unmodified peptide along with the +72 Da and +144 Da modification states. MS/MS analysis of the ion at m/z 570.29 (Fig.4B) identified a tryptic HA peptide (residues 148–159) with carbamidomethylation of Cys153. The similar MS/MS fragmentation patterns of the peptides at +72 Da (m/z 606.30, Fig.4C) and +144 Da (m/z 642.31, Fig.4D) confirmed the same peptide backbone sequence and the mass shift initiating at the fragment ion of y6 indicates His155 as the specific residues that was modified by either a single or double BPL molecules. The MS/MS spectrum did not provide definitive evidence for the structure of the bis-alkylated side chain, therefore it is reasonable to assume the linear side chair as found by Uittenbogaard 7. His155 is originally a positively charged basic residue, and localized at the specific antigenic site Ca2 within the receptor-binding pocket (Fig.5). The polarity of this amino acid was reduced by BPL, and the extended length of side-chain may mask the antigenic epitopes of HA.
Figure 4.
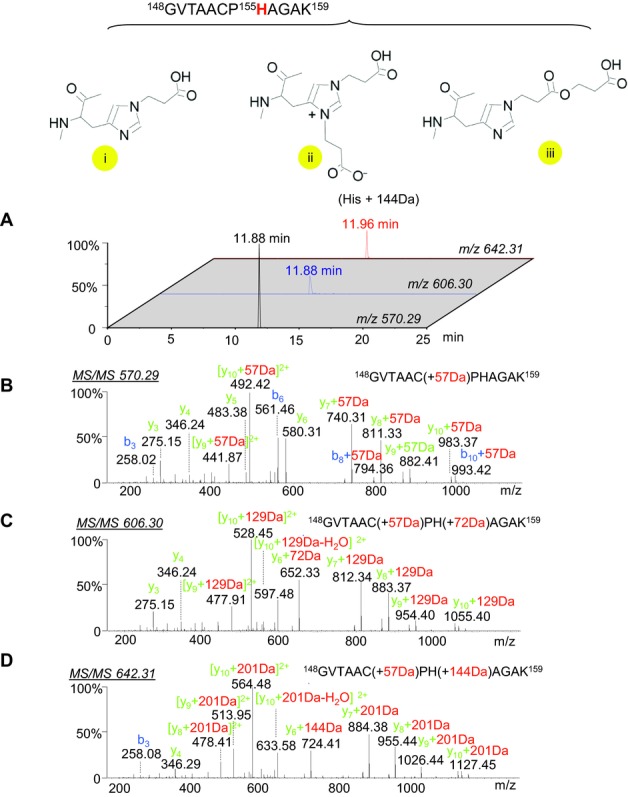
Determination of the double BPL-modified histidine-containing peptides of HA with a mass increase of +144 Da. (A) Combined extracted ion chromatogram of m/z 570.29, m/z 606.30, and m/z 642.31; (B–D) MS/MS spectra of the carbamidomethyl-cysteine-modified peptide at m/z 570.29, the peptide-containing a single BPL-modified histidine at m/z 606.30, and the peptide containing a double BPL-modified histidine at m/z 642.31.
Figure 5.
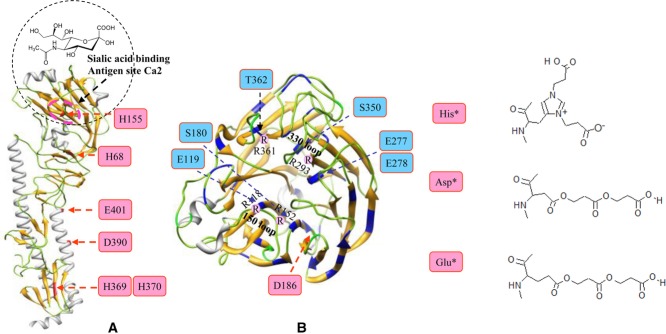
Structural assignment of the BPL-modified residues on the surface regions of influenza virus antigens. (A) hemagglutinin (A/California/04/2009 strain, PDB: 3LZG) and (B) neuraminidase-receptor (sialic acid) binding region (PDB: 3NSS). The single BPL-modified amino acids are highlighted in blue, and the double BPL-modified residues are highlighted in pink.
Using this strategy, other HA histidine residues (His68, His369, and His370) were also shown to be modified by both single and double BPL molecules, the observation supported by MS/MS analyses of the peptides from various enzymatic digests (Supporting Information Table 4). The protein 3D structure shows that His369 and His370 are positioned at the surface region of HA stem in the vicinity of the “fusion peptide” (Fig.5A). Recent study has shown that BPL treatment of HA inhibits the pH-induced conformational change associated with virus fusion 13, it is tempting to speculate that the modifications at His369 and His370 may play a role in the inhibition. In addition to histidine residues, initial database search results also identified the BPL-modified glutamic acids at positions Glu117, Glu241, Glu401, and Glu441 of HA (NIBRG-121xp). However, a thorough manual examination confirmed that these identities were all false positives except Glu401, mainly caused by mis-assignment of the charge states of the peptide ions as shown in Supporting Information Table 4.
Similar to the vaccine derived from the strain NIBRG-121xp, the vaccine from the strain NYMC-X181A has well-conserved histidine modifications at His68, His155, His369, and His370 of HA, but also a bis-alkylated site at aspartic acid residue, Asp390. The MS/MS spectrum of a triply charged ion at m/z 750.05 (Supporting Information Fig. 6A) contained peaks matching the N-terminal fragments up to b6 (384STQNAI389) and the C-terminal fragments up to y12 (391EITNKVNSVIEK402) of the peptide 384–402 without modification. Therefore the mass increase of 144 Da must be localized at the Asp390 residue suggesting a unique residue double BPL modification. Although these modifications at Asp390 and Glu 401 are of interest, they are located within HA α-helical stem regions and are expected to have little effect on protein function.
In contrast, the protein sequence of NA was rarely modified by bis-alkylation of BPL, with Asp186 being the only site identified in the NIBRG-121xp vaccine. MS/MS analysis of the doubly charged ion at m/z 694.31 (Supporting Information Fig 6B) confirmed the exact location of the +144 Da modification, which initiates at fragment ion b6 in the peptide 181–191 (ASACHDGINWL), corresponding to bis-alkylated BPL at Asp186. Despite there being a large number available, arginine residues of proteins do not appear to extensively react with BPL. Asp186 and other single BPL-modified residues (Glu119, Ser180, Glu277, Glu278, Ser350, and Thr372) surrounding the 150 and 330 loop regions are in close proximity to the critical sites of Arg118, Arg152, Arg293, and Arg361 residues (Fig.5B), yet they remain unmodified. However, the modifications that do occur result in an increase in negatively charged side chains near positively charged arginine residues. This could potentially weaken the intramolecular electrostatic interactions of hydrogen bonds and salt bridges around the active site 27,34,35, and consequently impact on the stability and enzymatic activity of NA. This may explain earlier observations that BPL inactivation reduced the NA activity tenfold or more from the human pandemic influenza virus 8. Based on the structural characteristics and polarity of BPL derivatives, our analyses thus identified the key residues in the proteins that may be responsible for a partial loss of the immunogenicity in HA and enzymatic activity in NA.
4 Concluding remarks
Influenza viral genes are translated to ten protein components including HA, NA, polymerase subunits (PB1, PB2, and PA), nucleoprotein (NP), matrix proteins M1 and M2, nonstructural proteins NS1 and NS2. HA and NA are the two major membrane glycoproteins, the primary immunogens, that the immune system responds to influenza virus infection and vaccination. Vaccine quality and efficacy can be affected by the extent of chemical modifications of these proteins caused by formaldehyde or BPL treatments. Currently established proteomic methods have been applied to determine the full-length protein sequence of influenza virus antigens in the complex vaccine samples and to identify their inactivation sites and other related modifications. Unlike formaldehyde, BPL does not react with arginine residues such that positively charged arginine residues are well preserved for receptor binding. We demonstrated that cysteines of HA and NA were not modified by BPL during vaccine inactivation, and presume they are stabilized and/or protected by inter- or intramolecular protein disulfide bridges, oxidization, or acylation (palmitoylation or stearoylation) in the folded HA homotrimers and NA tetramers. Our analyses reliably identified the exact sites of BPL modifications of HA and NA in two vaccines derived from the seed strains NIBRG-121xp and NYMC-X181A. The findings help to explain the effects of BPL inactivation which may cause partial loss of the immunogenicity of influenza vaccines by steric hindrance from the adjacent double BPL-modified His155 to the antigenic sites of HA. Also, enzymatic activity of NA was likely inhibited by charge-dependent reactions of BPL with surrounding residues around the catalytic cavity region of the protein, resulting in significant change of intra- and intermolecular interaction between the amino acids and the binding ligand. Taken together, proteomic analysis of BPL modifications can provide valuable insights into the structure and function of proteins in response to influenza virus inactivation. With the knowledge gained from this study, one would be able to design mutant strains to improve the antigenicity and immunogenicity of influenza vaccines by avoiding the formation of long poly-propiolactone side chains on HA and NA upon BPL inactivation.
In terms of large datasets on analyzing membrane proteins, proteomic identification of nonspecific peptide digests still remains a technical challenge, due to “false positives” from automated database searches resulting from the mis-interpretation of peptide sequences and PTMs. Confident identification can be achieved by high-resolution accurate mass measurements, isotopic peak pattern analysis, and manual validation of the identified peptide sequences with MS/MS. Efforts to quantify these peptide pairs were also undertaken by integrating peak area of the extracted ion chromatograms. As an example, the oxidized level of cysteine-containing peptides was estimated to account for approximately 2–3% of the nonmodified counterparts. However, accurate quantification of BPL-modified species is difficult due to the shift of retention times on a RP C18 column, as well as the variation in ionization efficiencies of peptides with different modifications. As a consequence, further investigation to develop sensitive and accurate quantitation methods through incorporation of stable isotope labeling will be required for efficacy measurement, evaluation, and quality control of inactivating viruses during vaccine production.
Acknowledgments
The authors have declared no conflict of interest.
Glossary
- BPL
β-propiolactone
- FA
formic acid
- HA
hemagglutinin
- LTQ FT-ICR
linear ion-trap FT-ICR
- NA
neuraminidase
- PGC
porous graphitic carbon
- QqTOF
quadrupole TOF
- UPLC
ultra performance LC
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site
Supplementary
Supplementary
5 References
- 1.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu. Rev. Pathol. 2008;3:499–522. doi: 10.1146/annurev.pathmechdis.3.121806.154316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood JM, Robertson JS. From lethal virus to life-saving vaccine: developing inactivated vaccines for pandemic influenza. Nat. Rev. Microbiol. 2004;2:842–847. doi: 10.1038/nrmicro979. [DOI] [PubMed] [Google Scholar]
- 3.Chen JR, Ma C, Wong CH. Vaccine design of hemagglutinin glycoprotein against influenza. Trends Biotechnol. 2011;29:426–434. doi: 10.1016/j.tibtech.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Robertson JS, Nicolson C, Harvey R, Johnson R, et al. The development of vaccine viruses against pandemic A (H1N1) influenza. Vaccine. 2011;29:1836–1843. doi: 10.1016/j.vaccine.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 5.Nicolson C, Harvey R, Johnson R, Guilfoyle K, et al. An additional oligosaccharide moiety in the HA of a pandemic influenza H1N1 candidate vaccine virus confers increased antigen yield in eggs. Vaccine. 2012;30:745–751. doi: 10.1016/j.vaccine.2011.11.081. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson KG, Wood JM, Zambon M. Influenza. Lancet. 2003;362:1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uittenbogaard JP, Zomer B, Hoogerhout P, Metz B. Reactions of beta-propiolactone with nucleobase analogues, nucleosides, and peptides: implications for the inactivation of viruses. J. Biol. Chem. 2011;286:36198–36214. doi: 10.1074/jbc.M111.279232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonges M, Liu WM, van der Vries E, Jacobi R, et al. Influenza virus inactivation for studies of antigenicity and phenotypic neuraminidase inhibitor resistance profiling. J. Clin. Microbiol. 2010;48:928–940. doi: 10.1128/JCM.02045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuya Y, Regner M, Lobigs M, Koskinen A, et al. Effect of inactivation method on the cross-protective immunity induced by whole ‘killed’ influenza A viruses and commercial vaccine preparations. J. Gen. Virol. 2010;91(Pt 6):1450–1460. doi: 10.1099/vir.0.018168-0. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein MA, Tauraso NM. Effect of formalin, beta-propiolactone, merthiolate, and ultraviolet light upon influenza virus infectivity chicken cell agglutination, hemagglutination, and antigenicity. Appl. Microbiol. 1970;19:290–294. doi: 10.1128/am.19.2.290-294.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnsworth A, Cyr TD, Li C, Wang J, et al. Antigenic stability of H1N1 pandemic vaccines correlates with vaccine strain. Vaccine. 2011;29:1529–1533. doi: 10.1016/j.vaccine.2010.12.120. [DOI] [PubMed] [Google Scholar]
- 12.Budowsky EI, Smirnov YA, Shenderovich SF. Principles of selective inactivation of viral genome. VIII. The influence of β-propiolactone on immunogenic and protective activities of influenza virus. Vaccine. 1993;11:343–348. doi: 10.1016/0264-410x(93)90197-6. [DOI] [PubMed] [Google Scholar]
- 13.Desbat B, Lancelot E, Krell T, Nicolaï MC, et al. Effect of the β-propiolactone treatment on the adsorption and fusion of influenza A/Brisbane/59/2007 and A/New Caledonia/20/1999 virus H1N1 on a dimyristoylphosphatidylcholine/ganglioside GM3 mixed phospholipids monolayer at the air-water interface. Langmuir. 2011;27:13675–13683. doi: 10.1021/la2027175. [DOI] [PubMed] [Google Scholar]
- 14.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 15.Drisdel RC, Alexander JK, Sayeed A, Green WN. Assays of protein palmitoylation. Methods. 2006;40:127–134. doi: 10.1016/j.ymeth.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Creskey MC, Smith DG, Cyr TD. Strain identification of commercial influenza vaccines by mass spectrometry. Anal. Biochem. 2010;406:193–203. doi: 10.1016/j.ab.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Kordyukova LV, Serebryakova MV, Baratova LA, Veit MS. acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 2008;82:9288–9292. doi: 10.1128/JVI.00704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kordyukova LV, Serebryakova MV, Baratova LA, Veit M. Site-specific attachment of palmitate or stearate to cytoplasmic versus transmembrane cysteines is a common feature of viral spike proteins. Virology. 2010;398:49–56. doi: 10.1016/j.virol.2009.11.039. [DOI] [PubMed] [Google Scholar]
- 19.Air GM. Nucleotide sequence coding for the “signal peptide” and N terminus of the hemagglutinin from an Asian (H2N2) strain of influenza virus. Virology. 1979;97:468–472. doi: 10.1016/0042-6822(79)90358-1. [DOI] [PubMed] [Google Scholar]
- 20.Krokhin OV, Antonovici M, Ens W, Wilkins JA, et al. Deamidation of -Asn-Gly- sequences during sample preparation for proteomics: consequences for MALDI and HPLC-MALDI analysis. Anal. Chem. 2006;78:6645–6650. doi: 10.1021/ac061017o. [DOI] [PubMed] [Google Scholar]
- 21.Polevoda B, Sherman F. N-terminal acetylation of eukaryotic proteins. J. Biol. Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 22.Wang CC, Chen JR, Tseng YC, Hsu CH, et al. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc. Natl. Acad. Sci. U S A. 2009;106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison WS. Formation and reactions of sulfenic acids in proteins. Accts. Chem. Res. 1976;9:293–299. [Google Scholar]
- 24.Rehder DS, Borges CR. Cysteine sulfenic acid as an intermediate in disulfide bond formation and nonenzymatic protein folding. Biochemistry. 2010;49:7748–7755. doi: 10.1021/bi1008694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 26.Xu R, Ekiert DC, Krause JC, Hai R, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu X, Zhu X, Dwek RA, et al. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J. Virol. 2008;82:10493–10501. doi: 10.1128/JVI.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterfield M, Scrace G, Skehel J. Disulphide bonds of haemagglutinin of Asian influenza virus. Nature. 1981;289:422–424. doi: 10.1038/289422a0. [DOI] [PubMed] [Google Scholar]
- 29.Ward CW, Colman PM, Laver WG. The disulphide bonds of an Asian influenza virus neuraminidase. FEBS Lett. 1983;153:29–33. doi: 10.1016/0014-5793(83)80113-6. [DOI] [PubMed] [Google Scholar]
- 30.Varghese JN, Laver WG, Colman PM. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 A resolution. Nature. 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 31.Metz B, Kersten GF, Hoogerhout P, Brugghe HF, et al. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J. Biol. Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- 32.Metz B, Kersten GF, Baart GJ, de Jong A, et al. Identification of formaldehyde-induced modifications in proteins: reactions with insulin. Bioconjug. Chem. 2006;17:815–822. doi: 10.1021/bc050340f. [DOI] [PubMed] [Google Scholar]
- 33.Taubman MA, Atassi MZ. Reaction of beta-propiolactone with amino acids and its specificity for methionine. Biochem. J. 1968;106:829–834. doi: 10.1042/bj1060829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J. Biol. Chem. 2010;285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yen HL, Hoffmann E, Taylor G, Scholtissek C, et al. Importance of neuraminidase active-site residues to the neuraminidase inhibitor resistance of influenza viruses. J. Virol. 2006;80:8787–8795. doi: 10.1128/JVI.00477-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary
Supplementary


