Figure 1.
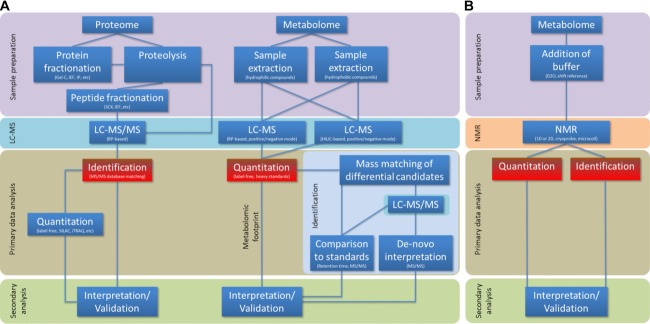
Conceptual differences between a typical proteomics workflow and possible metabolomics workflows. (A) A ‘shotgun’ proteomic discovery experiment will typically employ a pre-fractionation of the analyte pre- or post-proteolysis, followed by LC-MS/MS analysis. Identification of peptides/proteins is essential for both quantitation and interpretation. A metabolomic experiment requires a sample extraction compatible with the analytical workflow further downstream. A separation into hydrophilic and hydrophobic compounds (Supporting Information Fig. 2) can yield samples for HILIC and RP front-end separation. The quantitation of detected molecules builds the basis for further processing. Even without identification, a metabolomic footprint can be used for diagnostic purposes and differential analyses. (B) The NMR-based metabolite sample preparation and analysis is not limited towards compounds with physicochemical properties compatible with LC-MS. Minimal to no sample preparation is needed. However, NMR (as other powerful platforms for metabolomics such as GC-MS or TLC-GC-FID 115) is not a standard technique used in most proteomics laboratories and is considered less sensitive than MS.
