Abstract
PIWI-interacting RNAs (piRNAs) are a complex class of small non-coding RNAs that are mostly 24–32 nucleotides in length and composed of at least hundreds of thousands of species that specifically interact with the PIWI protein subfamily of the ARGONAUTE family. Recent studies revealed that PIWI proteins interact with a number of proteins, especially the TUDOR-domain-containing proteins, to regulate piRNA biogenesis and regulatory function. Current research also provides evidence that PIWI proteins and piRNAs are not only crucial for transposon silencing in the germline, but also mediate novel mechanisms of epigenetic programming, DNA rearrangements, mRNA turnover, and translational control both in the germline and in the soma. These new discoveries begin to reveal an exciting new dimension of gene regulation in the cell.
Keywords: PIWI, piRNA, TDRD, epigenetic, RNA decay, translational regulation
INTRODUCTION
Study of non-coding RNAs represents one of the most exciting frontiers of current biomedical research. Non-coding RNAs are commonly classified as long non-coding RNAs (lncRNAs) that are generally longer than 200 nucleotides (nt) and small non-coding RNAs (sncRNAs) that are mostly 20–35 nt [1]. Among sncRNAs, microRNAs (miRNAs) and small interfering RNAs (siRNAs) are generally 21 nt in length and have been extensively studied, whereas PIWI-interacting RNAs (piRNAs), mostly 24–32 nt in length, and other types of sncRNAs were discovered more recently and are thus less well characterized. Among all types of coding and non-coding RNAs, piRNAs are by far the most numerous, existing in at least hundreds of thousands of species in a multicellular organism, mostly in its germline. This number far exceeds the total number of all other types of known RNAs altogether. Furthermore, piR-NAs correspond to all types of genomic sequences (see below). These two salient features of piRNAs imply their potential function in diverse forms of gene regulation.
piRNAs are defined by their specific binding to the PIWI subfamily of ARGONAUTE (AGO)/PIWI family proteins. This is in contrast to miRNAs and siRNAs, both of which specifically bind to the AGO subfamily of the AGO/PIWI family proteins. PIWI proteins are required for piRNA biogenesis and function, and play a central role in germline development and gametogenesis (reviewed in [2]). Piwi mutants in Drosophila, C. elegans, mice, and zebrafish cause gametogenic defects such as failure in germline establishment, loss of germline stem cells (GSCs) meiotic arrest, and blockage in spermiogenesis, leading to sterility (reviewed in [3]). Because piRNAs are derived from the RNA transcripts of transposons, protein-coding genes, and specific intergenic loci [4–8], piRNAs are thought to act as sequence-specific guides of PIWI proteins to regulate the expression of genes and transposons at both transcriptional and post-transcriptional levels [9–11]. Most recently, PIWI proteins and piRNAs have also been found in somatic tissues [12]. Reviews on the PIWI–piRNA pathway have mostly been focused on its biogenesis and function in transposon silencing in the germline. Here, we will first give a brief update on these two topics with focus on PIWI proteins and their main interactors, TUDOR-domain-containing proteins. We will then review the regulatory function of the PIWI–piRNA pathway beyond transposon silencing both in germline and somatic cells.
AGO PROTEINS AND THEIR INTERACTING PROTEINS—AN OVERVIEW
The AGO protein family was first discovered as a gene family with stem cell function well conserved in both animal and plant kingdoms [13]. The AGO family proteins are defined by the presence of the highly conserved Piwi–Argonaute–Zwille (PAZ) and PIWI domains, as well as the less conserved N-terminal (N) and Middle (Mid) domains [14]. Structural studies revealed that the PAZ domain comprises an oligonucleotide-binding fold which anchors the two-nucleotide 3′-overhang of siRNAs resulted from digestion by RNase III (a step in the processing of siRNAs) [15–17], whereas the PIWI domain shows extensive homology to RNase H. In many AGO/PIWI proteins, such as the Drosophila Piwi protein (herein and hereafter, all-capitalized protein names, such as AGO and PIWI, are used as family and subfamily names, whereas first -letter-capitalized names are for individual proteins, such as Ago 1 and Piwi), the PIWI domain exhibits endonuclease activity, also known as the slicer activity [4]. However, this activity is not required for some AGO proteins, such as the Drosophila Piwi protein, for their function [18,19]. Based on phylogenetic analysis, the AGO/PIWI family is divided into two subfamilies: the AGO subfamily and the PIWI subfamily. In most organisms, PIWI subfamily proteins are mainly expressed in the germline, in contrast to AGO subfamily proteins that are ubiquitously expressed in all tissues (reviewed in [3]).
PIWI proteins interact with a number of proteins, such as Armitage (Armi) and Zucchini (Zuc) in Drosophila, to regulate the piRNA biogenesis (Table 1). Among these factors, the most predominant class is Tudor-domain-containing proteins (a.k.a. Tudor domain-related proteins, TDRDs). Their function and interaction with PIWI protein is becoming increasingly studied, whereas little is known about how other proteins interact with PIWI proteins. Therefore, this review highlights TDRD proteins as PIWI interactors.
Table 1.
List of PIWI-interacting proteins involved in the piRNA pathway.*
| TUDOR family proteins Flyhomolog | Mammalianhomolog | Proteindomain(s) | Interactingprotein(s) | Reference(s) |
|---|---|---|---|---|
| CG9684/CG9925 | Tdrd1/Mtr-1 | TUDOR(x2/x4), MYND | Miwi, Mili, Miwi2, Tdrd12 | [36,38,42,44,106,109] |
| PAPI | Tdrd2/Tdrkh | TUDOR, KH(x2) | Piwi, Ago3, Tral, Me31B, TER94, Miwi, Mili, Miwi2 | [33,35,42,108] |
| Kumo/Qin | Tdrd4/RNF17 | TUDOR(x5), RING, B-box | Piwi, Aub, Ago3, Vas, Spn-E, HP1, Miwi | [69,70,105] |
| Tej | Tdrd5 | TUDOR, Lotus | Aub, Vas, Spn-E | [41,45] |
| Tud | Tdrd6 | TUDOR (x11) | Aub, Ago3, Vls, Mili, Miwi, VASA, Tdrd1, Tdrd7 | [29,42,67,107,110,153] |
| CG8920 | Tdrd7/TRAP | TUDOR(x3), Lotus (x2) | Miwi,Tdrd1,Tdrd6, VASA | [42,43,110] |
| Spn-E | Tdrd9 | TUDOR, DEXDc, HELICc, HA2 | Aub, Vas, Tej, Miwi, Mili, Miwi2 | [37,40,42] |
| Yb(Femalesterile(1)Yb) | Tdrd12/ECAT8 | TUDOR, DEAD, Helicase, Helicase-like, ZnF | Piwi, Armi, Vret, Mili, Tdrd1 | [44,52–55] |
| BoYb | Tdrd12/ECAT8 | TUDOR, DEAD, Helicase, ZnF | Mili, Tdrd1 | [44,57] |
| SoYb | Tdrd12/ECAT8 | TUDOR (x2), DEAD, Helicase, ZnF | Mili, Tdrd1 | [44,57] |
| Vret | None | TUDOR (x2), RRM, MYND | Piwi, Aub, Ago3, Yb, Armi | [56,57] |
| Other known PIWI-interacting proteins | ||||
| Vas | VASA/MVH | DEAD, HELICc | Piwi, Aub, Tej, Spn-E, Miwi, Mili, Tdrd6 | [40,86,90,107,154] |
| Armi | MOV10,MOV10L1 | Helicase | Piwi, Yb, Vret, Miwi, Mili, Miwi2, Tdrd1 | [44,52–55,155,156] |
| Zuc | mitoPLD | PLD-family nuclease | Aub | [58,157–159] |
| Squ (Squash) | None | RNaseHII-like | Aub | [58] |
| Mael | Maelstrom | HMG-box | MTOC proteins, Miwi, Mili, VASA | [68,125,160] |
| Hen1 | HEN1 | dsRBD, SAM-methyltransferase | Piwi, Aub, Ago3 | [62,161] |
| Hsp83 | Hsp90 | Hsp90, ATPase | Piwi, Hop, Fkbp6 | [59,72,73,162] |
| Shu | Fkbp6 | FKBP, TPR | Piwi, Armi, Hsp90 | [59,60,162] |
| dPRMT5 | PRMT5/Capsuleen(Csul) | SAM-methyltransferase | Vls, Miwi, Mili, Miwi2 | [29,42,153] |
| Vls (Valois) | MEP50 | WD(x4) | dPRMT5, Tud, Miwi, Mili, Miwi2 | [42,153] |
Blue and red words indicate Drosophila and mammalian proteins, respectively.
B-box, B-box zinc finger domain; DEAD, DEAD-box helicase; DEXDc, DEAD-like helicase domain; dsRBD, double-stranded RNA-binding domain; FKBP, FK506-binding protein domain; HA2, helicase-associated domain; HELICc, helicase C-terminal domain; HMG-box, high mobility group box; KH, K homology domain; MBD, methyl-CpG-binding domain; MYND, zinc-finger myeloid-nervy-DEAF-1 domain; N.D., not determined; PAZ, PIWI/Argonaute/Zwille; PIWI, P-element-induced wimpy testes; PIWIL1–4, Piwi-like protein 1–4; PLD, phospholipase D; Pre-SET, pre-SET domain; RING, really interesting new gene finger domain; RRM, RNA recognition motif domain; SAM, S-Adenosyl methionine; SET, Su(var)3–9/Enhancer-of-zeste/Trithorax domain; TPR, tetratricopeptide-repeat domain; WD, WD-repeat domain; ZnF, zinc finger.
TDRD proteins belong to the TUDOR protein family, which received the name from the Drosophila tudor gene. Tudor was discovered in a genetic screen for maternal effect mutations that cause lethality or sterility in the progeny [20]. Subsequently, the Tudor domain has been found in proteins from a broad range of eukaryotes, including fission yeast, fungi, plants, and animals. Based on the sequence and structural similarity, Tudor domain, together with chromatin-binding (Chromo), malignant brain tumor (MBT), PWWP (conserved Proline and Tryptophan), and plant Agenet domains, comprises the Tudor domain ‘Royal Family’ [21]. Tudor domain has a ~60 amino acid core structure composed of four antiparallel β-strands that form a barrel-like structure with an aromatic binding pocket at the surface to accommodate methylated lysine/arginine ligands [21]. Tudor domain exists singly or in multiple copies, in the absence or in conjunction with other types of domains. The versatile protein domain architecture confers a diverse protein–protein/RNA interacting network and endows a wide variety of functions including RNA metabolism, spliceosome assembly, piRNA biogenesis, histone modification, and germline development to the TUDOR family members [20,22–32].
Recent studies have revealed that PIWI subfamily proteins, but not AGO proteins, are arginine-methylated by protein methyltransferase 5 (dPRMT5, also known as Capsuleen/Dart5) to form symmetrically dimethylated arginines (sDMAs) at their N-termini [29,33–35]. The sDMAs of PIWI proteins serve as binding motifs for TDRDs and play crucial roles in PIWI function [29,30,33–38]. Several TDRDs, such as Spindle-E (Spn-E), Tudor (Tud), Krimper (Krimp), and Tejas (Tej) in Drosophila [29,30,35,39–41], as well as Tdrd1, Tdrd2, Tdrd4, Tdrd5, Tdrd6, Tdrd7, Tdrd9, and Tdrd12 in mice [33,36–38,42–45], have recently been shown to be required for PIWI function.
PIWIs, TUDORS, AND OTHER PROTEINS IN piRNA BIOGENESIS
piRNAs was discovered and defined as a novel class of sncRNAs that bind to PIWI subfamily proteins in mammalian testes in 2006 [5–8]. In fact, the piRNA-like RNAs were identified even earlier in plants [46] and trypanosomes [47]. These RNAs contain sequences matching repetitive intergenic elements and thus are termed repeat-associated small RNAs (rasiRNAs). Subsequent studies in Drosophila ovaries further reported that PIWI proteins are associated with small RNAs identified as rasiRNAs [4,39,48]. Therefore, rasiRNAs are essentially piRNAs [48,49]. Genetic studies have demonstrated that the RNase III-like enzyme Dicer, which is necessary for processing miRNAs and siRNAs, is dispensable for piRNA biogenesis [39,48], and that piRNAs precursors are single-stranded RNAs. Furthermore, deep sequencing of small RNAs associated with Drosophila Piwi, Aubergine (Aub), and Argonaute 3 (Ago3) revealed a number of characteristics of piRNAs [4,9,48,50]. For example, in Drosophila, piRNAs are mainly derived from intergenic repetitive sequences in the genome, including transposable elements (TEs) and their remnants [4,9]. These loci are collectively called ‘piRNA clusters’ [48]. In addition, Piwi and Aub preferentially bind to piRNAs derived from the antisense strand of retrotransposon coding sequences that show a strong bias toward a U at the 5′ end (1U), whereas Ago3 preferentially binds sense-strand piR-NAs that tend to have an A at position 10 (10A) [48,50]. Moreover, piRNAs from opposite strands contain a 10 base pair (bp) overlap of complimentary sequence [48,50]. Based on these observations, two models for piRNA biogenesis have been proposed: the primary pathway and the ping-pong cycle (the secondary pathway) (Fig. 1).
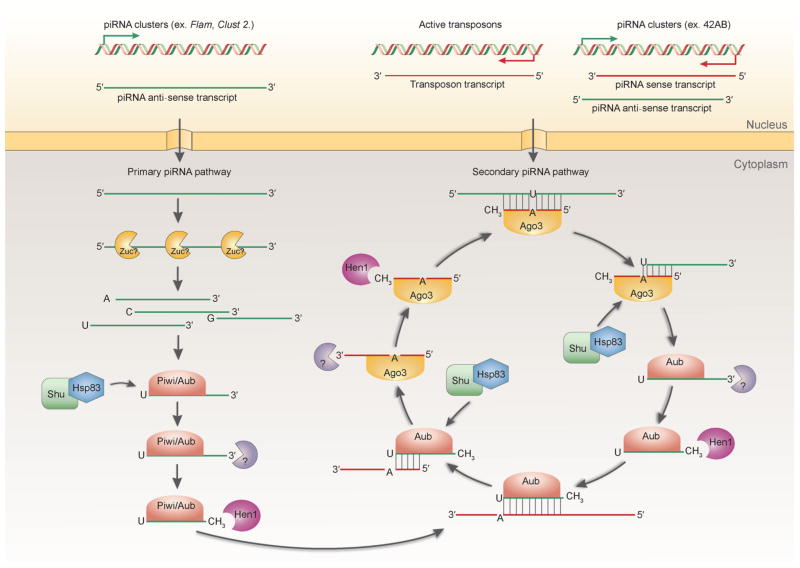
Figure 1.
piRNA biogenesis pathways in Drosophila. In the primary processing pathway in the germline, the antisense piRNA precursor transcripts produced from piRNA clusters are possibly processed by Zuc, a putative endonuclease [53,58]. The resulting piRNA intermediates with a U at the first nucleotide position are preferentially selected and loaded onto Piwi or Aub (In ovarian somatic cells, only Piwi is expressed and involved in the primary piRNA pathway). Hsp83 and its hypothesized co-chaperone Shu are required for the piRNA loading [59,60]. The 3′ end of piRNA intermediates is further trimmed by an unknown enzyme [61], and then methylated by the methyltransferase Hen1 [62,63] to mature the piRNAs and complete primary processing pathway. Aub and its bound mature antisense piRNAs can then enter the secondary processing pathway (the ping-pong cycle). piRNAs guide Aub to cleave the transposon transcripts and sense piRNA precursors produced from piRNA clusters [48,50]. The resulting products are then loaded onto Ago3 with the help of Shu and Hsp83 [59,60], and undergo 3′ end trimming and methylation to mature the sense piRNAs [61–63]. In turn, Ago3 and its associated mature sense piRNAs further cleave the target antisense piRNA precursors based on the sequence complementarity [48,50]. The resulting antisense piRNA intermediates are loaded onto Aub, with the assistance of Shu and Hsp83 [59,60], and further trimmed and methylated at the 3′ end as in the primary processing pathway [61–63]. The piRNA biogenesis pathways are well conserved across species such as C. elegans, fish, and mice (reviewed in [3]). Figure modified from [163].
The primary pathway
In Drosophila gonadal somatic cells, where Piwi, but not Aub or Ago3, is expressed [51], the single-stranded piRNA precursors are transcribed and then processed by a Dicer-independent machinery to generate piRNA intermediates [39,48]. These piRNA intermediates are delivered from the nucleus to the Yb body, the cytoplasmic ribonucleoprotein (RNP) complex comprising Piwi, RNA helicase Armi, and TDRD proteins Yb, Vreteno (Vret), and Sister of Yb (SoYb) [52–57]. The intermediate piRNAs are subsequently processed at the 5′ end possibly by Zuc, a putative endoribonuclease that localizes to mitochondria [53,58]. The intermediate piRNAs are loaded onto Piwi with the help of Shutdown (Shu) co-chaperone protein and Heat shock protein 83 (Hsp83) chaperone protein [59,60]. After loading onto the Piwi protein, the 3′ end of piRNAs is trimmed by an unknown enzyme [61] and then methylated by the methyltransferase Hen1 to form mature piRNAs [62,63]. The Piwi–piRNA complex is then transported into the nucleus to exert transposon silencing and gene regulation functions. Without piRNA loading, Piwi is not localized to the nucleus. These findings suggest that the Yb body not only generate mature primary piRNAs for Piwi, but also scrutinize the functional Piwi–piRNA complex before it enters the nucleus [52–55]. Similarly to gonadal somatic cells, germ cells also produce primary piRNAs that are loaded onto both Piwi and Aub. The primary piRNAs can further initiate the ping-pong cycle of piRNA biogenesis [48,64].
The secondary pathway—the ping-pong cycle
In Drosophila, the ping-pong cycle is primed by both maternally deposited piRNAs through germline transmission and zygotic primary piRNAs that are antisense to retrotransposon coding strands and loaded onto Aub [57,64–66]. In germ cells, Piwi shows steady-state localization to the nucleus [51], whereas Aub and Ago3 localize to the nuage and associate with Tudor [29,67]. Tudor might serve as a scaffold protein to recruit other piRNA biogenesis factors and initiate the ping-pong cycle. During the ping-pong amplification cycle, Ago3 binds to sense-strand piRNAs and catalyzes antisense-strand cleavage at an A:U bp that generates the 5′ end of anti-sense piRNAs [48,50]. The 5′ ends of the resulting cleavage products are loaded onto Aub with the help of Shu and Hsp83 [59] and then further trimmed by an unknown enzyme and methylated by Hen1 at the 3′ end to generate mature antisense piRNAs [62,63]. Shu and Hsp83 have been hypothesized but not been demonstrated to directly interact with each other for this function [59,60]. The antisense piRNAs then guide Aub to cleave sense-strand RNAs and generate the 5′ end of sense piRNA precursors that associate with Ago3 [48,50], again with the help of Shu and Hsp83 [59]. Further 3′ end processing by an unknown nuclease and methylation by Hen1 produce mature sense-strand piRNAs and complete the ping-pong cycle [62,63].
In addition to Aub, Ago3, and Tudor, recent studies have shown that many of the ping-pong cycle components localize to the nuage, such as the RNA helicase Armi, the DEAD-box helicase Vasa (Vas), and the HMG protein Maelstrom (Mael), as well as the TDRD proteins Spn-E, Kumo, Krimp, Vret, and Tej [40,41,67–70]. Their nuage localization suggests that the nuage is the cytoplasmic site where the ping-pong cycle takes place. Mutants of the nuage components such as vas, mael, armi, zuc, squ, krimp, and tej exhibit defects in piRNA production [40,41,58,71], supporting that the nuage is involved in the piRNA pathway. Despite these findings, the exact molecular functions of nuage and most of the nuage components remain largely to be investigated.
Although there are some distinguishing factors between the primary pathway and the ping-pong cycle, many proteins are involved in both. The common factors include the methyltransferase Hen1, the chaperone Hsp83, the co-chaperone Shu, and the TDRD protein Vret. While the knowledge about the precise roles of these proteins is still limited, mutations of any of them disrupt the normal function of PIWI proteins and piRNAs decrease dramatically [52–57,59,60,62,63,72,73].
Features of the Drosophila piRNA system have been identified in a number of other organisms, such as human, mice, rat, platypus, marsupials, zebrafish, planaria, silkworm, C. elegans, and Hydra. For example, signatures of the ping-pong cycle also exist in mice [5,6,74,75]. Moreover, piRNAs are also found in human and rat, with major clusters occurring in syntenic locations [6]. Zebrafish has also been demonstrated to have the piRNA ping-pong cycle [76,77]. Additionally, planaria piRNAs are in part organized in genomic clusters and share characteristic features with mammalian and fly piRNAs [78]. In C. elegans, 21U-RNAs, the piRNAs of nematodes, display 5′U bias and their precursors are not dsRNAs [79]. Furthermore, these 21U-RNAs complex with PRG-1, one of two C. elegans PIWI proteins, to regulate spermatogenesis [80,81]. In Hydra, two PIWI proteins, Hywi and Hyli, and piRNAs have been identified. Hywi- and Hyli-associated piRNAs show strong ping-pong signature. Furthermore, hywi function is shown to be essential in the somatic epithelial lineages [82]. This is the lowest eukaryote in which PIWI proteins and piRNAs have been found so far. Given that only AGO protein is present in fission yeast S. pombe but not baker’s yeast S. cerevisiae, it is clear that the separation between AGO and PIWI must occur in multicellular eukaryotes with germ cells.
THE PIWI–piRNA PATHWAY IN GERMLINE DEVELOPMENT
The PIWI–piRNA pathway has been intensively studied for their function in the germline. Several studies have shown that the PIWI–piRNA pathway is implicated in germline development, epigenetic regulation, transposon silencing, mRNA turnover, and translational control (Fig. 2). Importantly, these functions of PIWI proteins and the piRNA pathway have been demonstrated in a number of organisms, suggesting that this mechanism is evolutionarily conserved [4,8,34,39,48,50,64,76,83].
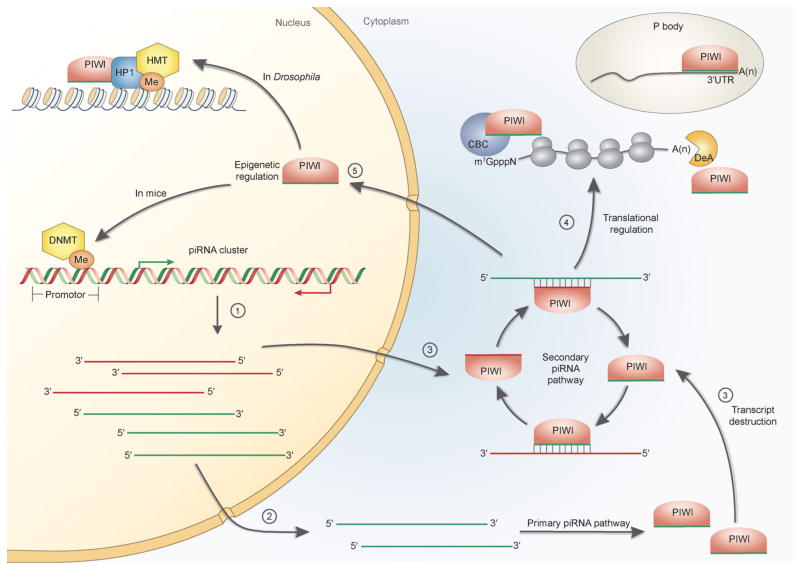
Figure 2.
Known gene regulation mechanisms mediated by the PIWI–piRNA pathway. PIWI proteins and piRNAs regulate the expression of genes and transposon at both transcriptional and post-transcriptional levels. (1) Sense and antisense piRNA precursor transcripts are produced in the nucleus. (2) Antisense piRNA precursor transcripts are transported to the cytoplasm and processed by the primary biogenesis pathway to generate mature sense piRNAs that associate with PIWI proteins. (3) The PIWI–antisense piRNA complexes mediate cleavage of sense piRNA precursors and transposon (and protein-coding) transcripts, which silences transposon and gene expression at the post-transcriptional level. The resulting sense transcripts are taken up by PIWI proteins responsible for sense piRNA binding. The PIWI–sense piRNA complexes then cleave antisense piRNA precursor transcripts to amplify the piRNA biogenesis cycle [48,50]. (4) The PIWI–piRNA complexes are involved in translational regulations by interacting with polysomes, mRNA cap-binding complex (CBC, in mice) [7,74,94], and mRNA deadenylase (DeA, in Drosophila) [133]. In addition, the PIWI–piRNA complexes are associated with P-body components [37,109,136,137] and piRNAs are mapped to the 3′UTR of mRNAs [131,132]. (5) The PIWI–piRNA complexes can enter the nucleus and regulate gene transcription through epigenetic mechanisms including heterochromatin formation [9,11,114–117] and DNA methylation [10,120] in the promoter region of target genes. 3′UTR, 3′ untranslated region; CBC, cap-binding complex; DeA, deadenylase; DNMT, DNA methyltransferase; HMT, histone methyltransferase; HP1, heterochromatin protein 1; Me, methylation.
In Drosophila, there are five AGO/PIWI proteins: Ago 1 and Ago 2 belong to the AGO subfamily, whereas Piwi, Aub, and Ago3 are in the PIWI subfamily [84]. Piwi, the founding member of the Drosophila AGO/PIWI family, was initially identified in a genetic screen for genes affecting the GSC maintenance [85]. piwi mutant males and females fail to self-renew their GSCs, resulting in germline degeneration and thus sterility [13,85]. Importantly, clonal analyses in adult fly ovaries have revealed that the function of Piwi for GSC maintenance resides in the somatic niche composed of terminal filaments and cap cells, while germline expression of Piwi controls the division of germ cells [13]. Furthermore, maternal Piwi is shown to be required for PGC formation [86]. In addition to Piwi, the other two Drosophila PIWI subfamily members, Aub and Ago3, are also involved in germline development. Genetic studies have demonstrated that Aub is essential for establishing PGCs [86,87]. Besides, the mutations of aub cause transposon derepression and DNA damage accumulation in germ cells that lead to sterility [39,50,88,89]. Similarly, ago3 mutant female flies are reported to be sterile. Homozygous ago3 mutant females lay fewer eggs than the wild-type or heterozygote sibling controls, and these eggs fail to hatch. Moreover, Ago3 may be necessary for GSC maintenance as ago3 mutant males do not contain Vas-positive germ cells adjacent to the hub [34].
PIWI proteins in other organisms are functionally conserved in germline development. There are three murine PIWI proteins, Miwi, Mili, and Miwi2, all of which are expressed during spermatogenesis [90–93]. Mili and miwi2 knockout mice show spermatogenic stem cell arrest [90,93,94]. Only few stem cells in the mili mutant can escape the arrest and produce spermatocytes that are then arrested at prophase I of meiosis [94]. In addition, the conditional inactivation of miwi2 has revealed that miwi2 is essential for male PGC reprogramming but is dispensable for postnatal male germline development and testicular function in mice [95]. In contrast, global knockout of miwi causes post-meiotic arrest of spermatogenic cells [92]. Additionally, Miwi forms RNP complexes that stabilize mRNAs essential for spermiogenesis [96]. PIWI proteins have also been characterized in other vertebrates. For example, zebrafish expresses two PIWI proteins: Ziwi (zebrafish Piwi) and Zili (ziwi-like). Although there is no clear block of germ cell formation in ziwi mutant fish, germ cells undergo increased apoptosis that results in loss of all germ cells [76]. Strikingly, in zili mutant fishes, the number of germ cells is severely decreased and germ cell differentiation is abolished, suggesting that zili is required for GSC maintenance [77]. In C. elegans, depletion of two piwi orthologs, prg-1 and prg-2, leads to meiotic defects [80]. Moreover, PRG-1 has been demonstrated to be essential for fertility [81]. Similar to PIWI proteins in Drosophila, mice, zebrafish, and C. elegans, PIWI proteins in jellyfish and Xenopus have also been reported to participate in germline development, indicating an evolutionarily conserved role of PIWI proteins in the germline [77,97–99].
Recent studies have identified a large group of TUDOR family members expressed in the germline, where they interact with sDMA-modified PIWI proteins to modulate PIWI functions. For example, the Drosophila Tudor, the founding member of the TUDOR protein family, contains 11 Tudor domains [100]. It is a component of the polar granule and is involved in germ plasm assembly and PGC formation [20,26,101]. In addition, Tudor associates with Aub and Ago3, specifically through the Tudor domains and the sDMAs, and regulates the piRNA pathway [29]. In tudor mutants, Aub- and Ago3-associated piRNAs are different from those in the wild-type control, suggesting that Tudor scrutinizes piRNAs and load them onto Aub and Ago3 [29]. In addition to the Tudor protein, several other TDRDs, including Partner of PIWIs (PAPI), Kumo/Qin, Tej, Spn-E, and Vret, also interact with PIWI proteins [35,40,41,52,53,55–57,69,70]. Additionally, a group of TDRDs including Brother of Yb (BoYb), SoYb, Krimp, and dSETDB1 have been demonstrated to participate in piRNA biogenesis [40,57,67,102], but their interacting PIWI partners have not yet been identified.
The crucial roles of TUDOR family proteins in germline development and PIWI functions are evolutionarily conserved. In mice, it has been reported that Tdrd1, Tdrd2, Tdrd4, Tdrd5, Tdrd6, Tdrd7, Tdrd8, Tdrd9, Tdrd10, SND1/Tdrd11, Tdrd12, and SetDB1 are expressed in the germline [33,44,103,104]. Among them, Tdrd1, Tdrd2, Tdrd4, Tdrd5, Tdrd6, Tdrd7, Tdrd9, and Tdrd12 have been demonstrated to be involved in nuage formation, spermatogenesis, and the piRNA pathway [33,38,43–45,105–110].
Recent advances in uncovering PIWI–TDRD interactions have demonstrated an elaborate interplay between these proteins in regulating transposon silencing, germ granule formation, and germ cell development (Table 1). Although more and more TUDOR family proteins have been discovered as PIWI protein interactors or the piRNA pathway components, the precise molecular functions of these proteins in the PIWI–piRNA pathway and how these proteins achieve optimal specificity for natural ligands await systematic investigation.
In Drosophila, many of the piRNA pathway genes such as Piwi, Aub, Armi, Zuc, and Squ were initially identified by screening for mutations that cause GSC loss, abnormal axis specification, defective gametogenesis, and sterility [13,34,51,58,71,87,111,112]. In mice, mili and miwi2 mutants also show defects in spermatogenic stem cell maintenance and a reduction of germ cells in the adult stage [93,94]. Subsequent studies further reported that the mutations of genes involved in DNA damage signaling are sufficient to repress some germline defects caused by the mutations of the piRNA pathway genes. These data suggest that the germline development defects in the piRNA pathway gene mutants may arise from DNA damage [88,93,112,113]. Additionally, upregulation of phosphorylated H2Ax (also called γ -H2Av in Drosophila), a marker for unrepaired DNA double-stranded breaks, has been observed in aub mutant fly ovaries and miwi2 mutant mouse testes [58,93,112,113]. These observations further support the idea that the mutations of the piRNA pathway genes cause DNA damage, which activates the DNA damage response and thus results in germline development defects.
THE PIWI–piRNA PATHWAY IN EPIGENETIC REGULATION
Recent studies have suggested that the PIWI–piRNA pathway is involved in epigenetic regulation through histone modification and DNA methylation (Fig. 2). In Drosophila, Piwi and Aub are found to act as regulators of position-effect variegation (PEV), in which the expression of a euchromatic gene is silenced to variable extent in different cells within the same tissue when it is localized in proximity to a heterochromatic region. This indicates that the piRNA pathway silences gene expression by promoting heterochromatin assembly [114]. Moreover, Piwi is a nuclear protein that physically interacts with Heterochromatin Protein 1a (HP1a) and colocalizes with HP1a on many sites along the chromosomes [9,11,115]. Flies lacking this interaction exhibit ineffective PEV that is caused by a loss of heterochromatin, suggesting that the Piwi–HP1a interaction is required for heterochromatin formation [115]. Besides the role in epigenetic silencing, the Piwi–piRNA complex can also function as an epigenetic activator at the subtelomeric region 3R-TAS1. It has been shown that in piwi mutants, 3R-TAS1 becomes heterochromatic, which indicates the role of Piwi in promoting the euchromatic state of this chromatin locus [9]. Additionally, the insertion of piRNA-complementary sequences into an ectopic site of Drosophila genome causes the recruitment of Piwi, HP1a, and Su(var)3–9 as well as H3K9me enrichment and reduced RNA polymerase II association at this ectopic site [11]. This evidence further supports the notion that piRNA is both necessary and sufficient to recruit Piwi and epigenetic factors to specific genomic loci. The interplay between Piwi, HP1, and H3K9me has also been demonstrated in C. elegans. PRG-1, the Piwi ortholog in C. elegans, can initiate transgenerational gene silencing in the germline by regulating H3K9me, HP1, and histone methyltransferases [116,117]. Importantly, in addition to histone modifications, the epigenetic functions of PIWI proteins can act through DNA methylations. In mili or miwi2 knockout mouse testes, the CpG methylation level in PGCs is reduced, with concordant transposon upregulation [83,118,119]. In addition, piRNAs are shown to direct DNA methylation on a non-transposon locus to regulate genomic imprinting in the mouse male germline [10]. Recent studies have also suggested that piRNAs influence synaptic plasticity of neuronal cells in Aplysia through DNA methylation on non-transposon genomic regions [120].
THE PIWI–piRNA PATHWAY IN TRANSPOSON SILENCING
Several genetic studies have reported that the PIWI–piRNA pathway genes play a critical role in transposon repression. For example, Piwi is required for silencing transposons within the 3amenco (3am) locus which is active in gonadal somatic cells [121]. In addition, the mutations of aub cause increased transposon activity [89,112,122–124]. Moreover, the localization of Aub and Ago3 to the nuage is mutually dependent, and mutations of either gene lead to defects in piRNA production and elevated levels of transposon transcripts [34]. Several piRNA pathway genes have also been reported to be involved in transposon regulation. For example, Mael, an HMG-box-containing protein localized to nuage, is required for Piwi-mediated transposon silencing [125,126]. Rhino (Rhi), an HP1 subfamily member, interacts with Cuto3 (Cu3) to regulate the transcription of 1/42AB piRNA cluster. The mutations of either cu3 or rhi cause defects in piRNAs biogenesis and transposon derepression [127,128]. Importantly, a large number of piRNAs have been mapped to the transposon-containing genomic loci [129]. These observations further support the model that PIWI proteins bind to piRNAs that direct them to silence harmful TEs in the germline [40,41,48,50,58,71]. Similar transposon silencing functions of PIWI proteins and piRNAs have also been demonstrated in other species. For example, mouse pre-pachytene piRNAs associated with Mili or Miwi2 that are expressed at the prenatal stage are mostly derived from repeated elements and transposons [83]. Furthermore, in the germ cells of mili or miwi2 knockout mice, transposons are dramatically upregulated [83,93]. In zebrafish, piRNAs are mainly derived from transposon sequences [76]. In Hydra, piRNA profiling analyses have suggested that transposons are the targets of the piRNA pathway [82,130]. These findings indicate that the piRNA pathway has a conserved function in silencing transposons across species.
THE PIWI–piRNA PATHWAY IN mRNA TURNOVER AND TRANSLATIONAL CONTROL
The slicer activity of Aub and Ago3 not only contributes to the amplification loop of piRNA production, but also silences transposons at the post-transcriptional level (Fig. 2). Furthermore, it has been shown that the 3′ untranslated regions (3′UTRs) of a broad set of protein-coding mRNAs are selected for piRNA production in Drosophila ovaries, mouse testes, and Xenopus eggs. These findings suggest that piRNAs may function in mRNA turnover for cellular regulation [131]. In support of this idea, the protein level of the Traffic Jam (Tj), whose 3′UTR generates abundant sense piR-NAs, is upregulated in piwi mutants [131,132]. Additionally, Aub is in the same complex with mRNA deadenylase CCR4, nanos (nos) mRNA, and piR-NAs that target the nos 3′UTR region in the bulk of the embryo. In aub and ago3 mutant embryos, the deadenylation and decay of nos mRNA are defective and the Nanos protein is ectopically accumulated [133]. Vas protein levels are also upregulated in aub and ago3 mutants, which is possibly caused by the depletion of piRNAs derived from the vas mRNA [34,134]. Several lines of evidence have demonstrated that the PIWI–piRNA pathway is closely related to processing bodies (P-bodies), cytoplasmic RNP complexes intensively engaging in translational control (reviewed in [135]). It has been shown that the piRNA pathway proteins such as Aub, Ago3, and Tud colocalize with P-body proteins in germ cells [136,137]. Importantly, the role of the PIWI–piRNA pathway in mRNA turnover and translational control has also been reported in other organisms. In mice, Miwi, and piRNAs are associated with mRNAs in polysomes and RNP fractions [7,74]. Furthermore, Miwi binds to eIF4E, an mRNA cap-binding protein functions in translational control [74]. Similarly, Mili also forms a complex with eIF3a, and interacts with the eIF4E and eIF4G-containing mRNA 5′cap-binding complex [94]. Besides, Miwi2, Tdrd9, and Mael are localized to piP-bodies [37,109], a variant of P-bodies (reviewed in [135]). It has also shown that Miwi associates and stabilizes mRNAs of genes required in the post-meiotic stages of spermatogenesis [96]. In C. elegans, mRNAs from spermatogenesis genes are downregulated in prg-1 mutants [80]. Together, these observations reveal an evolutionarily conserved role for PIWI proteins and piRNAs in post-transcriptional and translational control, a role that has been underinvestigated.
SOMATIC FUNCTION OF THE PIWI–piRNA PATHWAY
Increasing lines of evidence indicate that PIWI protein and piRNAs function not only in the germline, but also in somatic tissues. In Drosophila, maternal PIWI proteins are essential for the maintenance of chromatin structure and cell cycle progression during early embryogenesis—the first phase of somatic development [138]. Moreover, Drosophila Piwi binds to chromosomes in somatic tissues such as salivary glands and is responsible for epigenetic effects at the binding sites [9,11,114,115,139], but these results await more lab to repeat and follow up. In Hydra, piRNAs are mainly mapped to non-transposon genes and Hywi is essential for somatic epithelial lineages [82]. In planaria, the two PIWI proteins, SMEDWI-2 (S. mediterranea Piwi2) and SMEDWI-3 (S. mediterranea Piwi3), are needed for the maintenance of neoblasts, the totipotent adult stem cells for tissue regeneration [140,141]. In Aplysia, piRNAs mediate epigenetic DNA methylation which is required for synaptic plasticity in neuronal cells [120]. A similar mechanism has also been identified in mouse hippocampus, in which piRNAs may target non-transposon genes to control spine shape [142]. Mouse Mili and piRNAs are also found to be expressed in adult mouse mesenchymal stem cells [143]. In human, HIWI (human PIWI protein) is expressed in CD34(+) hematopoietic progenitor cells [144]. Furthermore, a large number of studies have reported that PIWI proteins are up-regulated in cancer cells and tissues, as reviewed in detail by [12]. For example, human Hili is expressed in prostate, breast, gastrointestinal, ovarian, and endometrial cancers [145]. In addition, human Hiwi is expressed in seminomas, and gastric, uterine cervical, breast, ovarian, and endometrial cancers [146–148]. Furthermore, piRNAs are detected in cancer cells [149–151]. These results indicate that cancer development may be linked to PIWI proteins and the piRNA pathway. In Drosophila, a study has demonstrated that ectopic expression of piRNA pathway genes contributes to the growth and development of MBT [152]. Moreover, inactivation of the germline genes vasa, piwi, or aub suppressed malignant tumor growth, demonstrating that germline traits are necessary for tumor growth, at least in Drosophila [152]. Although the functions of PIWI proteins and piRNAs in the somatic tissues remain largely unclear, these findings have pointed out that the PIWI–piRNA pathway exerts a broader-than-expected role beyond the germline.
CONCLUDING REMARKS
PIWI proteins and piRNAs have been long known as key players in germline development with a focus on transposon silencing. Recent effort in identifying other piRNA pathway factors leads to the discovery of TUDOR family proteins as a major class of PIWI interactors. Meanwhile the scope of PIWI–piRNA studies has been significantly expended from transposon silencing to other functions in gene regulation in both the germline and somatic tissues. Yet, there are still many intriguing and fundamental questions awaiting further investigations. For example, what are molecular mechanisms for each step of pathway of piRNA biogenesis? What are the exact mechanisms by which PIWI–piRNAs mediate epigenetic and post-transcriptional regulation? How do these mechanisms contribute to germline development such as germline specification, GSC maintenance, germ cell proliferation, differentiation, meiosis, and oocyte axis formation? What is the role of PIWI–piRNAs in somatic tissues and cancer cells? When these questions are addressed, we will be able to map the complex PIWI–piRNA regulatory network and apply this knowledge towards clinical applications such as the treatment of cancers.
Acknowledgments
FUNDING
Current work in the Lin lab on Piwi proteins and piRNAs is supported by the National Institutes of Health (DP1CA174418 and R01HD42012), the G. Harold & Leila Mathers Foundation, and an Ellison Medical Foundation Senior Scholar Award to H.L.
References
- 1.Perkel JM. Visiting ‘noncodarnia’. Biotechniques. 2013;54:301, 303–4. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 2.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–76. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–69. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saito K, Nishida KM, Mori T, et al. specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Gene Dev. 2006;20:2214–22. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- 6.Girard A, Sachidanandam R, Hannon GJ, et al. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 7.Grivna ST, Beyret E, Wang Z, et al. A novel class of small RNAs in mouse spermatogenic cells. Gene Dev. 2006;20:1709–14. doi: 10.1101/gad.1434406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 9.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–8. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe T, Tomizawa S, Mitsuya K, et al. Role for piR-NAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–52. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang XA, Yin H, Sweeney S, et al. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24:502–16. doi: 10.1016/j.devcel.2013.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353–9. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox DN, Chao A, Baker J, et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Gene Dev. 1998;12:3715–27. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405–12. doi: 10.1038/nature07755. [DOI] [PubMed] [Google Scholar]
- 15.Song JJ, Liu J, Tolia NH, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–32. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 16.Paukku K, Yang J, Silvennoinen O. Tudor and nuclease-like domains containing protein p100 function as coactivators for signal transducer and activator of transcription 5. Mol Endocrinol. 2003;17:1805–14. doi: 10.1210/me.2002-0256. [DOI] [PubMed] [Google Scholar]
- 17.Lingel A, Simon B, Izaurralde E, et al. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–9. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 18.Darricarrère N, Liu N, Watanabe T, et al. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc Natl Acad Sci USA. 2013;110:1297–302. doi: 10.1073/pnas.1213283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reuter M, Berninger P, Chuma S, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–7. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 20.Boswell RE, Mahowald AP. Tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 21.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, et al. The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 22.Brahms H, Meheus L, de Brabandere V, et al. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–42. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meister G, Eggert C, Bühler D, et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–4. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 24.Selenko P, Sprangers R, Stier G, et al. SMN tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 25.Huyen Y, Zgheib O, Ditullio RA, Jr, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 26.Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–70. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- 27.Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–83. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Fang J, Bedford MT, et al. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–51. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 29.Nishida KM, Okada TN, Kawamura T, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–31. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirino Y, Vourekas A, Sayed N, et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–8. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Gene Dev. 2010;24:636–46. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pek JW, Anand A, Kai T. Tudor domain proteins in development. Development. 2012;139:2255–66. doi: 10.1242/dev.073304. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Jin J, James DA, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA. 2009;106:20336–41. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Vagin VV, Lee S, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–21. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L, Qi H, Wang J, et al. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138:1863–73. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reuter M, Chuma S, Tanaka T, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol. 2009;16:639–46. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 37.Shoji M, Tanaka T, Hosokawa M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–87. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Saxe JP, Tanaka T, et al. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol. 2009;19:640–4. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagin VV, Sigova A, Li C, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 40.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:6714–9. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patil VS, Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol. 2010;20:724–30. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 42.Vagin VV, Wohlschlegel J, Qu J, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Gene Dev. 2009;23:1749–62. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka T, Hosokawa M, Vagin VV, et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc Natl Acad Sci USA. 2011;108:10579–84. doi: 10.1073/pnas.1015447108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandey RR, Tokuzawa Y, Yang Z, et al. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc Natl Acad Sci USA. 2013;110:16492–7. doi: 10.1073/pnas.1316316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yabuta Y, Hiroshi O, Takaya A, et al. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192:781–95. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Llave C, Kasschau KD, Rector MA, et al. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605–19. doi: 10.1105/tpc.003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Djikeng A, Shi H, Tschudi C, et al. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–30. [PMC free article] [PubMed] [Google Scholar]
- 48.Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 49.Zamore PD. RNA silencing: genomic defence with a slice of pi. Nature. 2007;446:864–5. doi: 10.1038/446864a. [DOI] [PubMed] [Google Scholar]
- 50.Gunawardane LS, Saito K, Nishida KM, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 51.Cox DN, Chao A, Lin H. Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–14. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- 52.Qi H, Watanabe T, Ku HY, et al. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 2011;286:3789–97. doi: 10.1074/jbc.M110.193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito K, Ishizu H, Komai M, et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Gene Dev. 2010;24:2493–8. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haase AD, Fenoglio S, Muerdter F, et al. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Gene Dev. 2010;24:2499–504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olivieri D, Sykora MM, Sachidanandam R, et al. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–17. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zamparini AL, Davis MY, Malone CD, et al. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–50. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Handler D, Olivieri D, Novatchkova M, et al. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–93. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pane A, Wehr K, Schüpbach T. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–62. doi: 10.1016/j.devcel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olivieri D, Senti KA, Subramanian S, et al. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol Cell. 2012;47:954–69. doi: 10.1016/j.molcel.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Preall JB, Czech B, Guzzardo PM, et al. Shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA. 2012;18:1446–57. doi: 10.1261/rna.034405.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawaoka S, Izumi N, Katsuma S, et al. 3′ end formation of PIWI-interacting RNAs in vitro. Mol Cell. 2011;43:1015–22. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- 62.Saito K, Sakaguchi Y, Suzuki T, et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Gene Dev. 2007;21:1603–8. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horwich MD, Li C, Matranga C, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–72. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 64.Malone CD, Brennecke J, Dus M, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–35. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grentzinger T, Armenise C, Brun C, et al. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012;22:1877–88. doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brennecke J, Malone CD, Aravin AA, et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagao A, Sato K, Nishida KM, et al. Gender-specific hierarchy in nuage localization of PIWI-interacting RNA factors in Drosophila. Front Genet. 2011;2:55. doi: 10.3389/fgene.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sato K, Nishida KM, Shibuya A, et al. Maelstrom coordinates microtubule organization during Drosophila oogenesis through interaction with components of the MTOC. Gene Dev. 2011;25:2361–73. doi: 10.1101/gad.174110.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anand A, Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 2012;31:870–82. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Xu J, Koppetsch BS, et al. Heterotypic piRNA ping-pong requires qin, a protein with both E3 ligase and Tudor domains. Mol Cell. 2011;44:572–84. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cook HA, Koppetsch BS, Wu J, et al. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–29. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- 72.Gangaraju VK, Yin H, Weiner MM, et al. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;43:153–8. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Specchia V, Piacentini L, Tritto P, et al. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–5. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 74.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci USA. 2006;103:13415–20. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe T, Takeda A, Tsukiyama T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Gene Dev. 2006;20:1732–43. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Houwing S, Kamminga LM, Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 77.Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–11. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friedlander MR, Adamidi C, Han T, et al. High-resolution profiling and discovery of planarian small RNAs. Proc Natl Acad Sci USA. 2009;106:11546–51. doi: 10.1073/pnas.0905222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruby JG, Jan C, Player C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 80.Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–7. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Batista PJ, Ruby JG, Claycomb JM, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Juliano CE, Reich A, Liu N, et al. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc Natl Acad Sci USA. 2014;111:337–42. doi: 10.1073/pnas.1320965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aravin AA, Sachidanandam R, Girard A, et al. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–7. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 84.Williams RW, Rubin GM. ARGONAUTE1 is required for efficient RNA interference in Drosophila embryos. Proc Natl Acad Sci USA. 2002;99:6889–94. doi: 10.1073/pnas.072190799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–76. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- 86.Megosh HB, Cox DN, Campbell C, et al. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16:1884–94. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 87.Harris AN, Macdonald PM. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–32. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- 88.Klattenhoff C, Bratu DP, McGinnis-Schultz N, et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 89.Vagin VV, Klenov MS, Kalmykova AI, et al. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004;1:54–8. [PubMed] [Google Scholar]
- 90.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–49. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 91.Kuramochi-Miyagawa S, Kimura T, Yomogida K, et al. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–33. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 92.Deng W, Lin H. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–30. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 93.Carmell MA, Girard A, van de Kant HJ, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 94.Unhavaithaya Y, Hao Y, Beyret E, et al. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–19. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bao J, Zhang Y, Schuster AS, et al. Conditional inactivation of Miwi2 reveals that MIWI2 is only essential for prospermatogonial development in mice. Cell Death Differ. 2014;21:783–96. doi: 10.1038/cdd.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vourekas A, Zheng Q, Alexiou P, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19:773–81. doi: 10.1038/nsmb.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tan CH, Lee TC, Weeraratne SD, et al. Ziwi, the zebrafish homologue of the Drosophila piwi: co-localization with vasa at the embryonic genital ridge and gonad-specific expression in the adults. Gene Expr Patterns. 2002;2:257–60. doi: 10.1016/s1567-133x(02)00052-2. [DOI] [PubMed] [Google Scholar]
- 98.Seipel K, Yanze N, Schmid V. The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int J Dev Biol. 2004;48:1–7. doi: 10.1387/ijdb.15005568. [DOI] [PubMed] [Google Scholar]
- 99.Wilczynska A, Minshall N, Armisen J, et al. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 2009;15:337–45. doi: 10.1261/rna.1422509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Talbot K, Miguel-Aliaga I, Mohaghegh P, et al. Characterization of a gene encoding survival motor neuron (SMN)-related protein, a constituent of the spliceosome complex. Hum Mol Genet. 1998;7:2149–56. doi: 10.1093/hmg/7.13.2149. [DOI] [PubMed] [Google Scholar]
- 101.Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 2005;15:281–91. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- 102.Rangan P, Malone CD, Navarro C, et al. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–9. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu K, Chen C, Guo Y, et al. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc Natl Acad Sci USA. 2010;107:18398–403. doi: 10.1073/pnas.1013106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Blackburn ML, Chansky HA, Zielinska-Kwiatkowska A, et al. Genomic structure and expression of the mouse ESET gene encoding an ERG-associated histone methyltransferase with a SET domain. Biochim Biophys Acta. 2003;1629:8–14. doi: 10.1016/s0167-4781(03)00155-6. [DOI] [PubMed] [Google Scholar]
- 105.Pan J, Goodheart M, Chuma S, et al. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development. 2005;132:4029–39. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chuma S, Hosokawa M, Kitamura K, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA. 2006;103:15894–9. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vasileva A, Tiedau D, Firooznia A, et al. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol. 2009;19:630–9. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saxe JP, Chen M, Zhao H, et al. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–85. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aravin AA, van der Heijden GW, Castañeda J, et al. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hosokawa M, Shoji M, Kitamura K, et al. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 111.Aravind L, Ponting CP. The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci. 1997;22:458–9. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 112.Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–42. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Girard A, Hannon GJ. The role of piRNAs in mouse spermatogenesis. J Soc Biol. 2007;201:411–8. doi: 10.1051/jbio:2007912. [DOI] [PubMed] [Google Scholar]
- 114.Pal-Bhadra M, Leibovitch BA, Gandhi SG, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–72. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 115.Brower-Toland B, Findley SD, Jiang L, et al. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Gene Dev. 2007;21:2300–11. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shirayama M, Seth M, Lee HC, et al. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012;150:65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ashe A, Sapetschnig A, Weick EM, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aravin AA, Sachidanandam R, Bourc’his D, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–99. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Gene Dev. 2008;22:908–17. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rajasethupathy P, Antonov I, Sheridan R, et al. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sarot E, Payen-Groschêne G, Bucheton A, et al. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–21. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Savitsky M, Kwon D, Georgiev P, et al. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Gene Dev. 2006;20:345–54. doi: 10.1101/gad.370206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pelisson A, Sarot E, Payen-Groschêne G, et al. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–60. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shpiz S, Kwon D, Uneva A, et al. Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol Biol Evol. 2007;24:2535–45. doi: 10.1093/molbev/msm205. [DOI] [PubMed] [Google Scholar]
- 125.Findley SD, Tamanaha M, Clegg NJ, et al. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:859–71. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- 126.Sienski G, Donertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–80. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klattenhoff C, Xi H, Li C, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–49. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pane A, Jiang P, Zhao DY, et al. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011;30:4601–15. doi: 10.1038/emboj.2011.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008;18:136–48. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lim RS, Anand A, Nishimiya-Fujisawa C, et al. Analysis of Hydra PIWI proteins and piRNAs uncover early evolutionary origins of the piRNA pathway. Dev Biol. 2014;386:237–51. doi: 10.1016/j.ydbio.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 131.Robine N, Lau NC, Balla S, et al. A broadly conserved pathway generates 3′UTR-directed primary piRNAs. Curr Biol. 2009;19:2066–76. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saito K, Inagaki S, Mituyama T, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–9. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- 133.Rouget C, Papin C, Boureux A, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–32. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nishida KM, Saito K, Mori T, et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–22. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eulalio A, Behm-Ansmant I, Schweizer D, et al. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol. 2009;186:333–42. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thomson T, Liu N, Arkov A, et al. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev. 2008;125:865–73. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mani SR, Megosh H, Lin H. PIWI proteins are essential for early Drosophila embryogenesis. Dev Biol. 2014;385:340–9. doi: 10.1016/j.ydbio.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–27. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 140.Palakodeti D, Smielewska M, Lu YC, et al. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–86. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reddien PW, Oviedo NJ, Jennings JR, et al. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–30. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 142.Lee EJ, Banerjee S, Zhou H, et al. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–9. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu Q, Ma Q, Shehadeh LA, et al. Expression of the Argonaute protein PiwiL2 and piRNAs in adult mouse mesenchymal stem cells. Biochem Biophys Res Commun. 2010;396:915–20. doi: 10.1016/j.bbrc.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sharma AK, Nelson MC, Brandt JE, et al. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97:426–34. doi: 10.1182/blood.v97.2.426. [DOI] [PubMed] [Google Scholar]
- 145.Lee JH, Schütte D, Wulf G, et al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15:201–11. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 146.Qiao D, Zeeman AM, Deng W, et al. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–99. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- 147.Liu X, Sun Y, Guo J, et al. Expression of hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer. 2006;118:1922–9. doi: 10.1002/ijc.21575. [DOI] [PubMed] [Google Scholar]
- 148.Li S, Meng L, Zhu C, et al. The universal overexpression of a cancer testis antigen hiwi is associated with cancer angiogenesis. Oncol Rep. 2010;23:1063–8. [PubMed] [Google Scholar]
- 149.Cheng J, Guo JM, Xiao BX, et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–5. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 150.Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15:563–8. doi: 10.1007/s12094-012-0966-0. [DOI] [PubMed] [Google Scholar]
- 151.Law PT, Qin H, Ching AK, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–73. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 152.Janic A, Mendizabal L, Llamazares S, et al. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–7. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- 153.Anne J, Mechler BM. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development. 2005;132:2167–77. doi: 10.1242/dev.01809. [DOI] [PubMed] [Google Scholar]
- 154.Kirino Y, Vourekas A, Kim N, et al. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem. 2010;285:8148–54. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Frost RJ, Hamra FK, Richardson JA, et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107:11847–52. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zheng K, Xiol J, Reuter M, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA. 2010;107:11841–6. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nishimasu H, Ishizu H, Saito K, et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–7. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- 158.Huang H, Gao Q, Peng X, et al. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20:376–87. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Watanabe T, Chuma S, Yamamoto Y, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20:364–75. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Costa Y, Speed RM, Gautier P, et al. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet. 2006;15:2324–34. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- 161.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Xiol J, Cora E, Koglgruber R, et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47:970–9. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 163.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–34. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]