Abstract
Noncoding RNAs (ncRNAs) have crucial roles in epigenetic, transcriptional, and post-transcriptional regulation. Recent studies have begun to reveal a role of ncRNAs in DNA replication. Here, we review the roles of ncRNAs in regulating different aspects of DNA replication in prokaryotic and eukaryotic systems. We speculate that ncRNAs might function to guide the origin recognition complex (ORC) to chromosomal DNA during replication initiation in higher eukaryotes.
Keywords: noncoding RNA, DNA replication, miRNA, ORC, primer
It has become increasingly apparent that noncoding RNAs (ncRNAs), now known to be transcribed from the majority of the genome, have crucial roles in various cellular functions including epigenetic, transcriptional, and post-transcriptional regulation of gene expression. Emerging evidence indicates that ncRNAs also participate in DNA replication, a process that is relatively conserved among different organisms. DNA replication comprises the loading and activation of helicase, the unwinding of double-stranded DNA, the establishment of replisomes, the migration of replication forks away from the origin, the converging of replication forks, and the disassembly of replisomes. Since the first discoveries of DNA replication mechanisms in the 1960s, RNA has been demonstrated to have integral roles in DNA replication, including priming for leading and lagging strand synthesis, and providing a template for telomere extension. This review focuses on recently discovered new functions of ncRNAs in the DNA replication machinery and in the regulation of DNA replication in both prokaryotic and diverse eukaryotic replication systems.
Noncoding RNAs in regulating primer formation during DNA replication
A clear illustration of the involvement of ncRNAs in DNA replication comes from bacterial plasmids. ColE1 and Marine RNA-based plasmids from Enterobacteriaceae and Vibrionaceae families use ncRNAs to initiate replication at the origin (ori) instead of initiator proteins. The ori region of the plasmids encodes two partially complementary RNAs transcribed from two opposite strands [1,2]. The longer form (250–500 nucleotides), called RNA II, is transcribed from the sense strand and forms a stable hybrid with the DNA template. This hybrid is then processed by RNAse H to generate a primer, enabling leading strand synthesis by the host DNA polymerase. The shorter form (68–108 nucleotides),called RNAI, is transcribed from the antisense strand and is complementary to the 5′ region of RNA II. It functions as a negative regulator of leading strand priming by forming an RNA I–RNA II duplex that prevents RNA II–DNA hybrid formation. The level of RNA I is proportional to the number of plasmids within the bacteria, thus constituting a negative feedback loop that leads to a dynamic regulation of plasmid replication in response to metabolic fluctuations (Figure 1). Another example of a ncRNA that regulates RNA priming during DNA replication has been demonstrated in the human BK polyomavirus. This virus exists episomally in human cells, and replicates its DNA by recruiting the host replication machinery [3]. A ncRNA named small replication-regulating RNA (srRNA) has been identified in mouse cells and binds simultaneously to both sense and antisense strands within the viral origin region, thereby blocking RNA primer synthesis by host DNA polymerase α and inhibiting viral replication (Figure 1).
Figure 1.
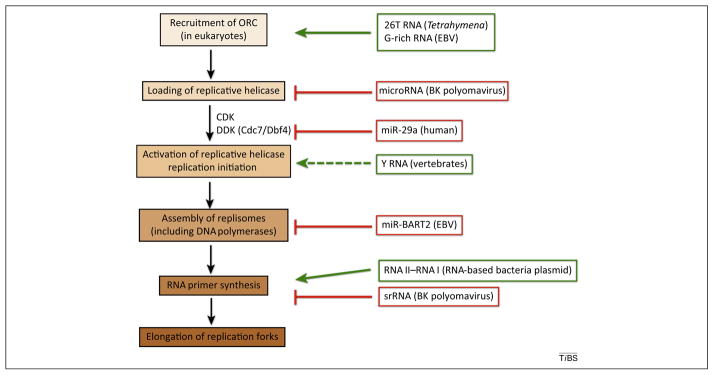
A summary of the known roles of noncoding RNAs in regulating the different steps of DNA replication. 26T RNA and G-rich RNA function to recruit the origin recognition complex (ORC) during DNA replication of T. thermophila and the Epstein-Barr virus (EBV), respectively. Y RNAs perform a conserved function to promote replication initiation in vertebrates. The BK polyomavirus and ColE1 and Marine RNA-based plasmids from Enterobacteriaceae and Vibrionaceae families use ncRNAs to regulate RNA priming events prior to DNA strand synthesis. MicroRNAs fine-tune the stability of replication helicase, DNA polymerase, and S phase kinase Cdc7/Dbf4, as demonstrated in the BK polyomavirus and in human cells. Abbreviation: srRNA, small replication-regulating RNA; CDK, cyclin-dependent kinase; DDK, Dbf4-dependent kinase. Unbroken green arrows illustrate promoting roles, whereas red blunted-ended lines illustrate inhibitory roles of ncRNA. Broken green arrow indicates a likely candidate of Y RNA.
Noncoding RNAs in ORC recruitment during DNA replication
A key initial step in DNA replication of eukaryotes is the binding of the origin recognition complex (ORC) to DNA. The ORC is composed of six subunits, ORC1-6, which serve as a landing platform for the assembly of DNA helicase and the subsequent establishment of replication forks. Although the ORC is conserved in eukaryotes, the mechanism of its recruitment to DNA varies in different species. In the budding yeast Saccharomyces cerevisiae, the ORC recognizes a specific DNA consensus sequence. In the fission yeast Schizosaccharomyces pombe, the ORC binds to AT-rich DNA owing to the presence of the AT-hook domain in one of its subunits, ORC4. In contrast to yeast, the ORC in higher eukaryotes lacks a sequence-specific DNA binding motif. It remains unclear what DNA-binding factors or chromatin environments direct ORC binding and determine the site of origin formation; one possibility is that ncRNAs might be involved.
So far, ncRNAs have been shown to mediate ORC recruitment in the protozoa Tetrahymena thermophila and in the Epstein-Barr virus (EBV). In T. thermophila, ribosomal DNA (rDNA) is amplified ~9000 times during development, and a ncRNA called 26T RNA mediates ORC recruitment to the origin during rDNA amplification [4] (Figure 1). 26T RNA spans the terminal 282 nucleotides of 26S rRNA, forms an integral part of the ORC, and base pairs with an essential cis-acting replication determinant at the rDNA origin. Mutations that perturb the base pairing process disrupt rDNA origin recognition by the ORC and the activation of the origin. Therefore, 26T RNA specifically ensures the efficient replication of rDNA and their massive amplification during T. thermophila development. EBV is a human virus and establishes latency in host cells after infection. Similar to BK polyomavirus, EBV uses host replication machinery, including the ORC, and replicates indistinguishably from the cellular chromosomes. EBV encodes a viral protein, EBNA1, which binds to the viral origin and recruits human ORC. The interaction between ORC and EBNA1 is mediated by a noncoding G-rich RNA, which forms a stable complex with both proteins and is predicted to form a G-quadruplex (G4) structure [5].
These examples of ncRNA-mediated ORC recruitment during DNA replication suggest a possible adaptation of this mechanism by higher eukaryotes. Recent genome-wide mapping of ORC1 in mammals has shown that ~72% of ORC1 are associated with active transcription start sites, among which only 46.5% are promoters of coding genes; however, the majority of the remaining 53.5% are transcription start site of ncRNAs [6]. The ORC has also been shown to interact with G-rich RNA in mammalian systems, which in turn mediates ORC recruitment to telomeres and AT-rich heretochromatin [7,8]. Furthermore, a domain within the ORC1 subunit has been mapped. This domain preferentially binds to G-rich RNA and single-stranded DNA, both of which form G4 structures [9]. Consistently, this ORC1 binding feature coincides with recent genome-wide data showing that G-rich repeated elements are present in 67–90% of replication origins in metazoans [10]. The ORC does not exhibit a preference for double-stranded DNA, therefore it is tempting to speculate that a G-rich/G4 RNA might act as a mediator to recruit the ORC to DNA replication origins in higher eukaryotes. One possibility is that G-rich RNA could guide the ORC to G-rich DNA elements by base pairing with the C-rich DNA strand, similar to the role of 26T RNA in T. thermophila replication. Alternatively, G-rich RNA could bridge the ORC with DNA binding factors that bind specifically to G-rich DNA elements, analogous to that during EBV replication.
Noncoding RNAs in the initiation of DNA replication
In vertebrates, a type of ncRNA called Y RNA has an important role during the initiation of chromosomal DNA replication (Figure 1). Y RNAs are small stem-loop RNAs of between 69 to 112 nucleotides in length, and are evolutionarily conserved among vertebrates. Although the exact mechanism of how Y RNAs promote replication initiation remains unclear, it has been shown that Y RNAs are recruited to chromatin by the ORC, where their interaction with pre-replication complex proteins and other initiation proteins is essential for their function. In addition, Y RNAs are only required for the initiation of DNA replication after the midblastular transition during vertebrate development, suggesting that Y RNAs function as a developmentally regulated layer of control over the evolutionarily conserved eukaryotic DNA replication machinery [11,12].
microRNAs fine-tune DNA replication
In addition to the above types of ncRNA, microRNAs, which target mRNA for transcript degradation and translational repression, also regulate eukaryotic DNA replication by fine-tuning the process (Figure 1). For example, in human cells, microRNA (miR)-29a targets the Cdc7/Dbf4 kinase, which is essential for the initiation of replication in S phase. Upon genotoxic stress, up-regulation of Cdc7/Dbf4 is accompanied by the down-regulation of miR-29a to maximize DNA damage repair and cell survival [13]. The microRNA miR-BART2, encoded by EBV, targets BALF5, the catalytic subunit of the viral DNA polymerase, to inhibit lytic replication and establish latency in the human host after infection [14]. Similarly, a microRNA from the BK polyomavirus targets large T antigen during the early course of infection, thereby controlling viral replication to establish persistence in the host [15]. The tight regulation of viral replication by the microRNA prevents the rapid death of the host, therefore conferring a selective advantage for viable viral progeny.
Perspective
It is clear that various types of ncRNA can regulate different aspects of DNA replication, including RNA primer formation, ORC recruitment to DNA, initiation of replication, as well as cellular levels of replication helicase, DNA polymerase, and S phase kinase (Figure 1). Integrating ncRNA into DNA replication presents several advantages. First, RNA couples DNA replication to cellular physiology. This is especially important for simple organisms such as bacterial plasmids, viruses, and T. thermophila. Second, the short half-life of RNA enables rapid regulation of DNA replication in response to environmental input such as metabolic fluctuations and genotoxic stress. This likely works together with protein-based sensing mechanisms in regulating cell cycle and DNA replication. Third, microRNAs confer fine-tuning of DNA replication at a global scale by regulating the expression of specific components within the replication machinery. Last, sequence-specific or G-rich ncRNA could bring replication proteins such as the ORC to specific DNA sequences by either base pairing with target DNA or bridging interactions between the ORC and certain DNA binding factors that render sequence-specific binding. Whether this mechanism operates in higher eukaryotes awaits further studies. Collectively, the emerging roles of ncRNA provide new opportunities for research on DNA replication. Future discoveries should reveal what is really beneath the tip of the iceberg.
Acknowledgments
This work was supported by a grant from the Connecticut Stem Cell Research Fund (10SCA05) to X.Q.G. and by a G. Harold & Leica Y. Mathers Award to H.L. We apologize to those authors whose exciting work we did not cite here owing to the strict 15-reference limitation.
References
- 1.Camps M. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat DNA Gene Seq. 2010;4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le Roux F, et al. Conserved small RNAs govern replication and incompatibility of a diverse new plasmid family from marine bacteria. Nucleic Acids Res. 2011;39:1004–1013. doi: 10.1093/nar/gkq852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tikhanovich I, et al. Inhibition of human BK polyomavirus replication by small noncoding RNAs. J Virol. 2011;85:6930–6940. doi: 10.1128/JVI.00547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohammad MM, et al. T. thermophila ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J. 2007;26:5048–5060. doi: 10.1038/sj.emboj.7601919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norseen J, et al. RNA-dependent recruitment of the origin recognition complex. EMBO J. 2008;27:3024–3035. doi: 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dellino GI, et al. Genome-wide mapping of human DNA-replication origins: levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013;23:1–11. doi: 10.1101/gr.142331.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomae AW, et al. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc Natl Acad Sci USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng Z, et al. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshina S, et al. Human origin recognition complex binds preferentially to G-quadruplex-preferable RNA and single-stranded DNA. J Biol Chem. 2013;288:30161–30171. doi: 10.1074/jbc.M113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cayrou C, et al. New insights into replication origin characteristics in metazoans. Cell Cycle. 2012;11:658–667. doi: 10.4161/cc.11.4.19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang AT, et al. Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci. 2011;124:2058–2069. doi: 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collart C, et al. The midblastula transition defines the onset of Y RNA-dependent DNA replication in Xenopus laevis. Mol Cell Biol. 2011;31:3857–3870. doi: 10.1128/MCB.05411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkley LR, Santocanale C. MicroRNA-29a regulates the benzo[a]pyrene dihydrodiol epoxide-induced DNA damage response through Cdc7 kinase in lung cancer cells. Oncogenesis. 2013;2:e57. doi: 10.1038/oncsis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth S, et al. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666–675. doi: 10.1093/nar/gkm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broekema NM, Imperiale MJ. miRNA regulation of BK polyomavirus replication during early infection. Proc Natl Acad Sci USA. 2013;110:8200–8205. doi: 10.1073/pnas.1301907110. [DOI] [PMC free article] [PubMed] [Google Scholar]


