Abstract
PTENP1 is a pseudogene of the PTEN tumor suppression gene (TSG). The functions of PTENP1 in clear-cell renal cell carcinoma (ccRCC) have not yet been studied. We found that PTENP1 is downregulated in ccRCC tissues and cells due to methylation. PTENP1 and PTEN are direct targets of miRNA miR21 and their expression is suppressed by miR21 in ccRCC cell lines. miR21 expression promotes ccRCC cell proliferation, migration, invasion in vitro, and tumor growth and metastasis in vivo. Overexpression of PTENP1 in cells expressing miR21 reduces cell proliferation, invasion, tumor growth, and metastasis, recapitulating the phenotypes induced by PTEN expression. Overexpression of PTENP1 in ccRCC cells sensitizes these cells to cisplatin and gemcitabine treatments in vitro and in vivo. In clinical samples, the expression of PTENP1 and PTEN is correlated, and both expressions are inversely correlated with miR21 expression. Patients with ccRCC with no PTENP1 expression have a lower survival rate. These results suggest that PTENP1 functions as a competing endogenous RNA (ceRNA) in ccRCC to suppress cancer progression.
Introduction
Renal cell carcinoma (RCC) is the third most common urological cancer after prostate and bladder cancer, accounting for about 3% of all human malignancies in adults (1, 2). With more than 200,000 individuals diagnosed with renal caner and more than 100,000 individuals who died from this type of cancer every year, RCC is the seventh most common cancer in women and the fifth in men (3). Statistics from the previous research on the incidence of RCC in most areas of the world are available (4, 5). The majority of RCC cases are of the clear-cell subtype (ccRCC), but approximately 10% of tumors are of the papillary subtype and approximately 5% the chromophobe subtype. The other histologic subtypes like collecting duct, transitional cell (urothelial cell) carcinoma, and medullary together account for approximately 5% to 10% of cases (6–8). Besides surgical intervention, RCC is not sensitive to chemotherapy or radiotherapy. The absence of biomarkers is responsible for late diagnosis and subsequent poor prognosis. Identification and characterization of genetic and biologic changes is necessary in understanding the pathogenesis of RCC and identifying new biomarkers.
Recent studies have shown that genetic events and histopathologically heterogeneous disorder play a major role in the development of ccRCC (9). Currently, there are at least 12 different genes associated with the development of kidney cancer: VHL, MET, FLCN, TSC1, TSC2, TFE3, TFEB, MITF, fumarate hydratase (FH), succinate dehydrogenase B (SDHB), succinate dehydrogenase D (SDHD), and PTEN (10, 11). Genetic analysis has shown that each subtype represents distinct tumor biology. Comprehensive genome analysis and studies involving several autosomal dominant syndromes characterized by the development of RCC have greatly contributed to the identification of the genes involved in the development of RCC (12). Noncoding RNAs (ncRNA), including miR-NAs, long noncoding RNAs (LncRNA), pseudogenes, etc., do not code for proteins and have recently been found to be pervasively transcribed in the genome. The noncoding transcripts range in length from 100 nt to approximately 100 kilobases (kb) and lack significant open reading frames. The great majority of ncRNAs are transcribed by RNA polymerase II (RNA pol II) and are polyadenylated (13, 14). It has been suggested that pseudogenes arise from protein-coding genes that lost the protein production function mostly due to mutation or aberrant duplication. These noncoding transcripts, including pseudogenes, were once considered useless transcription products with no functions. However, recent evidence increasingly links mutations and dysregulations of ncRNAs to diverse human diseases, including human cancers (15). NcRNAs have been validated to have important functions, including tumor suppressor-like (TSG-like) functions (16,17), in biologic processes. Recent studies have shown that some pseudogenes contain miRNA-binding elements and serve as competitive endogenous RNAs (ceRNA; refs. 16, 17), decoying that compete for miRNAs to regulate gene expression.
In human cancers, monoallelic mutation of PTEN without loss or mutation of the second allele is prevalent at presentation, whereas complete loss is observed at low frequencies with the exception of advanced cancers (18). It was reported that the PTENP1 (NM_023917), which was lost in many human cancers could increase PTEN abundance and showed tumor-suppressive activity (19, 20). Our previous research and other studies have reported that the ncRNA expression signatures of renal clear-cell carcinoma were revealed by microarray (21, 22) and found that the PTENP1 transcript was significantly downregulated in ccRCC. Here, we report that pseudogene PTENP1 serves as a ceRNA to modulate PTEN expression regulation by miR21. PTENP1 suppresses tumor growth, invasion, and metastasis in ccRCC. PTENP1 expression sensitizes chemotherapy treatment in human ccRCC cell lines. PTENP1 and PTEN expression is correlated with primary human ccRCC samples, and their expression is inversely correlated with miR21 expression. Lower PTENP1 and PTEN expression is correlated with worse clinical outcomes. These studies demonstrated that pseudogene PTENP1 plays critical roles in ccRCC progression and can potentially serve as a therapeutic target.
Materials and Methods
Patients and tumor samples
Written informed consent was obtained from all patients, and the study was approved by the Institutional Review Board of Huazhong University of Science and Technology, Tongji Medical College, Tongji Hospital (Hubei, China). Ninety-four patients with clear-cell carcinoma of kidney who received nephrectomy or partial nephrectomy were included in the study. The clinical information was retrieved from the medical records.
Cell culture and transfection
The human renal cell carcinoma cell lines 786-O, ACHN, and SN12PM6 were maintained in DMEM containing 10% FBS, OS-RC-2, and Caki-1 were cultured in RPMI1640 supplemented with 10% FBS. The human kidney proximal tubular epithelial cell line HK-2 was maintained in DMEM containing 10% FBS streptomycin at 37°C in a humidified atmosphere of 5% CO2. The cell lines were obtained from ATCC in March 2013 and authenticated by ATCC. Cells were transiently transfected with the indicated expression plasmid using FuGENE HD Transfection Reagent (Roche) according to the manufacturer’s instructions. The pcDNA3 empty vector was used as control. miR21 and negative control mimics, anti-miR21, and negative control inhibitors (Ribo-Bio Co. Ltd.) were transfected into cells, respectively, with X-tremeGENE siRNA Transfection Reagent (Roche) according to the manufacturer’s instructions.
Construction of expression plasmid and packaging of lentivirus
Oligonucleotides (5′-UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUCUGACA-3′) encoding miR21 precursor was subcloned into lentiviral vector pCDH (System Biosciences, Inc.) and verified by DNA sequencing. PTENP1 and PTEN were cloned into the same vector. Dicer shRNA (5′-CCGGGCCTCACTTGACCTGAAGTATCTCGAGATACTTCAGCGTCAAGTGAGGCTTTTTG-3′) or control scrambled shRNA were cloned into pCDH and the lenti virus was packed as above (23). Cells were transiently transfected with the indicated expression plasmid using FuGENE HD Transfection Reagent (Roche) according to the manufacturer’s instructions. For the gain of stable cell lines, lentivirus was packed with pPACKHl Lentivector Packaging Kit (System Biosciences, Inc.) and infected in renal cell lines, following the manufacturer’s instructions.
Luciferase reporter assay
PTENP1 3′-UTR containing the putative binding site of miR21, and its identical sequence with a mutation of the miR21 seed sequence were inserted between the restrictive sites Xhol and NotI of hluc+/hRluc luciferase reporter vector psiCHECK2 and validated by sequencing. The hluc+/hRluc luciferase reporter vectors of PTEN were constructed as above. Cells were seeded in 24-well plates and transfected with wild-type or mutated reporter vectors, or miR21 mimics, negative control mimics, anti-miR21 inhibitors, negative control inhibitors or vector pCDH-PTENP1, 24 hours after transfection, firefly and Renilla luciferase activities were consecutively measured, according to the Dual-Lucif-erase Assay Manual (Promega). The Renilla luciferase signal was normalized to the firefly luciferase signal for each individual analysis.
RNA isolation and quantitative reverse transcription-PCR
Total RNA was isolated from the indicated cell lines and renal cancer tissues by using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Total RNA (500 ng) was used to synthesize the first-strand cDNA by means of random primers and Superscript II reverse transcriptase (Invitrogen) following the manufacturer’s protocol. We prepared appropriate dilutions of each single-stranded cDNA for subsequent PCR by monitoring an amount of GAPDH as quantitative control. The quantitative real-time PCR reaction was set at an initial denaturation step of 10 minutes at 95°C, and 95°C (5 seconds), 61°C (30 seconds), and 72°C (30 seconds) in a total of 40 cycles with a final extension step at 72°C for 5 minutes. All experiments were performed in triplicate. The primers used were PTENP1 (long-form) 5′-TCAGAACATGGCATACACCAA-3′ (forward); 5′-TGATGACGTCCGATTTTTCA-3′ (reverse), PTEN 5′-GTTTACCGGCAGCATCAAAT-3′ (forward); 5′-CCCCCACTTTAGTGCACAGT-3′ (reverse).
Cell proliferation assay
ACHN, SN12PM6, and 786-0 cells were infected/transfected with lentiviral/plasmid for 72 hours/24 hours, then cells were digested and transferred to 96-well micro-plates, and replanted at a density of approximately 2,000 (ACHN)/3,000 (SN12PM6)/1,000 (786-O) cells per well at 12, 24, 48, and 72 hours after infection/transfection. All experiments were performed in triplicate. Cell proliferation was determined using the Cell Counting Kit-8 (Dojindo Laboratories) according to the manufacturer’s instructions.
Cell invasion and migration assays
Migration and invasion assays were performed using uncoated and Matrigel-coated Transwell inserts according to the manufacturer’s instructions. A density about 1 × 105 of ACHN cells or 5 × 104 of SN12PM6 or 2 × 104 of 786-O cells was suspended and then seeded in the top chambers of 24-well Transwell plates with FBS-free medium. Culture medium containing 10% FBS was deposited in the bottom chambers. After 12 to 18 hours of incubation at 37°C in a humidified 5% CO2 atmosphere for ACHN and SN12PM6 and 8 to 12 hours for 786-O, cells that migrated were stained by 0.5% crystal violet solution for 15 minutes and counted. For invasion, Transwell membranes were prepared with Matrigel for plating infected cells. After 24 hours for ACHN and SN12PM6 or 12 to 14 hours for 786-O, cells that invaded were stained by 0.5% crystal violet solution for 15 minutes and quantified by determining the total cell number derived from 5 randomly chosen visual fields per membrane at 400 × magnification. Each experiment was performed in duplicate.
Methylation-specific PCR
Genomic DNA was isolated using QIAamp DNA Mini Kit (Qiagen) and bisulfite modification of the genomic DNA was carried out using an Epitect Bisulfite Kit (Qiagen) according to the manufacturer’s instructions. Methylation-specific PCR (MSP) primers were designed with Methprimer. Unmethylated PTENP1 5′-TAGTGAGAATATTTGGATATAGGGC-3′ (forward); 5′-AATTTACTACACCGATTAACTCGTC-3′ (reverse), methylated PTENP1 5′-GAGAATATTTGGATATAGGGTGG-3′ (forward); 5′-AATTTACTACACCAATTAACTCAT-C-3′ (reverse).
Metastasis assay
The antimetastatic activity of PTENP1, PTEN, miR21, and PTENP1+miR21 were tested in the mouse ACHN lung metastasis model as described previously (24). ACHN cell was stably infected with these genes containing GFP label. Treated cells (2 × 105) were suspended in 100 µL of PBS and injected intravenously via the tail vein. Mice were sacrificed and lungs were resected 30 days later after injection. The incidence and volume of metastases were estimated by imaging the mice for bioluminescence using the Living Image software (Xenogen). The photon emission level was used to assess the relative tumor burden in the mice lungs. All animal studies were conducted under approved guidelines of the Animal Care and Use Committee of the Tongji Hospital (Wuhan, China).
Cell growth in vivo assay
Tumorigenesis in nude mice was determined as described previously (25). Five groups of four mice each were injected subcutaneously with prepared cells at a single site. Tumor onset was measured with calipers at the site of injection weekly at different times on the same day. Tumor volume was calculated using the formula, 0.5ab2, where "a" represents the larger and "b" represents the smaller of the two perpendicular indexes. Animals were sacrificed 41 days after injection. Nude mice were manipulated and cared for according to NIH Animal Care and Use Committee guidelines in the Experiment Animal Center of the Tongji Medical College of Huazhong University of Science and Technology. To evaluate the chemosensitivity effect of PTENP1, four groups of 6 mice each were injected subcutaneously with ACHN cells at a single site. After the injection when an appreciable tumor formed subcutaneously, cisplatin/gemcitabine or PBS were injected in Lenti-PTENP1 or Lenti-NC tumors. Among these groups, tumor onset was measured with calipers at the site of injection every 3 days by two trained laboratory staff at different times on the same day. Tumor volume was calculated using the formula, V = 0.5ab2, where "a" represents the larger and "b" represents the smaller of the two perpendicular indexes. Animals were sacrificed 39 days after injection. Nude mice were manipulated and cared for according to NIH Animal Care and Use Committee guidelines in the Experiment Animal Center of the Tongji Medical College of Huazhong University of Science and Technology (Wuhan, Hubei, China).
Statistical analysis
Statistical significance was determined by using the SPSS 17.0. The Fisher exact test was utilized to assess the significance between different proportions. Values are expressed as mean ± SEM unless otherwise indicated and t test was used to examine the difference between treatment and negative control group. The Spearman coefficient of rank correlation was also used to measure the correlation between methylation ratio and PTENP1 expression. Kaplan-Meier curves and the log-rank test were used for the survival analysis. The difference of tumor weight in tumorigenicity assay was assayed by t test. Significance was defined as P < 0.05.
Results
PTENP1 expression is downregulated in human ccRCC
We examined the expression of PTENP1 in 40 paired renal cancer tissue and adjacent renal tissue specimens, and a panel of 6 renal cell lines, including 5 cancerous cell lines (ACHN, 786-0, OS-RC-2, CaKi-1, and SN12-PM6) and control cell line HK-2, a human kidney proximal tubular epithelial cell line. The expression of PTENP1 is significantly lower in four ccRCC cell lines (ACHN, OS-RC-2, CaKi-1, and SN12-PM6) when compared with HK-2 cells (Fig. 1A). In paired clinical samples, the expression of PTENP1 is lower in tumor samples than adjacent normal samples in all 12 pairs (Fig. 1A and Supplementary Fig. S1). To determine whether the lower expression of PTENP1 is due to methylation, we carried out bisulfite modification of the genomic DNA from renal cancer tissues and cells as well as normal tissues. Methylation-specific PCR showed that the PTENP1 promoter was methylated in ccRCC cell lines and tissues (Fig. 1B). To further validate that the lower expression of PTENP1 is due to methylation, we treated the cancer cells with demethylation agent 5-aza-2′-deoxycytidine (5AZA.dc). Cells with 5AZA-C treatment showed higher PTENP1 expression than control treatment except 786-0 cells, which have high PTENP1 expression (Fig. 1C). These results suggested that the low PTENP1 expression in ccRCC is due to methylation. We then determined the expression of PTEN in clinical samples and found that PTEN expression is lower in cancer than in normal renal tissues and its expression is correlated with PTENP1 expression (Fig. 1D). Overexpression of PTENP1 (Supplementary Fig. S2) in ACHN and SN12PM6 cells upregulated PTEN expression (Fig. 1E) and activated AKT, a PTEN downstream target (Supplementary Fig. S3). These results indicated that PTEN expression is regulated by PTENP1 and that their expression is correlated.
Figure 1.
PTENP1 expression is downregulated in ccRCC. A, PTENP1 expression levels were evaluated by real-time PCR in nontumorigenic renal cell line HK-2, renal cancer cell lines, and paired case specimens. B, methylation-specific PCR was performed using primers specific for the unmethylated (U) or methylated (M) PTENP1 promoter region in the genomic DNA deriving from renal cell lines or renal cancer tissues. NC, unmethylated genomic DNA; PC, methylated genomic DNA. C, PTENP1 expression in 5 renal cancer cell lines after 5AZA-C or mock treatment. *, P < 0.05; **, P < 0.01. D, the expression of PTEN and PTENP1 in renal cancer tissues and normal control tissues. The expression of PTEN and PTENP1 is lower in cancer tissues than normal controls. The expression of PTEN and PTENP1 in cancer tissues includes positive correlation (P < 0.01; R2 = 0.6374). E, PTEN protein levels were determined by immunoblot analysis in renal cancer cell line ACHN and SN12PM6 after being infected with Lenti-PTENP1 or Lenti-NC.
miR21 regulates PTEN and PTENP1 in ccRCC
As it has been shown that PTEN expression is suppressed by miR21 in lung cancer (26, 27), we determined the miR21 expression in ccRCC cell lines and clinical samples. miR21 expression is upregulated in primary ccRCC samples in comparison with adjacent normal controls (Fig. 2A), as well as in ACHN, SN12PM6, Caki-1, and OS-RC-2 cells when compared with control HK-2 cells (Fig. 2A). To determine whether PTEN and PTENP1 are direct targets of miR21, we constructed reporter plasmids by cloning the wild-type 3′UTR or mutant 3′UTR of PTEN downstream of a luciferase gene. Cotransfection of the wild-type reporter plasmid and miR21 (Supplementary Fig. S4) in both ACHN and SN12PM6 cells resulted in a decrease of the luciferase signal when compared with the vector control (Fig. 2B). Expression of PTENP1 partially rescued the suppression of the luciferase signal by miR21 (Fig. 2B). Knockdown of miR21 increased the luciferase signal of wild-type reporter plasmid (Fig. 2B); overexpression of PTENP1 and knockdown of miR21 increased the luciferase signal further (Fig. 2B). In contrast, the luciferase signal of the mutant reporter plasmid is not affected by the expression of miR21 or knockdown of miR21 (Fig. 2B). To confirm the suppression of PTEN by miR21, we carried out immunoblotting of PTEN. miR21 expression suppressed PTEN expression in ACHN and SN12PM6 cells, whereas knockdown of miR21 increased PTEN expression (Fig. 2C). PTENP1 is a processed pseudogene residing at 9pl3.3. Its sequence is highly homologous to PTEN, with only 18 mismatches throughout the coding sequence and a high homology (~95%) region of 3′UTR. We identified a potential binding site of miR21 in the PTENP1 sequence (Fig. 3A) and constructed reporter plasmids containing the wild-type or mutant-binding site of PTENP1 downstream a luciferase gene. Similar to PTEN, the luciferase signal of the wild-type reporter was suppressed by miR21 (Fig. 3A), whereas the luciferase signal of the mutant reporter was not affected by miR21 expression (Fig. 3A). Similar to the regulation of PTEN by miR21, miR21 suppressed PTENP1 expression in ACHN and SN12MP6 cells (Fig. 3B). In primary ccRCC samples, the expression of PTEN (Fig. 3C) and PTENP1 (Fig. 3D) is inversely correlated with the miR21 expression. Taken together, these results demonstrated that both PTEN and PTENP1 are directly regulated by miR21, and their expression is inversely correlated with miR21 expression. PTENP1 expression relieves the suppression of PTEN expression by miR21, indicating PTENP1 is a competing endogenous RNA, competing with the binding of miR21 with PTEN, thus relieving the suppression of PTEN by miR21 in ccRCC.
Figure 2.
PTEN expression is regulated by miR21 in ccRCC. A, miR21 expression levels were evaluated by real-time PCR in nontumorigenic renal cell line HK-2, renal cancer cell lines and paired case specimens. B, luciferase signals of wild-type or mutant PTEN reporters in renal cancer cell line ACHN and SN12PM6. miR21 mimics or miR21 inhibitors or miR21 mimics and PTENP1 ormiR21 inhibitors and PTENP1 were cotransfected with the reporter in the cells (a). Sequences of PTEN 3′UTR, miR21, and mutant PTEN 3′UTR (b). The luciferase signals were suppressed by miR21 but partially rescued by PTENP1 expression. **, P < 0.01. C, PTEN expression in ACHN and SN12PM6 cells expressing miR21 mimics or miR21 inhibitors. PTEN expression is suppressed in cells expressing miR21 mimics, but increased in cells expressing miR21 inhibitors. PTENP1 expression rescued the suppression of PTEN expression by miR21.
Figure 3.
PTENP1 expression is regulated by miR21 inccRCC. A, luciferase signals of wild-type or mutant PTENP1 reporters in renal cancer cell line ACHN and SN12PM6. miR21 mimics or miR21 inhibitors were cotransfected with the reporter in the cells (a). Sequences of PTENP1 3′UTR, miR21, and mutant PTENP1 3′UTR (b). The luciferase signals were suppressed by miR21. B, PTENP1 expression in ACHN and SN12PM6 cells expressing miR21 mimics or miR21 inhibitors. PTENP1 expression is suppressed in cells expressing miR21 mimics, but increased in cells expressing miR21 inhibitors. C, correlation analysis of the expression of PTENP1 andmiR21 in ccRCC tissues. The expression of PTENP1 andmiR21 is inversely correlated. D, correlation analysis of the expression of PTEN and miR21 in ccRCC tissues. The expression of PTENP1 and miR21 is inversely correlated. **, P < 0.01.
PTENP1 suppresses cell proliferation and tumor growth
To elucidate the functions of PTENP1 and PTEN in ccRCC, we investigated their roles in cell proliferation and tumor growth. miR21 expression promoted cell growth in ACHN and SN12PM6 cells (Fig. 4A). In contrast, PTEN expression (Supplementary Fig. S5) suppressed cell growth in ACHN and SN12PM6 cells (Fig. 4B). Similar to PTEN, PTENP1 also suppressed cell growth in vitro (Fig. 4C and Supplementary Fig. S6). More importantly, PTENP1 expression partially reversed the cell growth promotion by miR21 (Fig. 4C). To confirm these results in vivo, we transplanted ACHN cells expressing a control vector, or miR21, or PTEN, or PTENP1, or miR21 and PTENP1 into mice. PTEN (Fig. 4D) and PTENP1 (Fig. 4D) suppressed tumor growth in vivo, whereas miR21 promoted tumor growth in vivo (Fig. 4D). PTENP1 expression reversed the tumor promotion phenotype induced by miR21 in vivo (Fig. 4D), partially recapitulating the phenotype induced by PTEN. These results indicated that PTENP1 serves as a competing endogenous RNA in ccRCC tumor growth.
Figure 4.
PTENP1 serves as a ceRNA in the regulation of tumor growth in RCC. A, cell proliferation in ACHN and SN12PM6 cells expressing miR21 or control. Cell viability was quantified at each time point. miR21 promoted cell growth. Data are plotted as the mean ± SEM of three independent experiments. B, cell proliferation in ACHN and SN12PM6 cells expressing PTEN or control. Cell viability was quantified at each time point. PTEN suppressed cell growth. C, cell proliferation in ACHN and SN12PM6 cells expressing vector control or PTENP1 ormiR21 ormiR21 and PTENP1. Cell viability was quantified at each time point. PTENP1 reversed the promotion of cell growth by miR21.D, we designed five groups in the animal models, and then ACHN cells expressing vector control or miR21 orPTEN,orPTENP1 orcoexpressingmiR21 andPTENPI were transplanted into mice. D, the size of transplanted tumors in different groups (a). Tumor weight of each mouse at the end of 41 days is indicated (b). Tumor volumes were determined at each time point (c). PTEN and PTENP1 suppressed tumor growth in vivo. miR21 promoted tumor growth in vivo. PTENP1 rescued the promotion of tumor growth by miR21 .These data mean PTENP1 serves as a ceRNA in the regulation of tumor growth in RCC. **, P < 0.01.
PTENP1 suppresses cell migration, invasion, and metastasis
It has been shown that PTEN is also involved in cell migration and invasion (24, 25). We determined the function of PTENP1 in cell migration and invasion assays. miR21 expression promoted migration and invasion in ACHN and SN12PM6 cells (Fig. 5A), whereas PTEN suppressed migration and invasion (Fig. 5B and C). Similar to PTEN, PTENP1 suppressed cell migration and invasion (Fig. 5D). Interestingly, PTENP1 expression reversed the increase of cell migration and invasion induced by miR21 (Fig. 5D). The quantification of the results was summarized in Fig. 5E. To investigate the function of PTENP1 in metastasis, we generated GFP-tagged ACHN cells and introduced control vector, or miR21, or PTEN, or PTENP1, or miR21 plus PTENP1 into these cells. These cells were then transplanted into mice and lung metastasis signals were determined by Xenogen bioluminescence system. Similar to the cell migration and invasion results, PTENP1 (Fig. 5F and G) and PTEN (Fig. 5F and G) suppressed tumor metastasis, whereas miR21 (Fig. 5F and G) promoted tumor metastasis. More importantly, PTENP1 expression reversed the metastasis promotion phenotype induced by miR21 expression (Fig. 5F and G), partially recapitulating the metastasis suppression phenotype by PTEN. As DICER is required for miRNA, including miR21 processing, we determined whether it is also required for PTENP1 functions. In contrast to PTENP1 functions in positive regulation of PTEN and suppression of cell proliferation, migration, and invasion (Fig. 1E–Fig. 5), PTENP1 overexpression in ACHN and SN12PM6 cells with DICER knockdown did not increase PTEN expression and had no effect in cell proliferation, migration, and invasion (Supplementary Fig. S7A–S7D), suggesting DICER is required for PTENP1 regulation of PTEN and its functions. As 5AZA-C treatment increased PTENP1 expression (Fig. 1C), we determined whether 5AZA-C induces similar phenotypes as PTENP1 overexpression. Similar to PTENP1 overexpression, 5AZA-C treatment in ACHN and SN12PM6 cells increased PTEN expression, suppressed cell proliferation, migration, and invasion (Supplementary Fig. S8A–S8E). These results indicated that 5AZA-C treatment in renal cancer cells resulted in phenotypes similar to those induced by PTENP1 overexpression. Taken together, these results suggested that PTENP1 serves as a ceRNA in ccRCC cell migration, invasion in vitro, and metastasis in vivo.
Figure 5.
PTENP1 serves as a ceRNA in the regulation of cell migration and invasion in vitro and metastasis in vivo. A, ACHN and SN12PM6 cells expressing miR21 or control were subjected to migration and invasion assay. Representative photographs were taken at ×200 magnification. C, the number of migrated and invaded cells was quantified in 4 random images from each group. miR21 promoted cell migration and invasion. B, ACHN and SN12PM6 cells expressing PTEN or control were subjected to migration and invasion assay. Representative photographs were taken at ×200 magnification. The number of migrated and invaded cells was quantified in 4 random images from each group (C). PTEN suppressed cell migration and nvasion. D, ACHN and SN12PM6 cells expressing control or miR21 orPTENPI ormiR21 and PTENP1 were subjected to migration and invasion assay. Representative photographs were taken at ×200 magnification. E, the number of migrated and invaded cells was quantified in 4 random images from each group. PTENP1 rescued the promotion of cell migration and invasion by miR21. F, GFP-tagged ACHN cells expressing vector control or miR21 or PTEN or PTENP1 or miR21 and PTENP1 were injected into mouse tail veins. G, GFP signals in lung metastasis were determined by Xenogen bioluminescence system. PTEN and PTENP1 suppressed metastasis, whereas miR21 promoted metastasis. PTENP1 rescued the promotion of metastasis by miR21. **, P < 0.01.
PTENP1 expression sensitizes renal cancer cells to cisplatin and gemcitabine
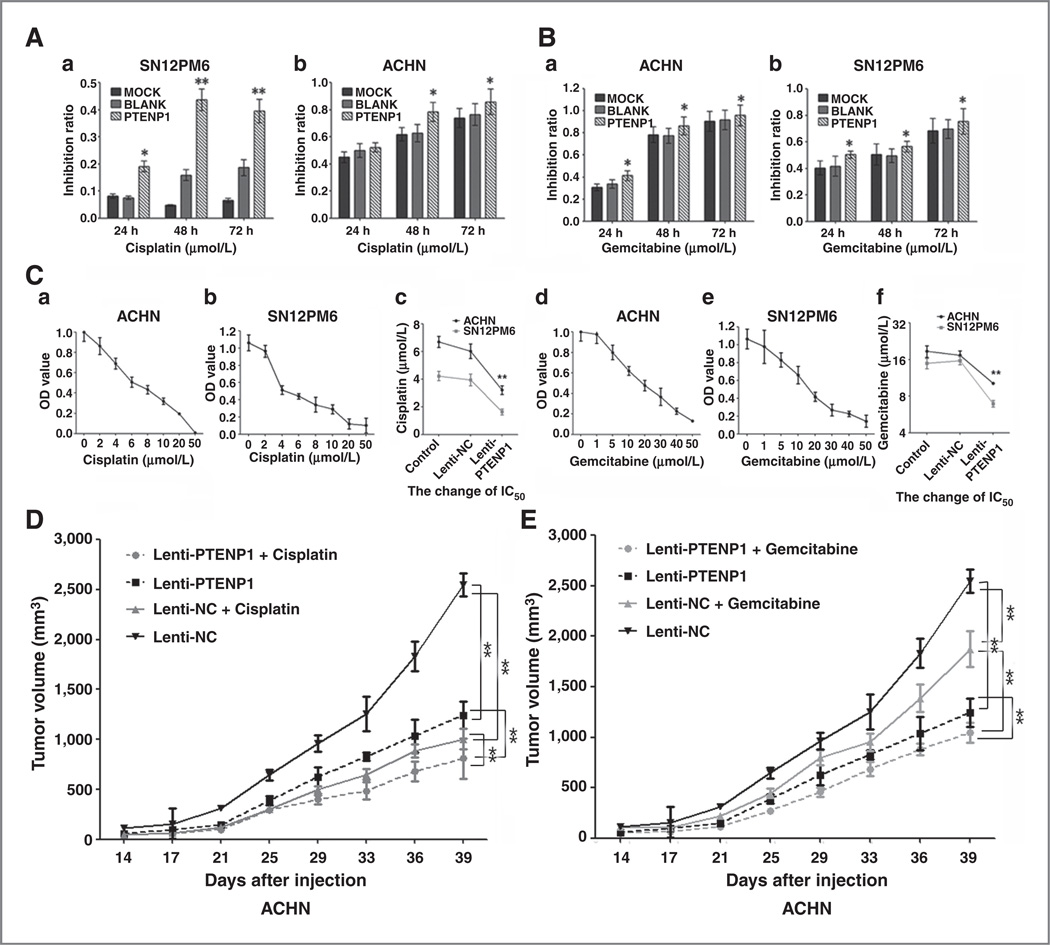
PTENP1 expression increased significantly in ACHN and SN12PM6 cells following cisplatin or gemcitabine treatment (Supplementary Fig. S9). To determine whether PTENP1 functions in chemosensitivity, we determined the cell growth inhibition in cells overex-pressing PTENP1 after drug treatment. Cell death was significantly increased in ACHN and SN12PM6 cells expressing PTENP1 following cisplatin (Fig. 6A, a and b) and gemcitabine (Fig. 6B, a and b) treatment. IC50 of these two chemotherapies decreased in the cells over-expressing PTENP1 (Fig. 6C, a–f). To determine the effect of PTENP1 in chemosensitivity in vivo, we transplanted ACHN cells expressing a vector control or PTENP1 into mice and treated with cisplatin or gemcitabine. PTENP1 expression reduced the tumor growth after the treatment of cisplatin (Fig. 6D) or gemcitabine (Fig. 6E) in vivo. These results indicated that PTENP1 increases sensitivity of ccRCC cells to cisplatin and gemcitabine.
Figure 6.
PTENP1 sensitizes renal cancer cells to chemotherapy. A, SN12PM6 cells expressing vector control or PTENP1 were treated with cisplatin (a) and ACHN cells expressing vector control or PTENP1 were treated with cisplatin (b). Cell viability was determined at 24, 48, and 72 hours after treatment. Cell death increased in cells expressing PTENP1 than vector control. Data are plotted as the mean ± SEM of three independent experiments. B, SN12PM6 cells expressing vector control or PTENP1 were treated with gemcitabine (a) and ACHN cells expressing vector control or PTENP1 were treated with gemcitabine (b). Cell viability was determined at 24, 48, and 72 hours after treatment. Cell death increased in cells expressing PTENP1 than vector control. Data are plotted as the mean ± SEM of three independent experiments. C, determination of IC50 in ACHN and SN12PM6 cells with cisplatin or gemcitabine treatment. Cell survival in ACHN and SN12PM6 cells follow the treatment of cisplatin (a and b) or gemcitabine (d and e) of various concentrations. IC50 in cells expressing PTENP1 is significantly lower than that in cells expressing vector control or control cells (c and f). D, ACHN cells expressing PTENP1 or vector control were transplanted into mice that were subjected to cisplatin treatment. Tumor volume was determined at each time point. E, ACHN cells expressing PTENP1 or vector control were transplanted into mice that were subjected to gemcitabine treatment. Tumor volume was determined at each time point. PTENP1 expression sensitizes ACHN cells to cisplatin or gemcitabine treatment in vivo. *, P < 0.05; **, P < 0.01.
Finally, we examined the significance of PTENP1 and PTEN expression in the clinical features of patients with ccRCC. The level of PTENP1 and PTEN expression was divided into two categories: negative and positive in the primary ccRCC samples. No remarkable difference of PTENP1 and PTEN expression was found in the distribution according to sex or age (Table 1). Significant differences in the distribution of the patients were observed in the clinic stage (P < 0.01; P < 0.01), Fuhrman grading (P < 0.01; P < 0.01) and lymph node metastasis (P < 0.01; P < 0.05; Table 1) in the groups according to PTENP1 or PTEN expression. In addition, we determined whether PTENP1/PTEN expression could be an important factor in the clinical outcomes of patients with ccRCC. The PTENP1 or PTEN-positive group showed significantly better overall survival than the negative group (Fig. 7 A and B), indicating PTENP1 or PTEN are potentially valuable biomarkers for the prognosis of ccRCC.
Table 1.
Clinical characteristics and outcome of 94 patients with renal clear cell cancer according to PTENP1 and PTEN gene expression
| PTEN |
PTENP1 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Biologic characteristics | n | Positive | Rate | x2 | P | Positive | Rate | x2 | P |
| Fuhrman nuclear grading | |||||||||
| I | 28 | 20 | 71.43% | 17.26 | <0.01 | 18 | 64.29% | 11.51 | <0.01 |
| II | 42 | 14 | 33.33% | 15 | 35.71 % | ||||
| III-IV | 24 | 4 | 16.67% | 6 | 25% | ||||
| Clinical stage | |||||||||
| I | 32 | 22 | 68.75% | 10.16 | <0.01 | 21 | 65.63% | 12.27 | <0.01 |
| II | 38 | 13 | 34.21% | 13 | 34.21 % | ||||
| III-IV | 24 | 3 | 12.5% | 5 | 20.83% | ||||
| Lymphatic metastasis | |||||||||
| Yes | 46 | 13 | 28.26% | 5.62 | <0.05 | 12 | 26.09% | 9.12 | <0.01 |
| No | 48 | 25 | 52.08% | 27 | 56.25% | ||||
Figure 7.
PTENP1 and PTEN expression is correlated with survival in RCC. Kaplan-Meier survival curves for patients with renal clear cell cancer according to the expression level of PTENP1 (A) and the expression level of PTEN (B).
Discussion
ccRCC is a genetic disease that is caused by a series of genetic alterations (9, 26). Comprehensive analysis of ccRCC clinical samples found that PTEN mutation occurs in some patients with ccRCC, suggesting its potential tumor suppressor functions (19, 20). miR21 was found to be overexpressed in ccRCC clinical samples (28–32). However, the functions of pseudogenes in ccRCC have not been characterized. In this study, we showed for the first time that pseudogene PTENP1 played critical roles in ccRCC progression. PTENP1 was shown to be deleted in human cancer (19, 20, 33–36). However, to our best knowledge, there is no report on the potential contribution of altered PTENP1 expression in ccRCC development and progression. We found that PTENP1 served as competing endogenous RNA for the regulation of PTEN expression by miR21. Our data in ccRCC cell lines and clinical samples indicated that PTENP1 promoter is hypermethylated. Lower expression of PTENP1 results in the suppression of PTEN by miR21. Our study indicated that the expression of tumor suppressor PTEN is significantly downregulated in ccRCC and PTEN has important functions in ccRCC progression, including tumor cell growth, invasion, and metastasis. These results underscore the significance of ceRNAs in ccRCC development. It is possible that additional ceRNAs are critical regulators of gene expression, which affects tumor progression. The identification of these ceRNAs is currently underway.
miR21 expression is upregulated in ccRCC and correlated with worse clinical outcome. It has been shown that miR21 targets KiSS-1 to regulate cell migration (37). Our study suggested that PTEN is a major direct target gene of miR21 in ccRCC development. Both 3′UTR of PTEN and PTENP1 sequences contain miR21-binding sites. PTEN and PTENP1 expressions are inversely correlated with miR21 expression in ccRCC clinical samples. In most cases, mRNAs serve as a critical rheostat in gene expression regulation. The identification of ceRNAs to regulate the control of miRNA in target gene expression added another layer of complexity in gene regulation. Our results suggested that intricate gene expression of a tumor suppressor is critical in ccRCC progression. These results have potential implications in the understanding of tumor development in other cancer types.
Our study in clinical samples demonstrated that the expression of PTENP1 and PTEN inversely correlate with worse overall patient survival. Patients who had high risk scores according to the PTENP1 and PTEN signature have shorter overall survival time compared with patients who had low risk scores, demonstrating its potential as biomarker for ccRCC prognosis and treatment. The expression of PTENP1 and PTEN can serve as prognostic markers independent of Fuhrman nuclear grading. In combination with the expression of other pseudogenes and noncoding RNAs, the expression level of PTENP1 and PTEN may provide additional value for the accurate prediction of clinical outcomes in patients with ccRCC.
In addition to the functions of PTENP1 in ccRCC development, our study also showed that PTENP1 expression sensitizes ccRCC cells to chemotherapies, suggesting PTENP1 potentially is a therapeutic target. Patients with ccRCC with low PTENP1 and PTEN expression potentially benefit from the therapies of PTENP1 expression increase in ccRCC cells. Our finding that PTENP1 promoter is hypermethylated suggested that demethylation agents can potentially be used as therapeutics in ccRCC in combination with other treatments.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by the National Natural Science Foundation of China (31072238, 31172441, 31372562, and 81170650; to H. Xu), the National Major Scientific and Technological Special Project for Significant New Drugs Development (2012ZX09303018; to H. Xu), and NCI R01CA148759 (to Q. Huang).
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors’ Contributions
Conception and design: W. Yao, J. Wang, Z. Ye, Q. Huang, H. Xu
Development of methodology: K. Gumireddy, J. Wang, H. Xiao, H. Li
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): K. Gumireddy, A. Li, H. Xiao, K. Tang
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): G. Yu, J. Wang, W. Xiao, K. Tang, Q. Huang
Writing, review, and/or revision of the manuscript: G. Yu, J. Wang, H. Li, Q. Huang, H. Xu
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): K. Chen
Study supervision: G Yu, Z. Ye, H. Xu
References
- 1.Schrader AJ, Sevinc S, Olbert PJ, Hegele A, Varga Z, Hofmann R. [Gender-specific characteristics and survival of renal cell carcinoma] Urologe A. 2008;47:1182–1186. doi: 10.1007/s00120-008-1832-0. [DOI] [PubMed] [Google Scholar]
- 2.Bukowski RM. Prognostic factors for survival in metastatic renal cell carcinoma: update 2008. Cancer. 2009;115:2273–2281. doi: 10.1002/cncr.24226. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Mathew A, Devesa SS, Fraumeni JF, Jr, Chow WH. Global increases in kidney cancer incidence, 1973–1992. Eur J Cancer Prev. 2002;11:171–178. doi: 10.1097/00008469-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 5.Sun M, Thuret R, Abdollah F, Lughezzani G, Schmitges J, Tian Z, et al. Age-adjusted incidence, mortality, and survival rates of stage-specific renal cell carcinoma in North America: a trend analysis. Eur Urol. 2011;59:135–141. doi: 10.1016/j.eururo.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z. 2004 WHO classification of the renal tumors of the adults. Eur Urol. 2006;49:798–805. doi: 10.1016/j.eururo.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 7.Storkel S, Eble JN, Adlakha K, Amin M, Blute ML, Bostwick DG, et al. Classification of renal cell carcinoma: Workgroup No. 1 Union Internationale Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC) Cancer. 1997;80:987–989. doi: 10.1002/(sici)1097-0142(19970901)80:5<987::aid-cncr24>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 8.Renshaw AA. Subclassification of renal cell neoplasms: an update for the practising pathologist. Histopathology. 2002;41:283–300. doi: 10.1046/j.1365-2559.2002.01420.x. [DOI] [PubMed] [Google Scholar]
- 9.Oosterwijk E, Rathmell WK, Junker K, Brannon AR, Pouliot F, Finley DS, et al. Basic research in kidney cancer. Eur Urol. 2011;60:622–633. doi: 10.1016/j.eururo.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maher ER. Genomics and epigenomics of renal cell carcinoma. Semin Cancer Biol. 2012;23:10–17. doi: 10.1016/j.semcancer.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Linehan WM, Ricketts CJ. The metabolic basis of kidney cancer. Semin Cancer Biol. 2012;23:46–55. doi: 10.1016/j.semcancer.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junker K, Ficarra V, Kwon ED, Leibovich BC, Thompson RH, Oosterwijk E. Potential role of genetic markers in the management of kidney cancer. Eur Urol. 2012;63:333–340. doi: 10.1016/j.eururo.2012.09.040. [DOI] [PubMed] [Google Scholar]
- 13.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 14.Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, et al. Poly A- transcripts expressed in HeLa cells. PLoS ONE. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papa A, Chen M, Pandolfi PP. Pills of PTEN? In and out for tumor suppression. Cell Res. 2013;23:1155–1156. doi: 10.1038/cr.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poliseno L, Haimovic A, Christos PJ, Vega YSdMEC, Shapiro R, PavlickA etal. Deletion of PTENP1 pseudogene in human melanoma. J Invest Dermatol. 2011;131:2497–2500. doi: 10.1038/jid.2011.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS ONE. 2012;7:e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brito GC, Fachel AA, Vettore AL, Vignal GM, Gimba ER, Campos FS, et al. Identification of protein-coding and intronic noncoding RNAs down-regulated in clear cell renal carcinoma. Mol Carcinog. 2008;47:757–767. doi: 10.1002/mc.20433. [DOI] [PubMed] [Google Scholar]
- 23.Bu Y, Lu C, Bian C, Wang J, Li J, Zhang B, et al. Knockdown of Dicer in MCF-7 human breast carcinoma cells results in G1 arrest and increased sensitivity to cisplatin. Oncol Rep. 2009;21:13–17. [PubMed] [Google Scholar]
- 24.Parhar RS, Lala PK. Amelioration of B16F10 melanoma lung metastasis in mice by a combination therapy with indomethacin and inter-leukin 2. J Exp Med. 1987;165:14–28. doi: 10.1084/jem.165.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu GJ, Wu MW, Wang C, Liu Y. Enforced expression of METCAM/ MUC18 increases tumorigenesis of human prostate cancer LNCaP cells in nude mice. J Urol. 2011;185:1504–1512. doi: 10.1016/j.juro.2010.11.052. [DOI] [PubMed] [Google Scholar]
- 26.Yan-Nan B, Zhao-Yan Y, Li-Xi L, Jiang Y, Qing-Jie X, Yong Z. Micro-RNA-21 accelerates hepatocyte proliferation in vitro via PI3K/Akt signaling by targeting PTEN. Biochem Biophys Res Commun. 2013;443:802–807. doi: 10.1016/j.bbrc.2013.12.047. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Bai W, Zhang W. MiR-21 suppresses the anticancer activities of curcumin by targeting PTEN gene in human non-small cell lung cancer A549 cells. Clin Transl Oncol. 2013;16:708–713. doi: 10.1007/s12094-013-1135-9. [DOI] [PubMed] [Google Scholar]
- 28.Juan D, Alexe G, Antes T, Liu H, Madabhushi A, Delisi C, et al. Identification of a microRNA pan el for clear-cell kidney cancer. Urology. 2010;75:835–841. doi: 10.1016/j.urology.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 29.Lv L, Huang F, Mao H, Li M, Li X, Yang M, et al. MicroRNA-21 is overexpressed in renal cell carcinoma. Int J Biol Markers. 2013;28:201–207. doi: 10.5301/JBM.2013.10831. [DOI] [PubMed] [Google Scholar]
- 30.Zhang A, Liu Y, Shen Y, Xu Y, Li X. miR-21 modulates cell apoptosis by targeting multiple genes in renal cell carcinoma. Urology. 2011;78:e13–e19. doi: 10.1016/j.urology.2011.03.030. 474. [DOI] [PubMed] [Google Scholar]
- 31.Dey N, Das F, Ghosh-Choudhury N, Mandal CC, Parekh DJ, Block K, et al. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE. 2012;7:e37366. doi: 10.1371/journal.pone.0037366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bera A, Ghosh-Choudhury N, Dey N, Das F, Kasinath BS, Abboud HE, et al. NFkappaB-mediated cyclin D1 expression by microRNA-21 influences renal cancer cell proliferation. Cell Signal. 2013;25:2575–2586. doi: 10.1016/j.cellsig.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang CC, Huai L, Zhang CP, Jia YJ, Li QH, Chen YR, et al. [Study on expression of PTEN gene and its pseudogene PTENP1 in acute leukemia and correlation between them] Zhonghua Xue Ye Xue Za Zhi. 2012;33:896–901. [PubMed] [Google Scholar]
- 34.loffe YJ, Chiappinelli KB, Mutch DG, Zighelboim I, Goodfellow PJ. Phosphatase and tensin homolog (PTEN) pseudogene expression in endometrial cancer: a conserved regulatory mechanism important in tumorigenesis? Gynecol Oncol. 2012;124:340–346. doi: 10.1016/j.ygyno.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsit CJ, Zheng S, Aldape K, Hinds PW, Nelson HH, Wiencke JK, et al. PTEN expression in non-small-cell lung cancer: evaluating its relation to tumor characteristics, allelic loss, and epigenetic alteration. Hum Pathol. 2005;36:768–776. doi: 10.1016/j.humpath.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Ulger C, Toruner GA, AIkan M, Mohammed M, Damani S, Kang J, et al. Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate cancer-related genes in the HL-60 cell line. Cancer Genet Cytogenet. 2003;147:28–35. doi: 10.1016/s0165-4608(03)00155-9. [DOI] [PubMed] [Google Scholar]
- 37.Yoshioka K, Ohno Y, Horiguchi Y, Ozu C, Namiki K, Tachibana M. Effects of a KiSS-1 peptide, a metastasis suppressor gene, on the invasive ability of renal cell carcinoma cells through a modulation of a matrix metalloproteinase 2 expression. Life Sci. 2008;83:332–338. doi: 10.1016/j.lfs.2008.06.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.