Abstract
The complex milieu of inflammatory mediators associated with many diseases is often too dilute to directly measure in the periphery, necessitating development of more sensitive measurements suitable for mechanistic studies, earlier diagnosis, guiding therapeutic decisions, and monitoring interventions. We previously demonstrated that plasma samples from recent-onset Type 1 diabetes (RO T1D) patients induce a proinflammatory transcriptional signature in freshly drawn peripheral blood mononuclear cells (PBMCs) relative to that of unrelated healthy controls (HC). Here, using cryopreserved PBMC, we analyzed larger RO T1D and HC cohorts, examined T1D progression in pre-onset samples, and compared the RO T1D signature to those associated with three disorders characterized by airway infection and inflammation. The RO T1D signature, consisting of interleukin-1 cytokine family members, chemokines involved in immunocyte chemotaxis, immune receptors, and signaling molecules, was detected during early pre-diabetes and found to resolve post-onset. The signatures associated with cystic fibrosis patients chronically infected with Pseudomonas aeruginosa, patients with confirmed bacterial pneumonia, and subjects with H1N1 influenza all reflected immunological activation, yet each were distinct from one another and negatively correlated with that of T1D. This study highlights the remarkable capacity of cells to serve as biosensors capable of sensitively and comprehensively differentiating immunological states.
Keywords: Type 1 diabetes, Cystic Fibrosis, Influenza, Gene expression profiling, Biomarker, Inflammation
INTRODUCTION
Many diseases arise from uncontrolled inflammatory processes and become evident after the progression of significant tissue and organ damage. These diseases include auto-inflammatory diseases such as Type 1 diabetes (T1D), multiple sclerosis, and rheumatoid arthritis, as well as bacterial and viral infections. A need remains for discovery and evaluation of biomarkers that can sensitively detect and differentiate inflammatory processes, define immunological mechanisms of action that can guide selection of appropriately targeted therapies, and sensitively monitor changes in the inflammatory state associated with disease progression or responses to therapeutic intervention. Fortunately, advances in genomic technologies now offer unprecedented opportunities to not only gain new mechanistic insights into inflammatory processes, but to improve diagnosis and treatment. While many studies have examined transcript levels in tissue biopsies, peripheral blood remains the most accessible human tissue and represents a practical, minimally invasive surrogate biopsy material.
The most common blood-based genomics strategy has been to directly profile transcripts of peripheral blood mononuclear cells (PBMCs) or purified immunocyte subsets of cases and controls. When applied to infectious disease, clinically useful, pathogen-specific transcriptional patterns have been associated with various bacterial, viral, fungal, and protozoan infections1, 2. These responses possess species-level or even strain-level specificity and arise because different microbial pathogens exhibit unique pathogen-associated molecular patterns, which interact with specific host pattern recognition receptors (such as the toll-like receptors, TLRs)3, 4 to trigger different signal transduction pathways and unique transcriptional programs5–8. These differential responses have enabled the detection of specific transcriptional signatures in PBMCs from individuals harboring various infections including viruses9–12, Gram-negative and Gram-positive bacteria11, 13, and eukaryotic parasites such as Plasmodium14.
Transcriptional signatures of patient PBMCs also reflect the inflammatory mechanisms utilized by various autoimmune diseases15–17. These include systemic lupus erythmatosus, rheumatoid arthritis (where direct profiling of PBMC has been found useful for classifying disease and predicting response to infliximab)18–21, multiple sclerosis22–24, inflammatory bowel disease (where PBMC profiles have been described that distinguish Crohn’s disease from ulcerative colitis25), psoriasis26, dermatomyositis27, 28, and systemic onset juvenile idiopathic arthritis29, 30.
A less common strategy has been to use patient serum or plasma to induce gene expression in healthy, unrelated third-party PBMCs. This approach, where the cells are used as reporters that sensitively respond to soluble disease-associated factors present in the periphery, has been used by Pascual et al.,29 to study the inflammatory state associated with systemic onset juvenile idiopathic arthritis and we have applied it to T1D31–33. We previously determined that culturing plasma of recent onset (RO) T1D patients with freshly drawn unrelated, healthy PBMCs induces a unique innate inflammatory transcriptional signature31. This signature includes many genes regulated by interleukin (IL)-1, a cytokine known to co-stimulate T cells and cause pancreatic β-cell death in vitro. The RO T1D signature is distinct from that induced by the plasma of unrelated healthy controls (HCs) or long-standing (LS) T1D patients (>10 years post-onset), suggesting that the RO T1D signature is induced by factors related to active autoimmunity31. In longitudinal samples collected from progressors to diabetes, this T1D signature was evident as much as 5 years prior to onset and prior to the emergence of auto-antibodies toward islet cell antigens, which are currently considered to be the best predictor of progression to diabetes31. Our application of this approach to studies of T1D in the BioBreeding rat disease model also revealed an onset-associated signature partially dependent on IL-1. Blocking the IL-1 receptor by treating BioBreeding rats with IL-1 receptor antagonist (IL-1RA) delayed onset and, in part, normalized the disease signature, suggesting that induced transcriptional signatures are not only mechanistically informative, but may possesses utility in monitoring immunotherapeutic intervention32.
A limitation in our past studies, as well as many other studies that have utilized cell-based assays, is the reliance on freshly isolated PBMCs. Despite utilizing PBMCs drawn from multiple healthy blood donors, we found that the plasma samples from a given subject cohort induced relatively homogenous profiles31 with modest donor cell-specific effects. Commercially available cryopreserved PBMCs are a relatively new option for cell-based assays. These cells are collected via aphaeresis from highly characterized blood donors, enabling the harvesting of billions of cells during a single draw. Rapid processing/cryopreservation under optimized and controlled conditions results in high viability and retained functionality. In this report we extend our initial studies by using a cryopreserved PBMC of a single donor to 1) examine a larger cohort of pediatric RO T1D patients and unrelated HC, 2) study the development of the signature during the progression of T1D, and 3) evaluate the disease-specificity of the technique by examining for the first time transcriptional signatures induced by plasma collected from patients with conditions characterized by pathologic pulmonary inflammation: cystic fibrosis (CF), bacterial pneumonia, and H1N1 influenza.
RESULTS
Cross-sectional analysis of RO T1D and unrelated HC samples
At diagnosis of T1D, an estimated 60–90% of β cell mass is either dysfunctional or destroyed. Upon initiation of insulin therapy 25–100% of new onset patients experience a transient remission with functional restoration of the residual mass, termed the “honeymoon period”, which lasts months to years34–37. This immunologically active time is clinically significant because 1) it offers a window for therapeutic intervention aimed at preserving remaining β cell mass; and 2) as shown by our previous studies31, 33, it is a period where it is still possible to measure processes related to β cell autoimmunity. Therefore, our studies of plasma induced transcriptional signatures have primarily focused on the first 7 months post onset.
In order to reproduce and expand the observations of our initial report31, plasma drawn from 47 RO T1D patients (mean age 9.97 ± 2.89 years, Supplemental Table 1) and 44 unrelated HC subjects (mean age 14.98 ± 4.13 years, Supplemental Table 1) were used to induce gene expression in commercially-supplied cryopreserved PBMC of a single draw of a single healthy blood donor (UPN727). All RO T1D subjects were positive for ≥1 autoantibody and to avoid inducing gene expression due to factors related to hyperglycemia, samples were collected after stabilization on exogenous insulin from subjects that exhibited histories of good glycemic control.
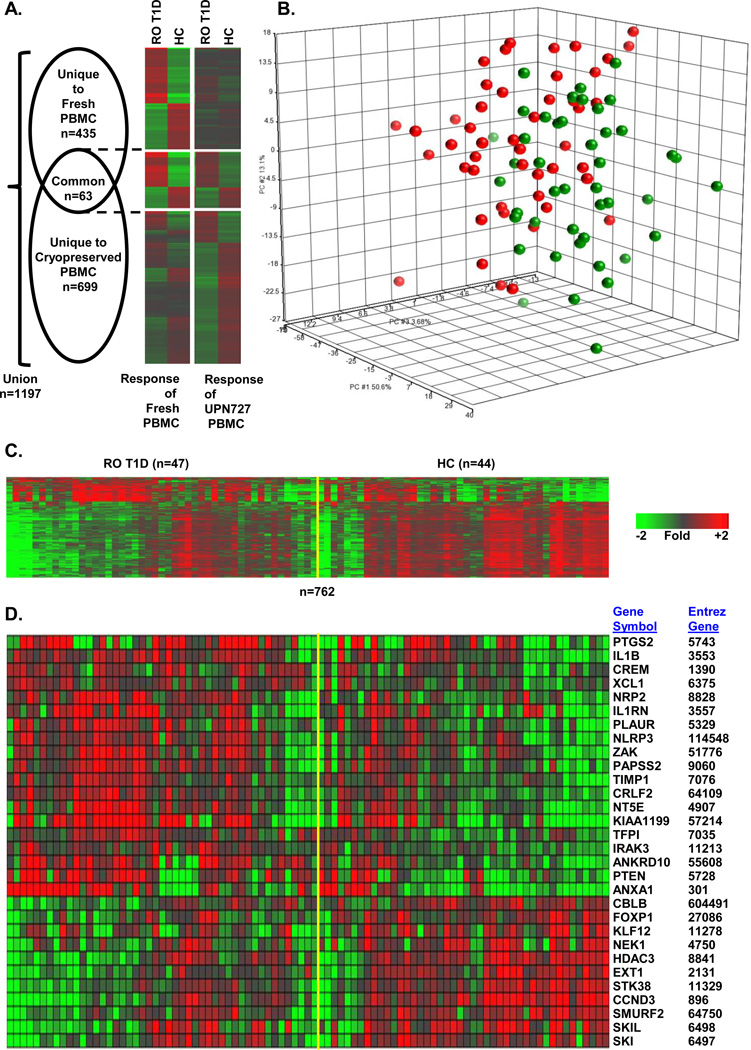
We identified 762 probe sets that were differentially regulated when UPN727 cells were co-cultured with plasmas of the RO T1D or HC cohorts (|log2 ratio|>0.263, 1.2-fold; FDR<0.2; ANOVA p<0.036); these represented 622 unique UniGenes (Supplemental Table 2). The relationship of this dataset to those genes most significantly regulated following co-culture of plasma collected from 12 RO T1D and 12 HC subjects with fresh PBMCs from six healthy blood donors in our previous report31 (498 probe sets |log2 ratio|>0.5; FDR<0.2) is illustrated in Figure 1A. The Pearson correlation coefficient between the union of these two datasets is 0.59; however, if the analysis is restricted to the 63 common probe sets, the Pearson correlation coefficient is 0.93. Both principal component analysis (PCA; Figure 1B) and one-way hierarchical clustering (Figure 1C) using the 762 probes differentially expressed in UPN727 cells revealed heterogeneity within the RO T1D and HC cohorts. While the majority of individual samples within the two cohorts induced distinct signatures, a subset of samples did not, resulting in the overlap detected by both clustering methods.
Figure 1.
Cross-sectional analysis of RO T1D patients identifies an inflammatory signature relative to unrelated HC subjects. (A) Venn diagram and one-way hierarchical clustering (probe sets only) for each component of the Venn diagram illustrate the relationship between the mean expression of probe sets regulated when fresh cells were previously cultured with plasma from 12 RO T1D and 12 HC subjects (498 probe sets |log2 ratio|>0.5; FDR<0.2) versus culturing UPN727 cells with plasma from 47 RO T1D and 44 HC subjects (n=762; (|log2 ratio|>0.263, 1.2-fold; FDR<0.2; ANOVA p<0.036). (B) PCA using 762 differentially regulated probe sets RO T1D vs HC) in cross-sectional studies. Green spheres, HCs; red spheres, RO T1D subjects. (C) One-way hierarchical clustering (probe sets only) of the RO T1D and HC expression profiles using the 762 regulated probe sets. (D) Relative expression levels of selected, well-annotated genes reflective of innate immune activity in RO T1D patients relative to HC subjects; additional well-annotated genes appear in Figure 3C. Fold of change is expressed relative to the mean of all samples.
Genes significantly over-expressed (n=186 probe sets) and under-expressed (n=576 probe sets) when UPN727 PBMCs were cultured with RO T1D or HC plasma were independently evaluated for biological pathway enrichment using DAVID to identify regulated Gene Ontology Biological Processes. Representative pathway terms appear in Table 1 (a complete list is provided in Supplemental Table 2) and selected probe sets are illustrated in Figure 1D. Ontological analysis of the 186 probe sets up-regulated by RO T1D plasma revealed 41 significant Gene Ontology Biological Processes (p<0.01; FDR<0.10), including immune response, chemotaxis, positive regulation of cytokine production, cell surface receptor signal transduction, and regulation of IL-6 production. These categories included genes encoding immune signaling molecules and receptors including the IL-1 cytokine family members IL1, IL1RN, IL1R1, and IL1R2, the IL-10 family member IL24, and the chemokines CXCL1, CXCL2, CXCL3, and CXCL5, which are involved in neutrophil chemotaxis38. RO T1D plasma induced cyclooxygenase-2 or COX-2 (PTGS2) and prostaglandin E synthase (PTGES). Genes for several immune receptors were also significantly up-regulated by RO T1D plasma, including TLR2, FCAR, LILRA3, and TREM1.
Table 1.
Enrichment of gene ontology (GO) terms in differentially expressed probe sets.a
| A. Probe sets regulated by RO T1D versus HC plasma | ||||
| GO Identifierb | Biological Process | nc | p value | FDR (%) |
| ↑ GO:0006955 | Immune response | 21 | 3.28E-7 | 5.40E-4 |
| ↑ GO:0006935 | Chemotaxis | 11 | 7.47E-7 | 1.22E-3 |
| ↑ GO:0001819 | Positive regulation of cytokine production | 7 | 1.10E-4 | 0.18 |
| ↑ GO:0009617 | Response to bacterium | 9 | 1.40E-4 | 0.23 |
| ↑ GO:0043405 | Regulation of MAP kinase activity | 7 | 1.08E-3 | 1.76 |
| ↑ GO:0007166 | Cell surface receptor linked signal transduction | 25 | 1.25E-3 | 2.03 |
| ↑ GO:0032680 | Regulation of TNF production | 4 | 2.29E-3 | 3.70 |
| ↑ GO:0032675 | Regulation of IL-6 production | 4 | 3.53E-3 | 5.64 |
| ↓ GO:0050672 | Negative regulation of lymphocyte proliferation | 6 | 1.31E-3 | 2.20 |
| ↓ GO:0016481 | Negative regulation of transcription | 24 | 1.75E-3 | 2.93 |
| ↓ GO:0030509 | BMP signaling pathway | 6 | 4.47E-3 | 7.34 |
| ↓ GO:0045892 | Negative regulation of transcription, DNA-dependent | 19 | 4.77E-3 | 7.81 |
| ↓ GO:0042130 | Negative regulation of T cell Proliferation | 5 | 4.92E-3 | 8.05 |
| ↓ GO:0016568 | Chromatin modification | 16 | 5.27E-3 | 8.60 |
| B. Probe sets regulated by CF versus HC plasma | ||||
| GO Identifierb | Biological Process | nc | p value | FDR (%) |
| ↑ GO:0006955 | Immune response | 104 | 3.34E-12 | 6.06E-9 |
| ↑ GO:0042110 | T cell activation | 35 | 1.77E-10 | 3.22E-7 |
| ↑ GO:0002521 | Leukocyte differentiation | 34 | 2.83E-9 | 5.14E-6 |
| ↑ GO:0042113 | B cell activation | 17 | 3.33E-4 | 0.60 |
| ↓ GO:0009617 | Response to bacterium | 39 | 5.02E-8 | 9.30E-5 |
| ↓ GO:0001819 | Positive regulation of cytokine production | 24 | 9.46E-6 | 0.02 |
| ↓ GO:0043405 | Regulation of MAP kinase activity | 28 | 2.48E-4 | 0.45 |
| ↓ GO:0007166 | Cell surface receptor linked signal transduction | 25 | 1.25E-3 | 2.03 |
| C. Probe sets regulated by bacterial pneumonia versus HC plasma. | ||||
| GO Identifierb | Biological Process | nc | p value | FDR (%) |
| ↑ GO:0006955 | Immune response | 74 | 6.72E-8 | 1.20E-4 |
| ↑ GO:0010942 | Positive regulation of cell death | 47 | 1.27E-4 | 0.23 |
| ↑ GO:0034620 | Cellular response to unfolded protein | 8 | 1.64E-4 | 0.29 |
| ↑ GO:0007050 | Cell cycle arrest | 16 | 1.36E-3 | 2.41 |
| ↑ GO:0045582 | Positive regulation of T cell differentiation | 8 | 2.64E-3 | 4.62 |
| ↓ GO:0043066 | Negative regulation of apoptosis | 55 | 3.18E-11 | 5.83E-8 |
| ↓ GO:0007596 | Blood coagulation | 22 | 2.26E-7 | 4.14E-4 |
| ↓ GO:0050715 | Positive regulation of cytokine secretion | 7 | 1.27E-3 | 2.31 |
| D. Probe sets regulated by pre-H1N1 versus active H1N1 plasma | ||||
| GO Identifierb | Biological Process | nc | p value | FDR (%) |
| GO:0009615 | Response to virus | 24 | 3.91E-25 | 6.46E-22 |
| GO:0051607 | Defense response to virus | 5 | 4.03E-05 | 6.64E-2 |
| GO:0030330 | DNA damage response, signal transduction by p53 class mediator | 4 | 2.13E-3 | 3.45 |
| GO:0032647 | Regulation of interferon-α production | 3 | 2.65E-3 | 4.29 |
| GO:0032649 | Regulation of interferon-γ production | 4 | 3.22E-3 | 5.19 |
Analysis restricted to GO biological processes with ≥3 UniGenes were detected per category, p<0.01,and FDR<10%.
Arrow denotes up- or down-regulation. Due to the relatively low numbers of genes identified in the H1N1 studies, up- and down-regulated genes were not separated for analysis with DAVID.
Total probe sets detected.
Ontological analysis of the 576 probe sets down-regulated by RO T1D plasma identified 27 significant Gene Ontology Biological Processes (p<0.01; FDR<10%) that were functionally opposed to the terms enriched in the up-regulated subset, such as negative regulation of mononuclear cell proliferation, negative regulation of transcription, BMP signaling pathway, negative regulation of T cell proliferation (Table 2 and Supplemental Table 3). Relative to HC plasma, culture with RO T1D plasma led to the under-expression of genes such as KLF12, SKIL (a TGF-β-induced gene that negatively regulates TGF-β signaling), the histone deacetylase HDAC3, FOXP1 (a transcription factor that is important in maintaining immune quiescence), and ZEB2 (a human zinc finger transcription factor and transcriptional repressor that regulates T-cell activity). Overall, the RO T1D and HC profiles reflect inflammatory versus immune-regulatory processes, respectively, suggesting that RO T1D sera possesses higher levels of pro-inflammatory mediators such as IL-1 and lower levels of anti-inflammatory factors.
Table 2.
Cytokine/chemokine levels in RO T1D (n=17) and unrelated HC (n=15) plasma samples.
| Cytokine | RO T1D (pg/ml) |
Unrelated HC pg/ml |
Fold RO T1D/HC |
Lower detection limit (pg/ml) |
|---|---|---|---|---|
| Eotaxin | 104.2 ± 10.3 | 79.8 ± 25.1 | 1.3 | >3.2 pg/ml |
| Granulocyte/macrophage CSF | 25.9 ± 9.1 | 7.90 ± 4.3 | 3.3 | >3.2 pg/ml |
| Interferon-α2 | 73.9 ± 7.3 | 54.5 ± 7.2 | 1.4 | >3.2 pg/ml |
| Interferon-γ | 9.8 ± 4.7 | 5.5 ± 2.6 | 1.8 | >3.2 pg/ml |
| IL-1a | 59.8 ± 24.1 | 23.4 ± 11.0 | 2.6 | >3.2 pg/ml |
| IL-1b | 7.8 ± 2.4 | 6.4 ± 2.3 | 1.2 | >3.2 pg/ml |
| IL-2 | 2.3 ± 1.2 | 0.8 ± 0.2 | 2.9 | >3.2 pg/ml |
| IL-3 | 0.0 ± 0.0 | 0.2 ± 0.7 | 0.0 | >3.2 pg/ml |
| IL-4 | 9.0 ± 4.0 | 2.1 ± 1.8 | 4.3 | >3.2 pg/ml |
| IL-5 | 1.7 ± 0.6 | 0.5 ± 0.2 | 3.2 | >3.2 pg/ml |
| IL-6 | 8.0 ± 2.9 | 6.2 ± 2.2 | 1.3 | >3.2 pg/ml |
| IL-7 | 14.1 ± 2.7 | 10.0 ± 2.7 | 1.4 | >3.2 pg/ml |
| IL-8 | 14.7 ± 3.4 | 15.4 ± 4.0 | 1.0 | >3.2 pg/ml |
| IL-10 | 11.8 ± 2.9 | 11.6 ± 4.1 | 1.0 | >3.2 pg/ml |
| IL-12p40 | 83.4 ± 22.85 | 58.9 ± 16.6 | 1.4 | >3.2 pg/ml |
| IL-12p70 | 9.5 ± 6.7 | 1.6 ± 0.7 | 6.0 | >3.2 pg/ml |
| IL-13 | 9.5 ± 4.7 | 9.5 ± 4.8 | 2.5 | >3.2 pg/ml |
| IL-15 | 7.4 ± 2.2 | 7.5 ± 2.4 | 1.0 | >3.2 pg/ml |
| IL-17 | 4.8 ± 1.6 | 6.8 ± 3.1 | 0.7 | >3.2 pg/ml |
| IP-10 | 221.8 ± 26.2 | 242.8 ± 35.4 | 0.9 | >3.2 pg/ml |
| Monocyte chemoattractant protein -1 | 252.8 ± 14.1 | 214.9 ± 22.3 | 1.2 | >16.0 pg/ml |
| Macrophage inflammatory protein-1a | 12.6 ± 4.0 | 13.0 ± 4.5 | 1.0 | >16.0 pg/ml |
| Macrophage inflammatory protein-1b | 27.2 ± 6.5 | 13.4 ± 5.3 | 2.0 | >16.0 pg/ml |
| TNFa | 9.3 ± 1.0 | 6.5 ± 0.6 | 1.4* | >3.2 pg/ml |
| TNFb | 30.6 ± 12.4 | 10.0 ± 4.5 | 3.1 | >3.2 pg/ml |
Data are means ± standard error (SE, pg/ml). Each sample was tested in duplicate using the Millipore BeadLyte cytokine assay kit.
p<0.05, two-tailed t-test.
Finally, we employed ToppGene39, 40 to further explore the functional commonality in the originally reported response of multiple fresh PBMC to RO T1D and HC plasma compared to those of cryopreserved PBMC using the two datasets illustrated in Figure 1A. When we used the 498 probe sets differentially expressed in fresh cells as the training set, we detected 431/762 (56.6%) significant (p<0.01) functionally related probe sets in cryopreserved cells. Overall, the cross-sectional analysis of the RO T1D and HC samples demonstrates that a pro-inflammatory signature is induced in cryopreserved PBMCs that is functionally concordant with that previously observed in fresh cells31.
Direct detection of inflammatory mediators in RO T1D and HC plasma samples
In a continuing effort to account for the induced signatures, cytokine levels were measured by multiplex ELISA in a subset of 17 RO T1D and 15 HC subjects examined in the expression studies (Table 2). Higher levels of several cytokines were detected in RO T1D plasma compared to HCs, including IL-1α, which is consistent with the induction of IL-1-mediated gene expression. However, with the exception of TNFα (RO T1D: 9.3 ± 1.0 pg/ml; HC: 6.5 ± 0.6 pg/ml; p<0.05), none of these differences reached statistical significance with the number of subjects analyzed. Furthermore, correlations between the intensity of the signature among individual samples (Figure 1B and 1C) and mediator levels, antibody levels, gender, age of onset, or time to post-onset sample collection could not be made.
Analysis of Long-Standing (LS) T1D and Pre-Diabetes
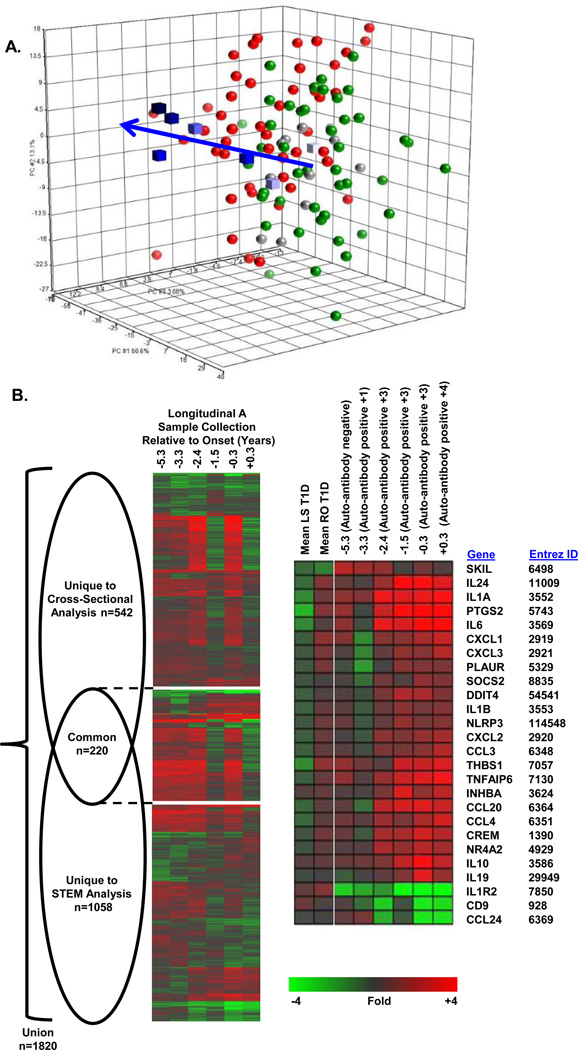
Using cryopreserved UPN727 PBMC as reporter cells, we analyzed sera of 11 LS T1D subjects, all of whom were >10 years post-onset; 10/11 possessed measurable titers for at least one islet cell auto-antibody. As reflected by PCA of the 762 probe sets differentially expressed in the cross-sectional RO T1D and HC samples (Figure 2A), LS T1D plasma induced transcription similar to that induced by HC plasma. Of the 762 probe sets differentially regulated between the RO T1D and HC cohorts, only 84 probe sets (11%) met our thresholds (|log2 ratio|>0.263, 1.2-fold; FDR<0.2) when comparing the LS T1D and HC groups. Consistent with our previous report31, the signature associated with RO T1D is not induced by samples collected from established T1D patients, supporting the hypothesis that the inflammatory signature induced by RO T1D plasma arises from factors related to active autoimmune processes.
Figure 2.
Analysis of T1D progression in Longitudinal Subject A. (A) PCA using the 762 probe sets regulated by RO T1D (n=47) vs HC (n=44) plasma identified in the cross-sectional studies (|log2 ratio|>0.263; FDR<0.2). Green spheres, HC; red spheres, RO T1D; grey spheres, LS T1D (>10 years post-onset); blue cubes, Longitudinal Subject A series (lightest to darkest blue indicates sample order, −5.3, −3.3, −2.4, −1.5, −0.3, +0.3 years relative to onset, respectively). Arrow shows progression to RO T1D. (B) Venn diagram and one-way hierarchical clustering (probe sets only) for each component of the Venn diagram illustrate the relationship between the probe sets identified in the STEM analysis of the Longitudinal Subject A series versus the cross-sectional analyses of the RO T1D and HC samples. The signatures share a significantly nonrandom (p<10−51, Χ2 test), commonly regulated intersection of 220 probe sets (Supplemental Table 2). Relative expression levels are shown for selected, well-annotated genes related to inflammatory processes that were significantly identified by the STEM analyses. Additional well-annotated genes are shown in Figure 3C.
We also used cryopreserved UPN727 PBMC to evaluate longitudinal samples collected from a prospectively monitored sibling of a proband that progressed to T1D. This subject was diagnosed at the age of 21.7 years, and samples were collected at −5.3 years (auto-antibody negative), −3.3 years (+1 auto-antibody), −2.4 years (+3 auto-antibodies), −1.5 years (+3 auto-antibodies), −0.3 years (+3 auto-antibodies), and +0.3 years (+4 auto-antibodies) relative to onset (Supplemental Table 1). The response of cryopreserved PBMCs to the plasma of this longitudinal series was analyzed from the perspective of probe sets regulated by RO T1D and HC plasma in the cross-sectional studies, and with Short Time-Series Expression Miner (STEM), a software tool for the analysis of time-series gene expression data41.
Disease progression was evident in the PCA of the longitudinal plasma samples (Figure 2A). The relationship between the regulated genes identified in the cross-sectional analysis of RO T1D and HC samples (n=762 probe sets) compared to those identified by STEM analysis of this single longitudinal series (n=1,278 probe sets; STEM profiles with detection p <10−25 and a minimum 1.68-fold change between any two time points) is illustrated in Figure 2B; the signatures share a significantly nonrandom (p<10−51, Χ2 test), commonly regulated intersection of 220 probe sets. The Pearson correlation coefficients (calculated using the RO T1D:HC log2 ratio for the cross-sectional samples and the last:first time point log2 ratio for the longitudinal series) for the union and intersection of these two datasets were 0.55 and 0.78, respectively. The regulated probe sets uniquely regulated in the longitudinal and cross-sectional analyses expectedly showed lower correlation, as 1) the inflammatory state measured at T1D onset differs from that at points earlier in disease progression, and 2) the signature of induced by samples prior to onset of T1D in the longitudinal analysis are distinct from the samples of unrelated HC examined in the cross-sectional analyses (Figure 2B and Supplemental Table 2).
STEM was also employed to examine the profiles generated when these longitudinally collected plasma samples were previously used to induce transcription in healthy, freshly drawn PBMCs from a single donor31. Among the 2,319 probe sets differentially expressed between the analyses of the longitudinal series using fresh versus cryopreserved cells, we identified a shared, significantly nonrandom (p<10−57, Χ2 test), correlative (Pearson correlation coefficient = 0.63), commonly regulated intersection of 154 probe sets. We again employed ToppGene39, 40 to examine the functional similarity in the responses of fresh and cryopreserved PBMCs to plasma samples from the longitudinal series. Using the 1,195 probe sets differentially expressed in fresh cells as the training set, we identified 885/1,278 (69.3%) significant (p<0.01), functionally related probe sets among those regulated in cryopreserved cells.
As with fresh PBMCs31, we observed an increase in the robustness and complexity of the signature with disease progression (Figure 2C). Genes such as such as IL1A, IL24, IL10, IL1R1, and PTGS2 exhibited increased induction while others such as CCL24 and SKIL showed lower induction by samples collected closer to onset. Importantly, induction of inflammatory genes was evident in the sample collected 5.3 years prior to onset, prior to autoantibody development, and overall the signature of all pre-onset samples was distinct from that of the mean response of the 44 unrelated healthy controls examined in the cross-sectional analysis. Finally, we examined the mean absolute fold-change for probe sets in the 2.5% tails of the log2 ratio distribution in the first and last samples of the series (the highest 1,365 and lowest 1,365 expressed probe sets). We observed a difference of 0.72 ± 0.22 in the UPN727 cells (40% plasma) versus 0.67 ± 0.23 in fresh PBMCs from a single donor (20% plasma). The responsiveness of the cryopreserved UPN727 cells under these conditions were on a par with that of the fresh PBMCs originally used to examine this longitudinal series31. The longitudinal studies further highlight the functional concordance of the plasma-induced transcriptional signatures generated in fresh and cryopreserved PBMCs.
The T1D signature is distinct from signatures associated with CF colonized with Pa, bacterial pneumonia, and H1N1 influenza
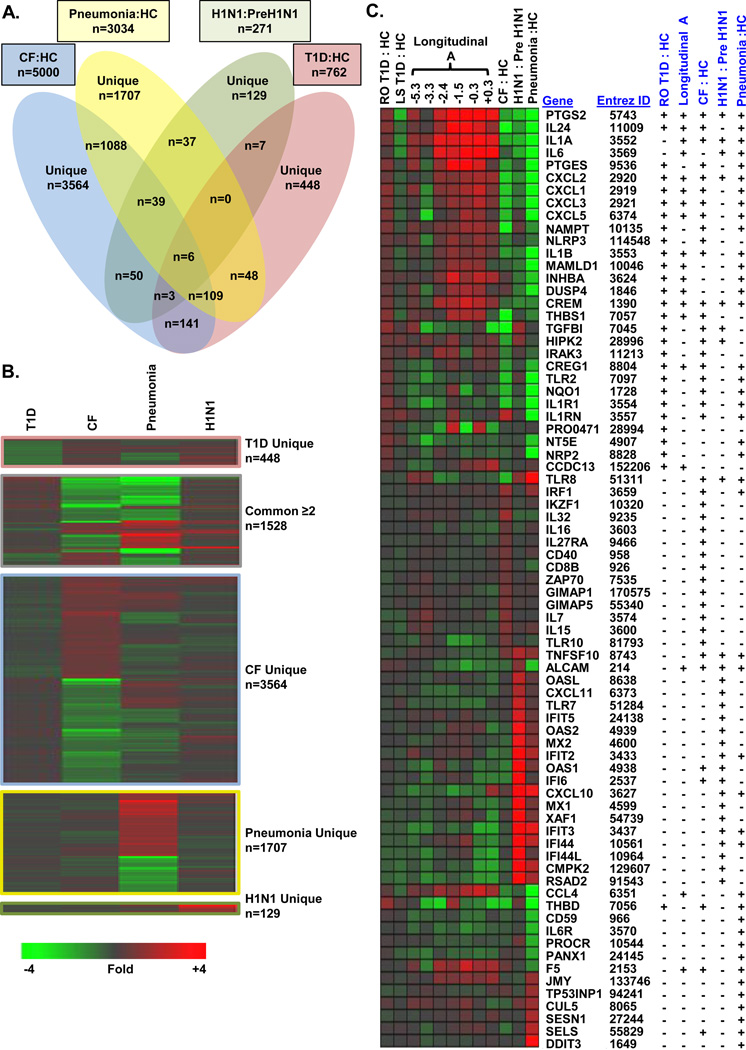
Next, we focused on the specificity of the signature induced by T1D plasma by evaluating samples from other inflammatory conditions. Plasma samples from 20 non-diabetic, pediatric CF patients chronically colonized with Pa and 24 age-matched unrelated HCs were used to induce transcription in UPN727 cells at a culture concentration of 20% (Supplemental Table 1). A robust signature consisting of 5,000 differentially expressed probe sets (|log2 ratio|>0.263, 1.2-fold; FDR<0.2; 3,404 unique UniGenes) was distinct from that induced by RO T1D plasma (Figure 3, Supporting Information Table 2). Ontological analysis of the 2,139 probe sets up-regulated by CF plasma vs HC plasma revealed 101 significantly enriched Gene Ontology Biological Processes (p<0.01; FDR<0.10); many of these annotations were related to immunological activation and T-cell/B-cell activation. CF sera induced transcription of INF regulatory factor 1 (IRF1), GIMAP1, GIMAP5, TLR10, IL32, CCL5, CD40, IKZF1, IKAROS family zinc finger 1 (IKZF1), IL15, and IL16. Analysis of the 2,861 probe sets down-regulated by CF plasma identified 238 significant Gene Ontology Biological Processes (p<0.01; FDR<10%). Many Biological Processes up-regulated by T1D sera were found down regulated by CF sera (Table 1B). Accordingly, many IL-1 regulated genes annotated within these categories representing functions of immune recognition and response were found to be down regulated by CF sera, including PTGS2, CCL2, IRAK3, IL1B, and IL1R1 (Figure 3). Selected pathway terms appear in Table 2B, and the complete list is provided in Supplemental Table 3.
Figure 3.
Distinctiveness of signatures associated with T1D, CF plus Pa colonization, and H1N1. (A) Venn diagram of the probe sets induced in UPN727 cells following exposure to the plasma from T1D patients (n=47) versus age-matched unrelated HCs (n=44; |log2 ratio|>0.263; FDR<0.20), CF patients harboring Pa (n=20) versus age-matched HCs (n=24; |log2 ratio|>0.263; FDR<0.20), patients with bacterial pneumonia (n=10) versus HCs (n=18; |log2 ratio|>0.5; FDR<0.20), and active versus pre-H1N1 infection (five subjects sampled during and before infection; |log2 ratio|>0.5; paired t-test p<0.05). (B) One-way hierarchical clustering (probe sets only) was conducted for each component of the Venn diagram using mean expression values for each dataset. (C) Well-annotated, differentially expressed genes related to immunological activation, signal transduction, or transcriptional regulation.
We examined 10 previously healthy, pediatric bacterial pneumonia patients with confirmed bacterial infection who had been admitted to the intensive care unit; eight required mechanical ventilation. Of the four inflammatory diseases examined in this investigation, the plasma of these critically ill children (at a culture concentration of 20%) induced the most robust signature (relative to 18 HCs; n=8,121; pneumonia:HC |log2 ratio|>0.263; FDR<0.20; 4,410 up-regulated probe sets and 3,711 down-regulated probe sets; 5,650 unique UniGenes; Supplemental Table 2). The transcriptional response of cryopreserved UN727 cells to sera collected from the bacterial pneumonia patients was distinct from that following exposure to T1D or CF sera. The ontological analysis was restricted to the 3,034 probe sets that had pneumonia:HC |log2 ratio|>0.5 and FDR<0.20 (2,144 unique UniGenes). Selected pathway terms appear in Table 1C; a complete list is provided in Supplemental Table 3.
Ontological analysis of the 1,515 probe sets up-regulated by plasma collected from bacterial pneumonia patients revealed 64 significant Gene Ontology Biological Processes (p<0.01; FDR<0.10), including cellular response to unfolded protein, cell cycle arrest, and positive regulation of T cell differentiation. Genes uniquely regulated by bacterial pneumonia patient plasma and annotated under these terms included DNA-damage-inducible transcript 3 (DDIT3), selenoprotein S (SELS; involved in the endoplasmic reticulum stress response and mediating inflammation), and the p53 target genes Sestrin1 (SESN1), cullin 5 (CUL5; an inhibitor of cellular proliferation), tumor protein p53 inducible nuclear protein 1 (TP53INP1), junction mediating and regulatory protein p53 cofactor (JMY), and others (Figure 3C). Ontological analysis of the down-regulated 1,519 probe sets identified 242 significant Gene Ontology Biological Processes (p<0.01; FDR<0.10), many of which were related to immune response modulation, negative regulation of apoptosis, chemotaxis, and blood coagulation. Genes annotated within these pathways included pannexin 1 (PANX1; involved with IL1-β release), IL6R, CCL4, the endothelial protein C receptor (PROCR), protectin (CD59; a complement regulatory protein), coagulation factor V (F5), thrombomodulin (THBD), and others (Figure 3).
Lastly, samples collected from 5 middle-aged subjects (mean age 49.60 ± 2.07 years) before and during H1N1 infection were also analyzed. Analysis of cultures exposed to pre-H1N1 versus symptomatic H1N1 plasma revealed a signature of 271 significantly regulated probe sets representing 182 unique UniGenes (255 over-expressed probe sets; 16 under-expressed probe sets; Figure 3; |log2 ratio|>0.5; paired t-test p<0.05). This signature was distinct from the signatures induced with plasma from RO T1D patients, CF patients colonized with Pa, and previously healthy patients with bacterial pneumonia. We used DAVID to identify 32 significantly enriched annotations (p<0.01; FDR<10%). As anticipated, numerous Biological Processes typically viewed as host responses to infection or immune defense were induced by H1N1 plasma, including response to virus, defense response to virus, and regulation of α and γ INF production. Selected pathway terms are tabulated in Table 1D, and the complete list appears in Supplemental Table 3.
Notably, sera collected during H1N1 infection induced transcription of numerous IFN-regulated genes, including INF-induced protein with tetratricopeptide repeats 1 (IFIT1), IFIT5, IFITM1, and IFITM3. Members of this gene family are induced in response to type I and type II INF, dsRNA, and endotoxin42, 43. Sera of H1N1-infected individuals induced expression of other IFN-regulated genes including IFI6 and MX1, and INF-stimulated exonuclease gene 20kDa (ISG20), which encodes an anti-viral ssRNA-specific exonuclease. Also induced by H1N1 sera were 2',5'-oligoadenylate synthetase 3 (OAS3) and OASL, which are INF-induced enzymes that bind dsRNA and polymerize ATP into 2'-5' linked oligomers of adenosine (pppA(2'p5'A)n). H1N1 sera also regulated numerous genes related to pathogen recognition, including TLR7 and TLR8, which recognize viral RNA. Interestingly, several genes differentially expressed in response to H1N1 sera also exhibited significant, opposite induction by T1D sera, including the transcription factor cAMP response element modulator (CREM), IL1A, PTGS2, IL6, INHBA, and the nuclear receptor subfamily 4 group A member 2 (NR4A2).
Figure 3 illustrates the uniqueness of each inflammatory state and how the genes induced under the various conditions are regulated. Correlation coefficients between the signatures induced by each of the inflammatory states were calculated using the probe sets significantly regulated by each condition as well as the union of all significantly regulated probe sets of the four conditions (Table 3). The T1D signature was negatively correlated with the signatures of CF patients chronically colonized with Pa, bacterial pneumonia patients, and subjects with H1N1 influenza. The signatures associated with CF colonized with Pa compared to the bacterial pneumonia patients showed the highest, albeit modest, level of overall correlation (0.19), further demonstrating the disease-specificity of the approach.
Table 3.
Pairwise Pearson correlation coefficients for the signatures induced by different inflammatory states.
| n=a | T1D | CF with Pa colonization |
Bacterial pneumonia |
H1N1 | |
|---|---|---|---|---|---|
| T1D | 762 | 1.00 (1.00)b | −0.72 (−0.59) | −0.47 (−0.23) | −0.42 (−0.15) |
| CF/Pa colonization | 5000 | 1.00 (1.00) | 0.23 (0.19) | −0.03 (−0.03) | |
| Bacterial pneumonia | 3034 | 1.00 (1.00) | 0.39 (0.16) | ||
| H1N1 | 271 | 1.00 (1.00) |
The total number of regulated probe sets meeting the following thresholds: RO T1D:HC |log2 ratio|>0.5, FDR<0.20; CF:HC |log2 ratio|>0.263, FDR<0.20; pneumonia:HC |log2 ratio|>0.5, FDR<0.20; H1N1:pre-H1N1 |log2 ratio|>0.5; paired t-test p<0.05).
Pearson correlation coefficients for genes in the union of the probe sets differentially regulated under all four conditions (n=7,376) are shown in parentheses.
DISCUSSION
Autoimmunity and β-cell destruction occur asymptomatically for months to years prior to onset of T1D, providing an excellent opportunity for early diagnosis and intervention. The evaluation of auto-antibody status in combination with HLA genotyping can reliably predict susceptibility to T1D44. However, auto-antibodies are thought to appear late during pre-diabetes, their levels can be transient, and not all auto-antibody positive subjects progress to T1D onset45. Furthermore, the humoral response is considered secondary to the initiation of autoimmune processes and it is generally accepted that auto-antibodies are markers of β-cell destruction but may not directly mediate T1D development46–48. Thus, a need remains for the discovery and evaluation of minimally invasive biomarkers that are amenable to repeated measurement and capable of detecting early T1D in a mechanistically informative, disease-specific manner49.
A complex but dilute cytokine milieu is associated with T1D50–55. While these mediators are difficult to measure directly via present ELISA-based approaches, our studies have shown that they are sufficient to drive transcription in a reporter cell system31–33. To overcome the inherent limitations associated with cell-based assays that rely on freshly drawn responder PBMCs, we previously explored the use of lymphoid and myeloid cell lines as potential PBMC surrogates33. We found them able to differentially respond to RO T1D versus HC sera however the induced profiles of each cell line were distinct from one another and from fresh PBMC, and less biologically informative.
Here, we expanded the observations of our initial report31 using commercially supplied cryopreserved PBMCs from a single donor as the responder cell population. Such cells are collected in high numbers through aphaeresis and are rapidly processed to maximize post-thaw viability. The use of these cells allows for control of HLA type, gender and immune reactivities, reduces data heterogeneity introduced by using responder PBMCs from multiple donors or draws of the same individual, eliminates the need of repeated recruitment of volunteer blood donors, and simplifies laboratory workflow by obviating the need for monocyte/lymphocyte isolation from whole blood for each assay. As a prelude to this study, we compared the response of cryopreserved PBMC drawn from 5 different donors to RO T1D and HC plasma (data not shown). While PBMCs of all the donors tested were able to differentially respond to RO T1D and HC plasma, UPN727 PBMC were selected as they most closely mimicked the mean response of freshly isolated PBMC of multiple donors used previously and the highest number of cryopreserved cells were available31. We observed >90% post-thaw viability with UPN727 cells based upon trypan blue exclusion, and multi-parameter flow cytometry has shown that the relative abundances of natural killer cells, CD8+ T cells, CD4+ T cells, B cells, and monocytes do not significantly differ from the normal expected ranges for fresh PBMCs (Supplemental Table 4).
When analyzing the transcription induced in UPN727 cells by plasma of 47 RO T1D and 44 HC subjects we identified 762 significantly regulated probe sets. The analysis of these samples required multiple assays where good inter and intra assay reproducibility were observed. Specifically, in the duplicate analysis of 5 independent RO T1D and HC samples, we observed low mean intra-assay coefficients of variation (0.029±0.030 for the entire array and 0.034±0.036 for the 762 probe set RO T1D:HC signature). When analyzing the same sample in 5 independent assays we observed low mean inter-assay CVs (0.051±0.041 for the entire array and 0.095±0.062 for the 762 probe set RO T1D:HC signature). In these analyses, the intra and inter assay correlation coefficients for the 762 probe set RO T1D:HC signature was 0.94 and 0.71, respectively.
PCA and hierarchical clustering revealed heterogeneity within the RO T1D and HC cohorts (Figure 2). We attribute this to the possibility that greater inter-subject variability was captured in this analysis which possessed a larger number of subjects compared to our initial report31. Furthermore, some of the less distinct RO T1D signatures likely arose through post-onset resolution of the autoimmune response similar to that observed the LS T1D cohort. Ongoing studies with this bioassay show that resolution of the immune response occurs during the first year post-onset (data not shown).
In this study, using cryopreserved cells, we re-examined a single longitudinal series studied in our previous report31, confirming temporal changes in the inflammatory state preceding T1D onset. Furthermore, a concordant response was induced in the cryopreserved PBMCs (40% plasma) that exhibited an overall fold-change similar to that of freshly drawn PBMCs (20% plasma). The elimination of inter-individual heterogeneity is an advantage of longitudinal analyses compared to cross-sectional studies. This benefit is highlighted by the observation that, in general, for any regulated gene, the fold-change detected in the longitudinal study tended to be larger than that observed between cohorts in the cross-sectional study. For example, at onset a >10-fold change in IL-6 and PTGS2 expression was observed in the longitudinal analysis versus a <2-fold mean change observed in the cross-sectional study. The maximal individual fold-changes in IL-6 and PTGS2 relative to the mean HC response were 2.25-fold and 2.97-fold, respectively. Thus, a baseline sample collected before disease initiation is likely the best control sample for a given individual in the study of disease progression through T1D onset. Since significantly regulated genes identified in cross-sectional studies of samples collected at onset may not necessarily be the most informative early biomarkers of T1D, longitudinal studies of pre-onset samples are needed to identify the earliest indices of immunological activation and/or loss of immune regulation. The transcripts most consistently regulated from disease initiation through onset in most/all subjects will reflect the most common pathways associated with diabetogenesis and will represent the most reliable biomarkers for predicting T1D onset. Biobanks and associated databases such as the TrialNet Natural History Study56 and The Environmental Determinants of Diabetes in the Young (TEDDY)57 will greatly facilitate such efforts. Importantly, our longitudinal analyses of progressors to T1D studies conducted thus far have identified differentially expressed genes not identified in cross-sectional studies, and vice versa, clearly indicating that both analysis strategies are necessary.
In our larger study sample, genes known to be influenced by IL-1 were again over-represented in the transcriptional signatures of cells exposed to RO T1D plasma. These genes included IL1B, cyclooxygenase type 2 (PTGS2), prostaglandin E synthase (PTGES), IL-1R1, CXCL1, CXCL2, CXCL3, CXCL5, PLAUR, CREM, and others. This observation is, in general, consistent with the elevated plasma IL-1α levels detected in the RO T1D cohort by multiplex cytokine analysis (Table 2) as well as other functional genomics-based investigations of T1D that have implicated innate immunity and IL-1 in T1D pathogenesis58, 59. In addition to regulating T-cell activity, IL-1 promotes β-cell dysfunction through the mitogen-activated protein kinase and NFκB pathways, leading to endoplasmic reticulum and mitochondrial stress and cell death60–62. Blockade of the IL-1 receptor with IL-1RA can protect β cells from the downstream consequences of IL-1 exposure63. The clinical efficacy of blocking IL-1 action has been observed in many inflammatory diseases, including rheumatoid arthritis and Type 2 diabetes mellitus29, 64. Furthermore, genetic or pharmacological abrogation of IL-1 action has been reported to delay and/or reduce T1D incidence in rodent models32, 65, 66. These findings underlie the rationale for recent efforts to evaluate therapeutic targeting of IL-1 in T1D67–69, where we believe this approach may hold utility in monitoring such interventions and yielding additional insight.
Similar to studies that have identified disease-specific signatures from the transcriptomes of PBMC or immunocyte subpopulations, we find that plasma-induced transcription offers a mechanistically informative and disease-specific read out. Relative to T1D, the cytokine milieus associated with CF, bacterial pneumonia, and H1N1 drove distinct transcriptional programs. The innate, IL-1 related components of the T1D signature were oppositely regulated in these three inflammatory states and the overall signatures associated with these disorders negatively correlated with the T1D signature. While multiplex analysis of peripheral cytokines was not carried out in the samples from CF patients with chronic Pa colonization, bacterial pneumonia, or H1N1, the number of regulated transcripts and fold-change in induced gene expression are consistent with these conditions being more robust inflammatory pathologies of larger tissues. It is important to note that although the genes differentially regulated among the other conditions have varying roles in pathologic pulmonary inflammation, as a group these genes participate in immune recognition and response, phagocytosis, and matrix degradation.
Overall, these studies validate the remarkable capacity of cryopreserved cells to serve as disease-specific biosensors that are capable of sensitively and comprehensively capturing the plasticity of the immune system and differentiating the diverse range of inflammatory processes that underlie human disease. Ongoing studies include analyses of our methodology to other auto-inflammatory disorders that are mechanistically more related to T1D; we anticipate that these investigations will more rigorously test the disease-specificity of our approach within the context of autoimmunity.
MATERIALS AND METHODS
Subjects and subject characterization
T1D patients were recruited through the Diabetes Clinic at Children’s Hospital of Wisconsin and diabetes was defined according to World Health Organization criteria70. Samples of normoglycemic RO T1D patients (n=47; mean age 9.97 ± 2.89 years, mean glycosylated hemoglobin (HgA1c) fraction 7.47 ± 1.20%) were collected after stabilization on exogenous insulin 2–7 months after diagnosis. Samples from LS T1D patients (n=11; mean age 28.20 ± 9.29 years, mean HgA1c fraction 7.91 ± 1.34%) were collected from well-controlled normoglycemic subjects that were ≥10 years post-onset. The recruitment criteria for unrelated Caucasian HCs (n=44; mean age 14.98 ± 4.13 years) included fasting blood glucose levels <100 mg/dl, no familial history of any autoimmune/autoinflammatory disorder, <25 years of age, and negativity for islet auto-antibodies at the 99th percentile71. All HC and T1D study subjects were free of known infection at the time of sample collection and had peripheral blood drawn aseptically and collected in acid citrate dextrose solution A or K+EDTA anti-coagulant at Children’s Hospital of Wisconsin by trained phlebotomists. Blood components were immediately separated by Ficoll-Histopaque (Sigma Aldrich, St. Louis, MO) density gradient centrifugation, and plasma was stored at −80 °C until use. GAD, IA2, IAA, and ZNT8 autoantibody titers were determined as described71. HLA-DQB1 genotyping in T1D and HC subjects was performed via direct sequencing of the second exon with the SeCore DQB1 Locus Sequencing Lit (Invitrogen Life Technologies, Grand Island, NY) per the manufacturer’s instructions. HLA-DQA1-DQB1 haplotypes were inferred using reported European Caucasian haplotype frequencies72. Subject characteristics are shown in Supplemental Table 1A.
The plasma of 20 pediatric CF patients (mean age 14.87 ± 5.54 years) and 24 age-matched, unrelated HCs (mean age 15.17 ± 5.25 years; Supplemental Table 1B) was aseptically collected in acid citrate dextrose solution A or K+EDTA anti-coagulant. The diagnosis of CF was documented in the medical record based on the results of the pilocarpine iontophoresis sweat test (sweat chloride ≥60 mEq/lL). Clinical serum marker levels and lung function measurements confirmed that the values were performed at baseline and not during a pulmonary exacerbation defined as a forced expired volume change of greater than 15% predicted. At the time of sample collection, all patients were screened to identify microbiological flora; all subjects harbored mucoid Pseudomonas aeruginosa (Pa), as Pa was cultured from sputum or oropharygneal swab for 6 consecutive months73, 74. All subjects were homozygous for the F508del CFTR mutation and were pancreatic insufficient, determined by stool elastase level, but none were diabetic (fasting blood glucose <120 mg/dl). Subject characteristics appear in Supplemental Table 1B.
The plasma of 10 children (mean age 9.37 ± 6.72 years) with community-acquired pneumonia admitted to the Pediatric Intensive Care Unit at the Children’s Hospital of Wisconsin was aseptically collected in K+EDTA anticoagulant. Community-acquired pneumonia was defined using previously published guidelines75 and included: 1) acute illness (<14 days of symptoms), 2) the presence of a new chest radiographic infiltrate or consolidation confirmed by a radiologist, and 3) clinical features compatible with pneumonia including one of the following three features: fever >37.8 °C, hypothermia <36 °C, peripheral white blood count >10,000/µl or <4,500/µl or >15% immature neutrophils; plus two of the following three criteria: tachypnea (respiratory rate >2 standard deviations from the mean for age), dyspnea, or hypoxemia (pulse oximetry ≤94% on room air on initial evaluation without a known mixing heart lesion). Plasma was collected within 24 hours of admission to the Pediatric Intensive Care Unit and analyzed as described below. These children all had confirmed bacterial pneumonia; 8/10 required mechanical ventilation for acute respiratory failure. These subjects were compared to a cohort of unrelated HCs (mean age 12.92 ± 3.89 years). Subject characteristics are shown in Supplemental Table 1C.
We also examined plasma from five healthy middle-aged (mean age 49.60 ± 2.07 years), HLA-A2.1 blood donors. Samples were collected prior to influenza infection, and at the onset of flu-like symptoms, subjects were re-drawn and H1N1 status was ascertained by PCR-based testing of a nasal swab. The blood samples were collected as part of a protocol approved by the Institutional Review Board of the Blood Center of Wisconsin. The characteristics of these subjects appear in Supplemental Table 1D.
This study was approved by the Institutional Review Board of the Children’s Hospital of Wisconsin and informed consent was obtained from subjects or parents/legal guardians.
PBMC cultures and gene expression analysis
Commercial cryopreserved PBMCs from a Caucasian HLA-A2 male donor were thawed and washed per the manufacturer’s protocol (UPN727, Cellular Technology Ltd., Shaker Heights, OH). Gene expression was accomplished by culturing PBMCs at 37 °C in 5% CO2 with either 20% or 40% of autologous, HC, RO T1D, LS T1D, longitudinal pre-T1D, CF, pneumonia, or H1N1 plasma. Cultures were prepared in a Costar 24-well plate (Corning) using 500,000 cells/well and in RPMI 1640 medium supplemented with 100 U/ml penicillin and 100 µg/ml streptomycin in a total volume of 500 µl. After culture (9 hours), total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies). Using purified total RNA (100 ng), cRNA was synthesized and amplified/labeled using the Affymetrix Express Kit, then fragmented and hybridized to the GeneChip Human Genome U133 plus 2.0 array in accordance with the Affymetrix GeneChip expression analysis technical manual (Affymetrix, Santa Clara, CA). After hybridization, arrays were washed and stained with Affymetrix fluidics protocol FS450_0001 and scanned with a 7G Affymetrix GeneChip Scanner. Image data were analyzed with Affymetrix Expression Console™ 1.1.2 software and normalized with Robust Multichip Analysis (www.bioconductor.org) to determine signal log ratios.
The statistical significance of differential gene expression was determined though ANalysis Of VAriation (ANOVA) and false discovery rates (FDR) using Partek Genomics Suite 6.5. Hierarchical clustering was conducted with Genesis76. Pathway analysis was performed with the Database for Annotation, Visualization, and Integrated Discovery (DAVID)77, 78. Longitudinal samples collected from progressors to T1D were analyzed with Short Time-series Expression Miner version 1.3.8 (STEM), software for the analysis of time-series gene expression data (3-8 time points)41. Datasets were also analyzed with ToppGene39, 40, which prioritizes genes based on functional similarity using information on gene expression, protein domains and interactions, transcription-factor binding sites, miRNAs, ontologies, human disease, and mouse phenotypes, drug-gene associations, and literature co-citation. The data generated in this investigation are MIAME compliant79 and have been deposited in the NCBI Gene Expression Omnibus80, accessible through GEO Series accession numbers GSE35725 (T1D data), GSE35713 (CF data), GSE35716 (pneumonia data), and GSE35712 (H1N1 data).
Direct detection of inflammatory mediators
Plasma from RO T1D and HC subjects were assayed with the BeadLyte cytokine assay kit (Millipore, Billerica, MA) per the manufacturer's protocol and a Bio-Plex Luminex 100 XYP instrument. Concentrations for Eotaxin, GM-CSF, interferon (IFN) α2, IFNγ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17, IP-10, MCP-1, MIP-1a, MIP-1b, TNFα, and TNFβ were calculated with the Bio-Plex Manager 4.1 software; a 5-parameter curve-fitting algorithm was applied for standard curve calculations.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank the patients and families who participated in this study. The authors acknowledge the physicians, nurses, and staff of Children’s Hospital of Wisconsin and The Max McGee National Research Center for Juvenile Diabetes who assisted in subject recruitment and sample collection/processing. This work was supported by the Juvenile Diabetes Research Foundation International (1-2008-1026 and 5-2012-220 to M.J.H.); the National Institutes of Health (R01AI078713 to M.J.H, DP2OD007031 to H.L., R01DK080100 to X.W., U19AI062627 to J.G., and 1-UL1-RR031973 the Clinical and Translational Science Institute of Southeast Wisconsin); Genentech (IST grant to H.L.); and The Children’s Hospital of Wisconsin Foundation.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3(4):281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 2.Lewis CC, Yang JY, Huang X, Banerjee SK, Blackburn MR, Baluk P, et al. Disease-specific gene expression profiling in multiple models of lung disease. Am J Respir Crit Care Med. 2008;177(4):376–387. doi: 10.1164/rccm.200702-333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medzhitov R, Janeway C., Jr Innate immune recognition: mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 4.Elson G, Dunn-Siegrist I, Daubeuf B, Pugin J. Contribution of Toll-like receptors to the innate immune response to Gram-negative and Gram-positive bacteria. Blood. 2007;109(4):1574–1583. doi: 10.1182/blood-2006-06-032961. [DOI] [PubMed] [Google Scholar]
- 5.Underhill DM, Ozinsky A. Toll-like receptors: key mediators of microbe detection. Curr Opin Immunol. 2002;14(1):103–110. doi: 10.1016/s0952-7915(01)00304-1. [DOI] [PubMed] [Google Scholar]
- 6.Nau GJ, Schlesinger A, Richmond JF, Young RA. Cumulative Toll-like receptor activation in human macrophages treated with whole bacteria. J Immunol. 2003;170(10):5203–5209. doi: 10.4049/jimmunol.170.10.5203. [DOI] [PubMed] [Google Scholar]
- 7.Toshchakov V, Jones BW, Perera PY, Thomas K, Cody MJ, Zhang S, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nat Immunol. 2002;3(4):392–398. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 8.Huang Q, Liu D, Majewski P, Schulte LC, Korn JM, Young RA, et al. The plasticity of dendritic cell responses to pathogens and their components. Science. 2001;294(5543):870–875. doi: 10.1126/science.294.5543.870. [DOI] [PubMed] [Google Scholar]
- 9.Lempicki RA, Polis MA, Yang J, McLaughlin M, Koratich C, Huang DW, et al. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis. 2006;193(8):1172–1177. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
- 10.Thach DC, Agan BK, Olsen C, Diao J, Lin B, Gomez J, et al. Surveillance of transcriptomes in basic military trainees with normal, febrile respiratory illness, and convalescent phenotypes. Genes Immun. 2005;6(7):588–595. doi: 10.1038/sj.gene.6364244. [DOI] [PubMed] [Google Scholar]
- 11.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109(5):2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, Dung NT, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195(8):1097–1107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobsen M, Repsilber D, Gutschmidt A, Neher A, Feldmann K, Mollenkopf HJ, et al. Candidate biomarkers for discrimination between infection and disease caused by Mycobacterium tuberculosis. J Mol Med. 2007;85(6):613–621. doi: 10.1007/s00109-007-0157-6. [DOI] [PubMed] [Google Scholar]
- 14.Ockenhouse CF, Hu WC, Kester KE, Cummings JF, Stewart A, Heppner DG, et al. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006;74(10):5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197(6):711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crow MK, Kirou KA, Wohlgemuth J. Microarray analysis of interferon-regulated genes in SLE. Autoimmunity. 2003;36(8):481–490. doi: 10.1080/08916930310001625952. [DOI] [PubMed] [Google Scholar]
- 17.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100(5):2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards CJ, Feldman JL, Beech J, Shields KM, Stover JA, Trepicchio WL, et al. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Mol Med. 2007;13(1–2):40–58. doi: 10.2119/2006-000056.Edwards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM, et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007;66(8):1008–1014. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lequerre T, Gauthier-Jauneau AC, Bansard C, Derambure C, Hiron M, Vittecoq O, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8(4):R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batliwalla FM, Baechler EC, Xiao X, Li W, Balasubramanian S, Khalili H, et al. Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun. 2005;6(5):388–397. doi: 10.1038/sj.gene.6364209. [DOI] [PubMed] [Google Scholar]
- 22.Achiron A, Feldman A, Mandel M, Gurevich M. Impaired expression of peripheral blood apoptotic-related gene transcripts in acute multiple sclerosis relapse. Ann N Y Acad Sci. 2007;1107:155–167. doi: 10.1196/annals.1381.017. [DOI] [PubMed] [Google Scholar]
- 23.Achiron A, Gurevich M, Snir Y, Segal E, Mandel M. Zinc-ion binding and cytokine activity regulation pathways predicts outcome in relapsing-remitting multiple sclerosis. Clin Exp Immunol. 2007;149(2):235–242. doi: 10.1111/j.1365-2249.2007.03405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh MK, Scott TF, LaFramboise WA, Hu FZ, Post JC, Ehrlich GD. Gene expression changes in peripheral blood mononuclear cells from multiple sclerosis patients undergoing beta-interferon therapy. J Neurol Sci. 2007;258(1–2):52–59. doi: 10.1016/j.jns.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 25.Burczynski ME, Peterson RL, Twine NC, Zuberek KA, Brodeur BJ, Casciotti L, et al. Molecular classification of Crohn's disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn. 2006;8(1):51–61. doi: 10.2353/jmoldx.2006.050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoeckman AK, Baechler EC, Ortmann WA, Behrens TW, Michet CJ, Peterson EJ. A distinct inflammatory gene expression profile in patients with psoriatic arthritis. Genes Immun. 2006;7(7):583–591. doi: 10.1038/sj.gene.6364334. [DOI] [PubMed] [Google Scholar]
- 27.Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57(5):664–678. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- 28.Baechler EC, Bauer JW, Slattery CA, Ortmann WA, Espe KJ, Novitzke J, et al. An interferon signature in the peripheral blood of dermatomyositis patients is associated with disease activity. Mol Med. 2007;13(1–2):59–68. doi: 10.2119/2006-00085.Baechler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J Exp Med. 2005;201(9):1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allantaz F, Chaussabel D, Stichweh D, Bennett L, Allman W, Mejias A, et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J Exp Med. 2007;204(9):2131–2144. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Jia S, Geoffrey R, Alemzadeh R, Ghosh S, Hessner MJ. Identification of a Molecular Signature in Human Type 1 Diabetes Mellitus Using Serum and Functional Genomics. J Immunol. 2008;180(3):1929–1937. doi: 10.4049/jimmunol.180.3.1929. [DOI] [PubMed] [Google Scholar]
- 32.Kaldunski M, Jia S, Geoffrey R, Basken J, Prosser S, Kansra S, et al. Identification of a serum-induced transcriptional signature associated with type 1 diabetes in the BioBreeding rat. Diabetes. 2010;59(10):2375–2385. doi: 10.2337/db10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia S, Kaldunski M, Jailwala P, Geoffrey R, Kramer J, Wang X, et al. Use of transcriptional signatures induced in lymphoid and myeloid cell lines as an inflammatory biomarker in Type 1 diabetes. Physiol Genomics. 2011;43(11):697–709. doi: 10.1152/physiolgenomics.00235.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Belle V, Pelckmans K, Van Huffel S, Suykens JA. Improved performance on high-dimensional survival data by application of Survival-SVM. Bioinformatics. 2011;27(1):87–94. doi: 10.1093/bioinformatics/btq617. [DOI] [PubMed] [Google Scholar]
- 35.Abdul-Rasoul M, Habib H, Al-Khouly M. 'The honeymoon phase' in children with type 1 diabetes mellitus: frequency, duration, and influential factors. Pediatr Diabetes. 2006;7(2):101–107. doi: 10.1111/j.1399-543X.2006.00155.x. [DOI] [PubMed] [Google Scholar]
- 36.Bonfanti R, Bazzigaluppi E, Calori G, Riva MC, Viscardi M, Bognetti E, et al. Parameters associated with residual insulin secretion during the first year of disease in children and adolescents with Type 1 diabetes mellitus. Diabet Med. 1998;15(10):844–850. doi: 10.1002/(SICI)1096-9136(199810)15:10<844::AID-DIA679>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 37.Bonfanti R, Bognetti E, Meschi F, Brunelli A, Riva MC, Pastore MR, et al. Residual beta-cell function and spontaneous clinical remission in type 1 diabetes mellitus: the role of puberty. Acta Diabetol. 1998;35(2):91–95. doi: 10.1007/s005920050110. [DOI] [PubMed] [Google Scholar]
- 38.Moser B, Wolf M, Walz A, Loetscher P. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 2004;25(2):75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Xu H, Aronow BJ, Jegga AG. Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics. 2007;8:392. doi: 10.1186/1471-2105-8-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Terenzi F, White C, Pal S, Williams BR, Sen GC. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J Virol. 2007;81(16):8656–8665. doi: 10.1128/JVI.00322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Handley SA, Dube PH, Miller VL. Histamine signaling through the H(2) receptor in the Peyer's patch is important for controlling Yersinia enterocolitica infection. Proc Natl Acad Sci U S A. 2006;103(24):9268–9273. doi: 10.1073/pnas.0510414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32(4):468–478. doi: 10.1016/j.immuni.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu J, Yu L, Bugawan TL, Erlich HA, Barriga K, Hoffman M, et al. Transient antiislet autoantibodies: infrequent occurrence and lack of association with "genetic" risk factors. J Clin Endocrinol Metab. 2000;85(7):2421–2428. doi: 10.1210/jcem.85.7.6670. [DOI] [PubMed] [Google Scholar]
- 46.Atkinson MA, Eisenbarth GS. Type 1 diabetes: new perspectives on disease pathogenesis and treatment. Lancet. 2001;358(9277):221–229. doi: 10.1016/S0140-6736(01)05415-0. [DOI] [PubMed] [Google Scholar]
- 47.Devendra D, Liu E, Eisenbarth GS. Type 1 diabetes: recent developments. Bmj. 2004;328(7442):750–754. doi: 10.1136/bmj.328.7442.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skyler JS. Prediction and prevention of type 1 diabetes: progress, problems, and prospects. Clin Pharmacol Ther. 2007;81(5):768–771. doi: 10.1038/sj.clpt.6100179. [DOI] [PubMed] [Google Scholar]
- 49.Roep BO, Peakman M. Surrogate end points in the design of immunotherapy trials: emerging lessons from type 1 diabetes. Nat Rev Immunol. 2010;10(2):145–152. doi: 10.1038/nri2705. [DOI] [PubMed] [Google Scholar]
- 50.Hussain MJ, Peakman M, Gallati H, Lo SS, Hawa M, Viberti GC, et al. Elevated serum levels of macrophage-derived cytokines precede and accompany the onset of IDDM. Diabetologia. 1996;39(1):60–69. doi: 10.1007/BF00400414. [DOI] [PubMed] [Google Scholar]
- 51.Perez F, Oyarzun A, Carrasco E, Angel B, Albala C, Santos JL. Plasma levels of interleukin-1beta, interleukin-2 and interleukin-4 in recently diagnosed type 1 diabetic children and their association with beta-pancreatic autoantibodies. Rev Med Chil. 2004;132(4):413–420. doi: 10.4067/s0034-98872004000400002. [DOI] [PubMed] [Google Scholar]
- 52.Erbagci AB, Tarakcioglu M, Coskun Y, Sivasli E, Sibel Namiduru E. Mediators of inflammation in children with type I diabetes mellitus: cytokines in type I diabetic children. Clin Biochem. 2001;34(8):645–650. doi: 10.1016/s0009-9120(01)00275-2. [DOI] [PubMed] [Google Scholar]
- 53.Nicoletti F, Conget I, Di Marco R, Speciale AM, Morinigo R, Bendtzen K, et al. Serum levels of the interferon-gamma-inducing cytokine interleukin-18 are increased in individuals at high risk of developing type I diabetes. Diabetologia. 2001;44(3):309–311. doi: 10.1007/s001250051619. [DOI] [PubMed] [Google Scholar]
- 54.Nicoletti F, Conget I, Di Mauro M, Di Marco R, Mazzarino MC, Bendtzen K, et al. Serum concentrations of the interferon-gamma-inducible chemokine IP-10/CXCL10 are augmented in both newly diagnosed Type I diabetes mellitus patients and subjects at risk of developing the disease. Diabetologia. 2002;45(8):1107–1110. doi: 10.1007/s00125-002-0879-5. [DOI] [PubMed] [Google Scholar]
- 55.Dogan Y, Akarsu S, Ustundag B, Yilmaz E, Gurgoze MK. Serum IL-1beta, IL-2, and IL-6 in insulin-dependent diabetic children. Mediators Inflamm. 2006;2006(1):59206. doi: 10.1155/MI/2006/59206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mahon JL, Sosenko JM, Rafkin-Mervis L, Krause-Steinrauf H, Lachin JM, Thompson C, et al. The TrialNet Natural History Study of the Development of Type 1 Diabetes: objectives, design, and initial results. Pediatr Diabetes. 2009;10(2):97–104. doi: 10.1111/j.1399-5448.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- 57.The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes. 2007;8(5):286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 58.Padmos RC, Schloot NC, Beyan H, Ruwhof C, Staal FJ, de Ridder D, et al. Distinct monocyte gene-expression profiles in autoimmune diabetes. Diabetes. 2008;57(10):2768–2773. doi: 10.2337/db08-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaizer EC, Glaser CL, Chaussabel D, Banchereau J, Pascual V, White PC. Gene expression in peripheral blood mononuclear cells from children with diabetes. J Clin Endocrinol Metab. 2007;92(9):3705–3711. doi: 10.1210/jc.2007-0979. [DOI] [PubMed] [Google Scholar]
- 60.Bendtzen K, Mandrup-Poulsen T, Nerup J, Nielsen JH, Dinarello CA, Svenson M. Cytotoxicity of human pI 7 interleukin-1 for pancreatic islets of Langerhans. Science. 1986;232(4757):1545–1547. doi: 10.1126/science.3086977. [DOI] [PubMed] [Google Scholar]
- 61.Mandrup-Poulsen T. The role of interleukin-1 in the pathogenesis of IDDM. Diabetologia. 1996;39(9):1005–1029. doi: 10.1007/BF00400649. [DOI] [PubMed] [Google Scholar]
- 62.Mathis D, Vence L, Benoist C. beta-Cell death during progression to diabetes. Nature. 2001;414(6865):792–798. doi: 10.1038/414792a. [DOI] [PubMed] [Google Scholar]
- 63.Giannoukakis N, Rudert WA, Trucco M, Robbins PD. Protection of human islets from the effects of interleukin-1beta by adenoviral gene transfer of an Ikappa B repressor. J Biol Chem. 2000;275(47):36509–36513. doi: 10.1074/jbc.M005943200. [DOI] [PubMed] [Google Scholar]
- 64.Larsen CM, Faulenbach M, Vaag A, Volund A, Ehses JA, Seifert B, et al. Interleukin-1-receptor antagonist in type 2 diabetes mellitus. N Engl J Med. 2007;356(15):1517–1526. doi: 10.1056/NEJMoa065213. [DOI] [PubMed] [Google Scholar]
- 65.Thomas HE, Irawaty W, Darwiche R, Brodnicki TC, Santamaria P, Allison J, et al. IL-1 receptor deficiency slows progression to diabetes in the NOD mouse. Diabetes. 2004;53(1):113–121. doi: 10.2337/diabetes.53.1.113. [DOI] [PubMed] [Google Scholar]
- 66.Sandberg JO, Eizirik DL, Sandler S. IL-1 receptor antagonist inhibits recurrence of disease after syngeneic pancreatic islet transplantation to spontaneously diabetic non-obese diabetic (NOD) mice. Clin Exp Immunol. 1997;108(2):314–317. doi: 10.1046/j.1365-2249.1997.3771275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pickersgill LM, Mandrup-Poulsen TR. The anti-interleukin-1 in type 1 diabetes action trial--background and rationale. Diabetes Metab Res Rev. 2009;25(4):321–324. doi: 10.1002/dmrr.960. [DOI] [PubMed] [Google Scholar]
- 68.Sumpter KM, Adhikari S, Grishman EK, White PC. Preliminary studies related to anti-interleukin-1beta therapy in children with newly diagnosed type 1 diabetes. Pediatr Diabetes. 2011 doi: 10.1111/j.1399-5448.2011.00761.x. [DOI] [PubMed] [Google Scholar]
- 69.Sanda S, Bollyky J, Standifer N, Nepom G, Hamerman JA, Greenbaum C. Short-term IL-1beta blockade reduces monocyte CD11b integrin expression in an IL-8 dependent fashion in patients with type 1 diabetes. Clin Immunol. 2010;136(2):170–173. doi: 10.1016/j.clim.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 70.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 71.Woo W, LaGasse JM, Zhou Z, Patel R, Palmer JP, Campus H, et al. A novel high-throughput method for accurate, rapid, and economical measurement of multiple type 1 diabetes autoantibodies. J Immunol Methods. 2000;244(1–2):91–103. doi: 10.1016/s0022-1759(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 72.Klitz W, Maiers M, Spellman S, Baxter-Lowe LA, Schmeckpeper B, Williams TM, et al. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens. 2003;62(4):296–307. doi: 10.1034/j.1399-0039.2003.00103.x. [DOI] [PubMed] [Google Scholar]
- 73.Lee TW, Brownlee KG, Denton M, Littlewood JM, Conway SP. Reduction in prevalence of chronic Pseudomonas aeruginosa infection at a regional pediatric cystic fibrosis center. Pediatr Pulmonol. 2004;37(2):104–110. doi: 10.1002/ppul.10401. [DOI] [PubMed] [Google Scholar]
- 74.Johansen HK, Norregaard L, Gotzsche PC, Pressler T, Koch C, Hoiby N. Antibody response to Pseudomonas aeruginosa in cystic fibrosis patients: a marker of therapeutic success?--A 30-year cohort study of survival in Danish CF patients after onset of chronic P. aeruginosa lung infection. Pediatr Pulmonol. 2004;37(5):427–432. doi: 10.1002/ppul.10457. [DOI] [PubMed] [Google Scholar]
- 75.Chow AW, Hall CB, Klein JO, Kammer RB, Meyer RD, Remington JS. Evaluation of new anti-infective drugs for the treatment of respiratory tract infections. Infectious Diseases Society of America and the Food and Drug Administration. Clin Infect Dis. 1992;15(Suppl 1):S62–S88. doi: 10.1093/clind/15.Supplement_1.S62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 77.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 78.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4(10):R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 80.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.