Abstract
The process of plant speciation often involves the evolution of divergent ecotypes in response to differences in soil water availability between habitats. While the same set of traits is frequently associated with xeric/mesic ecotype divergence, it is unknown whether those traits evolve independently or if they evolve in tandem as a result of genetic colocalization either by pleiotropy or genetic linkage.
The self-fertilizing C4 grass species Panicum hallii includes two major ecotypes found in xeric (var. hallii) or mesic (var. filipes) habitats. We constructed the first linkage map for P. hallii by genotyping a reduced representation genomic library of an F2 population derived from an intercross of var. hallii and filipes. We then evaluated the genetic architecture of divergence between these ecotypes through quantitative trait locus (QTL) mapping.
Overall, we mapped QTLs for nine morphological traits that are involved in the divergence between the ecotypes. QTLs for five key ecotype-differentiating traits all colocalized to the same region of linkage group five. Leaf physiological traits were less divergent between ecotypes, but we still mapped five physiological QTLs. We also discovered a two-locus Dobzhansky–Muller hybrid incompatibility.
Our study suggests that ecotype-differentiating traits may evolve in tandem as a result of genetic colocalization.
Keywords: adaptation, drought, ecotype, physiology, pleiotropy, quantitative trait locus (QTL), reproductive isolation
Introduction
The process of speciation often occurs as a continuum over time through the accumulation of reproductive isolating barriers that prevent gene flow between new species (Dobzhansky, 1937; Clausen, 1951; Wu, 2001; Schemske, 2010; Nosil, 2012). A common intermediate stage in the process is the evolution of partially reproductively isolated ecotypes, which result from adaptation to different habitats (Turesson, 1922; Clausen, 1951; Grant, 1981; Lowry, 2012; Nosil, 2012). Reproductive isolation between ecotypes is often caused by ecologically based prezygotic isolation (Lowry et al., 2008; Nosil, 2012). However, postzygotic isolation can also occur between ecotypes as a result of chromosomal rearrangements or epistatic Dobzhansky–Muller interactions (Coyne & Orr, 2004).
In plants, soil water availability plays a dominant role in the distribution and productivity of species worldwide (Stebbins, 1952; Whittaker, 1975; Woodward et al., 2004). Thus, it is not surprising that ecotype formation in plants is frequently driven by divergent adaptations to habitats that differ in soil water availability (Clausen & Heisey, 1958; Porter, 1966; Lowry, 2012; Andrew et al., 2013). Adaptation to soil water availability can drive the evolution of divergence in physiology, development, morphology, phenology, life-history, and reproductive allocation strategies between plant ecotypes (Stebbins, 1952; Clausen & Heisey, 1958; Baker, 1972; Roux et al., 2006; Juenger, 2013). Ecotypes that are adapted to mesic habitats typically have leaves with greater area, more axillary branching, and are larger overall than ecotypes adapted to drier habitats (Clausen & Heisey, 1958; Kruckeberg, 1986; Rajakaruna, 2004; Lowry, 2012). These classic differences in morphology suggest a trade-off between above-ground growth, particularly of leaves, and the demand for water imposed by transpiration, which results from a greater transpiring surface area (Geber & Dawson, 1990; Dudley, 1996; Ackerly et al., 2000). Ecotypes occurring in dry habitats frequently evolve to develop fast and flower early, a life history that avoids periods of drought (Ludlow, 1989; McKay et al., 2003; Franks, 2011). Further, plants adapted to drier habitats typically have larger seeds than those adapted to wetter habitats, which reflects a trade-off between offspring provisioning and total reproductive output (Baker, 1972).
While there is often a common set of morphological and life-history traits that differentiate xeric and mesic ecotypes (Lowry, 2012) across plant species, very little information is known about the genetic basis of parallel evolution among ecotype-differentiating traits. One major unresolved question is whether the common set of traits involved in mesic vs xeric ecotype divergence evolves in tandem, as a result of colocalization of genetic effects to particular chromosomal regions, or whether these traits can evolve independently of each other. Recently, a study in the yellow monkeyflower, Mimulus guttatus, identified a major quantitative trait locus (QTL) that controlled the divergence in many traits differentiating xeric and mesic ecotypes (Hall et al., 2006; Lowry & Willis, 2010). This major QTL mapped to a large chromosomal inversion, which suggests that genetic linkage may play a role in the concerted evolution of many ecotype-differentiating traits (Kirkpatrick, 2010). Similar colocalizing QTLs were also found in a cross between xeric and mesic accessions of Arabidopsis thaliana (McKay et al., 2003, 2008; Lovell et al., 2013). One of these colocalizing QTLs appears to be the result of a mutation in FRIGIDA, and suggests that pleiotropy through mutations in major regulatory genes can be responsible for the divergence of multiple traits related to drought adaptations (Lovell et al., 2013). QTL colocalization has also been reported for traits involved in divergence between xeric and mesic ecotypes of Avena barbata (Latta & Gardner, 2009). Beyond these few examples, it is unknown how often colocalization of QTLs occurs for the common set of traits involved in xeric vs mesic ecotype divergence.
Here, we examine whether there is genetic colocalization of traits involved in the divergence of morphological, phenological, leaf physiological, and reproductive allocation traits between two ecotypes of the C4 perennial grass, Panicum hallii Vasey. We also evaluated potential mechanisms of reproductive isolation that maintain these ecotypes despite their overlapping geographic ranges. P. hallii is a highly self-fertilizing (Mean FIS = 0.895; Lowry et al., 2013) rangeland species that occurs across much of the southwestern USA and northern Mexico. There is a considerable population genetic structure among populations of P. hallii and among major biogeographical regions within its geographical range (Lowry et al., 2012, 2013). However, the greatest morphological divergence within P. hallii occurs between its two ecotypes, which have been previously classified by taxonomists as distinct varieties (Gould, 1975; Waller, 1976). P. hallii var. hallii (Scribn.) Waller is the most widespread ecotype and is typically located in water-limited (xeric) upland calcareous habitats (Gould, 1975; Waller, 1976). By contrast, P. hallii var. filipes (Scribn.) Waller is primarily found in mesic depressions and is more geographically restricted, most often found in the Rio Grande Valley and along the Gulf Coast Plain of Texas and Mexico (Gould, 1975; Waller, 1976; Hatch et al., 2003). The two ecotypes co-occur in areas where both wet and dry microsites are available (Waller, 1976), but are morphologically distinct (Lowry et al., 2013), especially in south Texas (Waller, 1976). Consistent with differential adaptation to wet and dry environments (Baker, 1972; Dudley, 1996; Ackerly et al., 2000; Picotte et al., 2007; Lowry et al., 2008; Maherali et al., 2009), var. hallii has smaller leaves, smaller overall plant size, earlier flowering time, and larger seeds than var. filipes (Waller, 1976; Lowry et al., 2013).
The primary aim of our study was to understand the genetic basis of multi-trait divergence and reproductive isolation between var. hallii and var. filipes through QTL mapping. To conduct QTL mapping in this system, we developed a linkage map using a new restriction site-associated DNA (RAD) genotyping method that utilizes IIB restriction enzymes (Wang et al., 2012). This map also allowed us to also evaluate the extent of chromosomal synteny between P. hallii and foxtail millet (Setaria italica), which is the closest relative of P. hallii with a published sequenced genome (Zhang et al., 2011; Bennetzen et al., 2012). Overall, we addressed the following four major questions: what genomic regions contribute to the morphological and leaf physiological trait variation/divergence within and among var. hallii and var. filipes? Do QTLs for multiple ecotype-differentiating traits cluster (colocalize) in particular chromosomal regions? What are the patterns of synteny between P. hallii and S. italica and is there evidence for colocalization of flowering time QTLs between those two species? What reproductive isolating barriers might account for maintenance of divergence between var. hallii and var. filipes?
Materials and Methods
Development of the F2 FIL2 × HAL2 mapping population
To better understand the genetic basis of divergence between Panicum hallii var. hallii and var. filipes, we created an F2 mapping population. The parents of the mapping populations were FIL2 and HAL2-11. FIL2 is a genotype derived from a natural var. filipes collection made near the Corpus Christi Botanical Gardens in South Texas (27.65°N, 97.40°W), and is the primary line being sequenced by the Joint Genome Institute (DOE) for var. filipes (Meyer et al., 2012). HAL2-11 is a one-generation selfed progeny of HAL2, which was derived from a natural collection made at the Lady Bird Johnson Wildflower Center (Austin, TX, USA; 30.19°N, 97.87°W), and is the primary line being sequenced for var. hallii. All of the F2 plants used in this study were derived as self-fertilized progeny from a single F1 hybrid, where FIL2 was the sire and HAL2-11 was the dam in the cross (see Supporting Information Methods S1 for details of the crossing methodology). We refer to HAL2-11 as HAL2 throughout the remainder of the manuscript because this lineage is highly homozygous and, thus, quite similar across generations.
Morphological phenotyping
We scarified F2 seeds with sandpaper and placed them on white paper towels moistened with water in Petri dishes sealed with parafilm on 18 July 2011. To synchronize germination the Petri dishes were placed in a refrigerator at 4°C, to stratify seeds. On 25 July 2011 the Petri dishes were moved to a growth chamber for germination using the same methods as Lowry et al. (2013). As seeds germinated they were transferred to a 60 : 40 mixture of Promix (Premier Tech Horticulture, Rivière-du-Loup, Québec, Canada): Turface (Profile Products, Buffalo Grove, IL, USA) in 4 inch square pots. The first seedlings were transplanted on 3 August 2011, and the last on 23 August 2011. Plants were then grown under 16 h days maintained by supplemental fluorescent lighting in the University of Texas glasshouses. Seedlings were arrayed in a fully randomized design and trays were rearranged weekly to minimize the effects of environmental heterogeneity across the experiment.
We measured four traits on the first flowering tiller at anthesis: tiller height, tiller thickness, flag leaf length, and flag leaf width. A flag leaf is the upper most leaf on a tiller and subtends the inflorescence. We also counted the number of tillers at anthesis. Flowering time was calculated as the date of transplantation subtracted from the date of first flower. Once the inflorescence of the first flowering tiller had fully expanded we measured the length of the inflorescence (first branch point to tip) and counted the number of flowers on the inflorescence. We calculated seed mass as the mean weight of 10 viable seeds collected from each F2 plant. We observed that many of the F2 plants had low levels of seed production or produced no apparent viable seed, and we scored sterility as a binary trait, with 1 indicating at minimum dozens of viable looking seeds (i.e. hard, dark colored seed coat) and a 0 indicating fewer than five viable looking seeds (i.e. soft, light colored seed coat).
Physiological phenotyping
We measured leaf physiological traits on 214 unique F2 lines, 18 clonally propagated FIL2, and 19 clonally propagated HAL2 individuals (see Methods S1 for details of environmental conditions). We aimed to determine efficiencies with respect to photosynthesis and leaf water use; higher photosynthetic capacity can help plants to maximize photosynthesis in dry environments, and differences in efficiency can be linked with leaf lifespan through variation in nutrient and structural investments (Wright et al., 2003; Osnas et al., 2013).
Survey measurements were made with two LI-6400XT portable photosynthesis systems (Li-Cor Inc., Lincoln, NE, USA) at ambient CO2 (reference CO2 controlled at 400 μmol mol−1; sample CO2, 386 ± 6 μmol mol−1 (mean ± SD)) to obtain values for net CO2 assimilation (A), stomatal conductance (gs), and leaf intercellular CO2 concentrations (ci). Maximization of A relative to gs will reduce ci, and all else being equal the capacity to maintain lower ci indicates a greater ratio of carbon fixation to water lost for a leaf (Farquhar & Richards, 1984). Quantum efficiency of photosystem II (ΦPSII) as well as its factors: photochemical quenching (qP) and the light adapted efficiency of energy harvesting by open PSII reaction centers (Fv′/Fm′), were measured concurrently with gas exchange using fluorometers integrated into the cuvette lids (LI-6400-40). These fluorescence parameters are indicators of the operating efficiency of PSII (Maxwell & Johnson, 2000), thus linear electron transport in the light reactions of photosynthesis; when incident light is similar, higher values for fluorescence parameters indicate that a leaf is making more effective use of absorbed light and, therefore, its investment in light harvesting capacity.
During gas exchange, a chlorophyll SPAD meter (Konica-Minolta SPAD 502; Konica-Minolta, Chiyoda, Tokyo, Japan) was used to measure each leaf adjacent to the section being measured for gas exchange, and a mean value was recorded. SPAD values are a dimensionless index of leaf chlorophyll content determined from the ratio of transmitted light with wavelengths 940 and 650 nm (Markwell et al., 1995), that is, a coarse indication of the investment in light harvesting capacity.
Before mapping of physiological traits, we independently excluded outliers for gas exchange, fluorescence, and SPAD among the 214 F2 individuals. When measuring gas exchange, estimates of gs in particular can be influenced by a variety of sources of measurement error, thereby influencing estimates of water-use efficiency. We therefore eliminated measurements of gas exchange that indicated substantial deviation from the expected relationships between A and gs. The top 5% (11) values for ci were removed; these values were >254 μmol mol−1, which is outside of the normal range for C4 photosynthesis (we would expect c. 150 μmol mol−1). We further removed gas exchange data representing the bottom 5% of residuals (7) from a regression of A on gs that included random effects of the machines used on both the slope and intercept. Fluorescence traits for one plant were eliminated from the dataset due to an unusually low Fv′/Fm′ (gas exchange data for this plant had already been eliminated due to unusually high ci). Seven SPAD values falling outside a 99% confidence interval around the mean (SPAD values 17.5–42.5) were removed. The same filters were applied to the FIL2 and HAL2 data, and gas exchange values for one FIL2 parent with ci > 254 μmol mol−1 were removed.
Subsequent to outlier exclusion, best linear unbiased predictors (BLUPs) for each F2 were generated for physiological traits, using mixed-effects models (lme4, Bates et al., 2013) to correct for the estimated random effects of the machine used and the day on which each measurement was taken.
Genotyping and analysis of RAD markers
Following the quantification of morphological traits, we collected tissue from all F2 plants and extracted high-quality DNA using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). To obtain markers for mapping, we prepared our DNA with a recently developed 2b restriction site–associated DNA (2b–RAD) genotyping method (Wang et al., 2012). This method utilizes type IIB restriction enzymes, which cut both upstream and downstream of the enzyme's target site and results in the production of RAD tags of uniform length. We used the AlfI restriction enzyme for preparation of our RAD libraries. We followed the reduction protocol of Wang et al. (2012) to target only 1/16th the AlfI sites across the genome for sequencing based on modified adaptors. We sequenced the RAD tags on both the Illumina and SOLiD (Applied Biosystems, Foster City, CA, USA) platforms in pools of barcoded individuals (range: 39–105 individuals per pool).
To create a reference genome for mapping of RAD tags, we conducted genome sequencing of the FIL2 P. hallii var. filipes accession with the Illumina GAIIx and Illumina HiSeq platforms (Illumina Inc., San Diego, CA, USA). Genomic DNA was extracted at the University of Texas at Austin and sequenced at the Department of Energy Joint Genome Institute (JGI) in Walnut Creek, CA, USA. DNA was prepared for sequencing with insert sizes ranging from 0.2 to 35 kbp and paired-end 76, 100, and 150 bp reads (Table S1). Preliminary scaffolds were assembled with Meraculous (Chapman et al., 2011), the JGI plant assembler, as part of a genome sequence project for P. hallii (used with permission). Raw sequence reads used in the Meraculous assembly can be found in the NIH sequence read archive (SRA) under project number SRP003932.
Trimmed and filtered reads (custom scripts: https://github.com/kmhernan/Publications) were mapped to the Meraculous assembly with SHRiMP mapping software (v2.2.3; David et al., 2011; global alignments with five significant hits per read) and pre-processed with the Picard software tools (http://picard.sourceforge.net/). Picard is a suite of utilities that are used to manipulate Sequence Alignment/Map (SAM) formatted files. To improve computational efficiency, we reduced the Meraculous assembly to include only scaffolds with at least five reads mapped across 25% of the population. We then applied the Genome Analysis Toolkit (GATK; McKenna et al., 2010) to conduct variant discovery, genotyping, and filtering at a large scale. We used GATK indel realignment, base quality recalibration (after an initial run to estimate high-quality SNPs), and performed genotyping across all samples simultaneously using standard filtering parameters (DePristo et al., 2011). We created an initial set of potential markers by filtering the variants resulting from GATK based on the following criteria: we converted individual genotypes with Phred-scaled genotype quality (GQ) of < 20 to Ns (i.e. missing data); we only retained sites with bi-allelic polymorphism across all individuals; we only retained sites that had been genotyped in both parents and were homozygous for different alleles in each parent (HAL2 vs FIL2); we eliminated sites with >75% missing data and sites that had segregation distortion of P < 0.00005 in a chi-squared test. Following these filtering steps, 803 potential markers remained for linkage map construction. See Fig. S1 for a flowchart of the genotyping pipeline.
Linkage map assembly and synteny analysis
We inferred linkage groups based on marker recombination fractions of 0.15, 0.1 and 0.05 using JoinMap 4 (Van Ooijen, 2006). We used the JoinMap regression mapping algorithm for linkage map construction with the following settings: pairwise recombination frequency < 0.4, LOD > 3, goodness of fit threshold = 5, ripple run after each marker placement, and the Kosambi mapping function to calculate genetic distances. Following this first round of linkage map construction, we removed markers that substantially affected the goodness of fit of the data (e.g. mean chi-squared contribution > 3). We purged markers to eliminate incorrect markers orders that we identified through visualization of recombination fractions with R/qtl (Broman et al., 2003).
We explored synteny with the foxtail millet genome (S. italica v9.0) and P. hallii linkage map to verify marker orders. We used the relative position of markers in the two genomes to identify the location of large-scale chromosomal rearrangements. See Fig. S2 for a flowchart of the synteny analysis pipeline. Briefly, we compared the position of foxtail millet exons with the position of homologous exons in the reduced P. hallii Meraculous genome assembly (tblastx–threshold 999). To identify most closely related homologous exons, we filtered out the ‘best’ BLAST hits. We then searched for the positions in the reduced Meraculous P. hallii assembly where a SNP fell within a region and had significant BLAST results to a foxtail millet exon. We extracted a 200-bp window around these SNPs and used BLAST to reciprocally compare them with the set of foxtail millet exons (tblastx–threshold 999). Finally, we extracted reciprocal ‘best’ hits from both BLAST outputs to identify syntentic positions in the two genomes.
QTL analyses
We conducted QTL mapping of morphological and physiological traits by stepwise fitting of multiple-QTL models in R/qtl (custom scripts: https://github.com/davidbryantlowry/panicumhalliiqtlmapping). Before model fitting, we calculated penalties for main effects and interactions for each trait through 1000 permutations of the scantwo function using Haley–Knott regression. We then used the stepwiseqtl function to conduct a forward/backward search using Haley–Knott regression for models with a maximum of six QTLs that optimized the penalized LOD score criterion. In addition to additive QTLs, the stepwiseqtl function considers models with all possible pairwise epistatic interactions. We calculated the 1.5–LOD drop interval of QTLs in the best-fit stepwise models using the lodint function. We also used the best-fit stepwise model for each trait to calculate the additive effect, dominance deviation, and percent of variance explained for each QTL with the makeqtl and fitqtl functions of R/qtl. Planting cohort was used as a covariate for QTL mapping of flowering time.
Results
Phenotypic variation and divergence
Consistent with previous studies (Waller, 1976; Lowry et al., 2013), there was considerable divergence in traits between var. hallii (HAL2) and var. filipes (FIL2). For example, FIL2 plants had on average 1.8-fold longer tillers, 1.6-fold longer leaves, and took twice as long to flower as HAL2 plants (Table 1). While FIL2 plants had 4.5-fold more flowers, their seeds were only 40% of the size of the seeds of the HAL2 plants. There were significant correlations across the F2 population for 26 out of 36 pairwise comparisons of morphological traits (Table 2). We also found evidence for a hybrid incompatibility between the varieties, as 45 out of 229 (19.7%) F2 had a high level of sterility or were completely sterile.
Table 1.
Means (SE) morphological trait values for FIL2, HAL2 and F2 Panicum hallii plants
| Trait | N | FIL2 | N | HAL2 | t | N | F2 | F2 range |
|---|---|---|---|---|---|---|---|---|
| Tiller height (cm) | 12 | 65.70 (2.21) | 30 | 35.61 (1.07) | −13.79**** | 246 | 46.88 (0.53) | 28.6–80.0 |
| Number of tillers | 12 | 19.08 (1.41) | 30 | 15.20 (0.68) | −2.78** | 246 | 15.40 (0.34) | 4–30 |
| Tiller width (mm) | 12 | 2.39 (0.08) | 30 | 1.70 (0.04) | −8.43**** | 246 | 1.86 (0.02) | 1.1–2.8 |
| Leaf length (cm) | 12 | 25.93 (1.86) | 30 | 15.97 (0.30) | −8.00**** | 245 | 20.28 (0.28) | 8.9–36.2 |
| Leaf width (mm) | 12 | 8.38 (0.39) | 30 | 5.45 (0.12) | −9.52**** | 246 | 6.01 (0.07) | 2.8–10.0 |
| Flowering time (d) | 12 | 85.08 (5.41) | 30 | 43.53 (1.02) | −11.14**** | 246 | 60.85 (0.95) | 36–105 |
| Inflorescence length (cm) | 9 | 24.02 (1.09) | 30 | 21.19 (0.36) | −3.22** | 243 | 22.27 (0.89) | 14.5–35.6 |
| Flower number | 9 | 527.67 (51.02) | 30 | 116.43 (6.63) | −13.85**** | 240 | 239.27 (5.94) | 72–533 |
| Seed mass (mg) | 8 | 0.58 (0.01) | 30 | 1.40 (0.01) | 34.83**** | 200 | 0.91 (0.01) | 0.65–1.24 |
N, number of replicates per parental lines or total number of F2 individuals (1 replicate per F2); t, t-statistic in test for divergence between parental lines
P < 0.01
P < 0.0001.
Table 2.
Significant pairwise morphological trait correlations in the FIL2 × HAL2 F2 population of Panicum hallii
| Tiller height | Tiller width | Tiller number | Leaf length | Leaf width | Flowering time | Inflorescence length | Flower number | |
|---|---|---|---|---|---|---|---|---|
| Tiller height | ||||||||
| Tiller width | 0.356 | |||||||
| Tiller number | 0.372 | 0.245 | ||||||
| Leaf length | 0.308 | 0.511 | 0.280 | |||||
| Leaf width | 0.423 | 0.618 | 0.458 | 0.790 | ||||
| Flowering time | 0.053 | −0.016 | −0.467 | −0.233 | −0.297 | |||
| Inflorescence length | 0.359 | 0.411 | 0.289 | 0.423 | 0.501 | −0.169 | ||
| Flower number | 0.472 | 0.510 | 0.440 | 0.478 | 0.624 | −0.250 | 0.524 | |
| Seed mass | −0.027 | 0.065 | 0.125 | 0.096 | 0.145 | −0.349 | 0.210 | 0.051 |
Significant pairwise Pearson product–moment correlations after Bonferroni correction at P < 0.05 are indicated in bold.
The distribution of morphological trait values in the F2 hybrids was generally bell-shaped, with a few traits, such as flowering time, having an extended tail in one direction (Fig. S3). The parental HAL2 and FIL2 means generally flanked the peak of the distribution of trait values (Figs 1a, S3). However, there was transgressive segregation for traits such as number of tillers and inflorescence length, where parental means were not as divergent (Fig. S3). Many of the extreme F2s in the distribution looked phenotypically similar to the parental phenotypes (Fig.1b).
Fig 1.
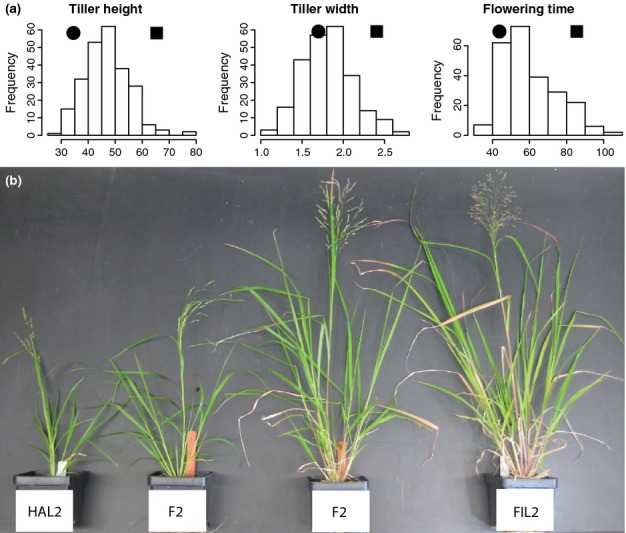
(a) The distribution of trait values in the F2 hybrid mapping population for tiller height, tiller width, and flowering time. The parental HAL2 (black circle) and FIL2 (black square) means are plotted above each distribution. (b) The divergence between Panicum hallii var. hallii (HAL2) and var. filipes (FIL2) was largely recovered in F2 individuals.
Most physiological traits were not significantly different between FIL2 and HAL2 (Table 3), and there were high levels of transgressive segregation in the F2 hybrids (Table 3; Fig. S4). Of the photosynthetic traits measured, only ci showed a significant difference between the parental lines, being lower in FIL2 than HAL2 (Table 3). This implied that although differences in A (slightly greater in FIL2, Table 3) and gs (smaller in FIL2, Table 3) were not significant, intrinsic water-use efficiency (A relative to gs) was greater in FIL2 than HAL2 (ci = ca−(A/gCO2) and gCO2 ≈ gs/1.6). This difference is indicative of greater photosynthetic capacity in FIL2. Fluorescence measurements indicated that FIL2 had a slightly higher ΦPSII, despite a lower fraction of PSII involved in photochemistry (Fv′/Fm′), a disparity linked with greater qP in FIL2 (Table 3). Since qP indicates turnover of electron carriers, which would be linked with demand for reducing equivalents by CO2 assimilation, this result further supports the indication of slightly greater photosynthetic capacity in FIL2. SPAD values indicated no significant difference in chlorophyll content between the parent lines (Table 3).
Table 3.
Means (SE) leaf physiological trait values for FIL2, HAL2 and F2 Panicum hallii plants
| Trait | N | FIL2 | N | HAL2 | LRT | N | F2 | F2 range |
|---|---|---|---|---|---|---|---|---|
| SPAD | 20 | 34.4 (0.64) | 18 | 32.7 (0.69) | 3 | 207 | 30 (0.44) | 18.9–40.5 |
| A (μmol CO2 m−2 s−1) | 19 | 15.1 (2.35) | 18 | 13.6 (2.23) | 0.87 | 196 | 12.8 (2.00) | 0.2–30.4 |
| gs (mol H2O m−2 s−1) | 19 | 0.091 (0.0112) | 18 | 0.102 (0.0112) | 0.77 | 196 | 0.082 (0.0077) | 0.019–0.184 |
| ci (μmol CO2 mol−1) | 19 | 114 (17) | 18 | 155 (17) | 8.97** | 196 | 127 (21) | 17–281 |
| qP | 20 | 0.57 (0.042) | 18 | 0.53 (0.042) | 0.99 | 213 | 0.53 (0.041) | 0.14–0.87 |
| Fv'/Fm' | 20 | 0.4 (0.0138) | 18 | 0.417 (0.0141) | 1.71 | 213 | 0.386 (0.0132) | 0.26–0.55 |
| ΦPSII | 20 | 0.235 (0.024) | 18 | 0.224 (0.0246) | 0.3 | 213 | 0.207 (0.0226) | 0.034–.408 |
N, number of replicates per parental lines or total number of F2 individuals (1 replicate per F2); LRT, Likelihood ratio test  , H0, no difference in parental means
, H0, no difference in parental means
P < 0.01. At ambient CO2: A, net CO2 assimilation rate; gs, stomatal conductance to water; ci, intercellular CO2 concentration; qP, photochemical quenching; Fv'/Fm', light adapted efficiency of energy harvesting by PSII; ΦPSII, quantum yield of PSII.
Genetic map
Our first round of linkage map construction in JoinMap utilized 782 out of the available 803 RAD-tag SNP markers. We recovered the nine linkage groups that were predicted from cytology (Gould, 1958; Waller, 1976). However, examination of this initial map revealed many marker order errors as well as poor markers with a high level of genotyping error. The genotyping errors appeared to be mostly the result of misidentification of heterozygote genotypes as homozygotes due to low sequence coverage at marker loci. After systematically purging poor markers using goodness of fit criteria (mean chi-squared contribution > 3) and close examination of recombination fraction plots, 278 remained for the second round of map construction. This linkage map had a combined length of 1252.1 cM and utilized recombination events from 257 F2 hybrids (Figs 2, S5). There was segregation distortion across many of the remaining markers, with 156 (56%) markers distorted at P < 0.05, 110 (40%) at P < 0.01, 66 (24%) at P < 0.001, and 36 (13%) at P < 0.0001.
Fig 2.
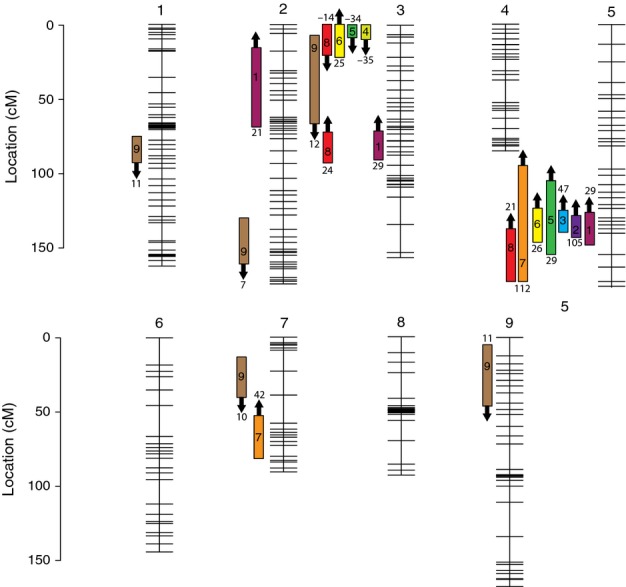
Genetic map of the FIL2 × HAL2 F2 hybrid population of Panicum hallii with the location of morphological trait quantitative trait loci (QTLs). Rectangular box indicates 1.5–LOD drop confidence intervals. Location of numbers within boxes is the location of QTL peaks. Arrow is the direction of additive effect, with an up arrow indicating that the FIL2 allele increases the trait value. Number above or below each QTL is the percentage of parental divergence (HAL2 vs FIL2) explained by the QTL. Traits: 1, tiller height; 2, number of tillers; 3, tiller width; 4, leaf length; 5, leaf width; 6, flowering time; 7, inflorescence length; 8, number of flowers; 9, seed mass. cM, centimorgans.
Comparisons of the marker location between P. hallii and their predicted location in the foxtail millet genome revealed a high level of conserved synteny with no clear evidence for translocations between chromosomes (Fig.3). However, we did observe two large-scale chromosomal inversions between the species on linkage groups 1 and 5 in dot plot comparisons between the physical map location of the foxtail millet genome and the linkage map location for P. hallii (Fig.4).
Fig 3.
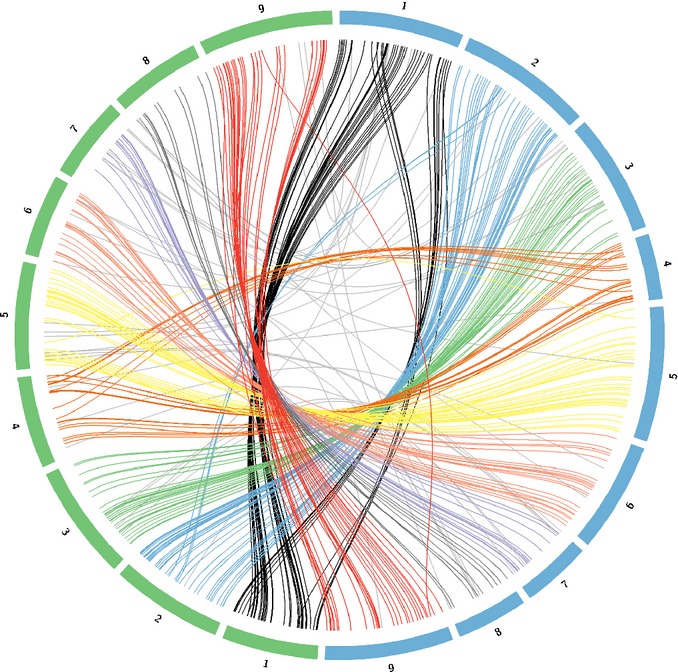
Synteny plot of marker positions from Panicum hallii genetic map (blue) with predicted homologous positions in the foxtail millet (Setaria italica) genome (green) suggests a high level of conserved synteny between the two species. Chromosome numbers are labeled 1–9 for both species.
Fig 4.
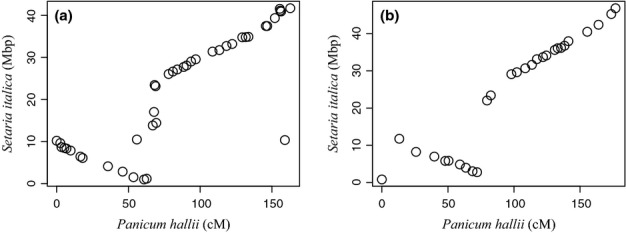
Comparisons of marker positions from Panicum hallii genetic map with predicted homologous positions in the foxtail millet (Setaria italica) genome (physical map) revealed large inversions between the two species on chromosomes (a) 1 and (b) 5. Mbps, mega base pairs; cM, centimorgans.
QTL analyses
Overall, we identified 28 QTLs from 14 phenotypic traits (nine morphological traits and five physiological traits) and hybrid sterility in the best-fit stepwise QTL models (Fig.2, Table 4). The vast majority of morphological QTLs (85%) had additive effects in the same direction as the parental divergence. See Tables S2 and S3 for R/qtl input files for QTL mapping.
Table 4.
Quantitative trait loci (QTLs) and their main effects from the best-fit models of stepwise analyses for the FIL2 × HAL2 F2 population of Panicum hallii
| Trait | Chr | Peak | 1.5–LOD interval | LOD | a | D | %var | %div |
|---|---|---|---|---|---|---|---|---|
| Tiller height | 2 | 38 | 20–71 | 5.387 | 3.172 | −0.4 | 6.695 | 21.083 |
| Tiller height | 3 | 77 | 73–85 | 9.361 | 4.338 | −2.045 | 12.096 | 28.833 |
| Tiller height | 5 | 140 | 124–148 | 10.731 | 4.43 | 0.436 | 14.057 | 29.445 |
| Number of tillers | 5 | 137 | 126–143 | 4.359 | 2.037 | −0.055 | 8.024 | 105.000 |
| Tiller width | 5 | 127 | 123–139 | 8.386 | 0.163 | 0.025 | 14.863 | 47.246 |
| Leaf length | 3 | 6 | 0–11 | 4.025 | −1.753 | −0.361 | 7.463 | −35.201 |
| Leaf width | 3 | 6 | 0–10 | 5.51 | −0.491 | −0.172 | 9.272 | −33.516 |
| Leaf width | 5 | 129 | 105–154 | 4.915 | 0.43 | −0.28 | 8.223 | 29.352 |
| Flowering time | 3 | 15 | 1–32 | 5.15 | 5.15 | −3.013 | 6.4 | 24.789 |
| Flowering time | 5 | 132 | 119–146 | 9.901 | 5.413 | −1.838 | 7.437 | 26.055 |
| Inflorescence length | 5 | 161 | 93–169 | 5.165 | 1.582 | 0.682 | 9.013 | 111.802 |
| Inflorescence length | 7 | 66 | 55–81 | 4.199 | 0.599 | 1.203 | 7.256 | 42.332 |
| Number of flowers | 3 | 18 | 0–27 | 3.912 | −29.391 | −6.714 | 5.748 | −14.294 |
| Number of flowers | 3 | 80 | 75–91 | 8.919 | 48.727 | −10.939 | 13.787 | 23.698 |
| Number of flowers | 5 | 148 | 136–169 | 6.059 | 42.351 | −1.959 | 9.098 | 20.596 |
| Seed mass | 1 | 83 | 75–90 | 5.506 | −0.046 | 0.014 | 8.629 | 11.220 |
| Seed mass | 2 | 152 | 131–161 | 3.989 | −0.03 | 0.035 | 6.139 | 7.317 |
| Seed mass | 3 | 14 | 9–68.907 | 5.155 | −0.049 | 0.008 | 8.045 | 11.951 |
| Seed mass | 7 | 26 | 15–39 | 4.34 | −0.039 | −0.012 | 6.707 | 9.512 |
| Seed mass | 9 | 19 | 9–47 | 5.633 | −0.047 | 0.008 | 8.842 | 11.463 |
| Sterility | 2 | 65 | 64–71 | 20.06 | −0.1 | 0 | 27.452 | na |
| Sterility | 7 | 29 | 16–39 | 4.89 | −0.101 | 0 | 5.708 | na |
| Sterility | 7 | 83 | 81–84 | 25.352 | −0.107 | 0 | 36.737 | na |
| SPAD | 3 | 1 | 0–13 | 4.073 | −1.425 | −1.339 | 8.724 | na |
| A (μmol CO2 m−2 s−1) | 3 | 94.4 | 89–107 | 3.844 | 2.818 | −1.528 | 8.808 | na |
| gs (mol H2O m−2 s−1) | 3 | 94.4 | 90–106 | 3.969 | 0.014 | −0.009 | 9.076 | na |
| ΦPSII | 3 | 94.4 | 88–107 | 4.54 | 0.035 | −0.017 | 9.564 | na |
| qP | 3 | 95 | 89–141 | 4.30 | 0.068 | −0.025 | 9.157 | na |
Chr, linkage group; Peak, location of the QTL peak; LOD, logarithm of odds; a, additive effect; D, dominance deviation; %var, percent of variance explained; %div, percent of parental divergence explained (only calculated for traits that showed significant differences between FIL2 and HAL2); A, net CO2 assimilation rate; gs, stomatal conductance to water; ΦPSII, quantum yield of PSII; qP, photochemical quenching; na, not applicable.
A large proportion (79%) of QTLs identified in our study had overlapping 1.5 LOD-drop confidence intervals with at least one other QTL. This is a high level of clustering, given that only 31% of the genome was occupied by the 1.5 LOD-drop of any QTL. Five morphological traits had overlapping confidence intervals on one region of linkage group 3, while seven morphological traits had overlapping confidence intervals on linkage group 5. Similarly, we mapped colocalizing physiological QTLs to a region of linkage group 3 for four physiological traits: A, gs, ΦPSII and qP (Table 4).
We mapped colocalizing QTLs for five traits to one region of linkage group 5 (Fig.2, Table 4). QTLs for infloresence length and flower number also had an overlapping confidence interval in this region, but the peak of these QTLs was offset from the other five. All five of the colocalizing QTLs had major phenotypic effects, explaining over 15% of the phenotypic divergence between the two ecotypes. This locus, or linked set of loci, accounted for 47% of the divergence between HAL2 and FIL2 in tiller width, 29% of the divergence in both tiller height and leaf width, 26% of the divergence in flowering time and 105% of the divergence in number of tiller. All seven of the QTLs that mapped to this region of linkage group 5 had additive effects in the same direction as the phenotypic divergence of the ecotypes, where the FIL2 allele made each trait larger.
The clustering of morphological QTLs on linkage group 3 was different from that of linkage group 5, as only QTLs for seed size and flowering time were in the same direction as the ecotype divergence. The three other QTLs in this region, for leaf length, leaf width and number of flowers, had additive effects in the opposite direction of the ecotype divergence. In other words, the FIL2 allele contributed to smaller trait values for these three traits.
Only six morphological QTLs did not have overlapping confidence intervals with other morphological QTLs (Fig.2). Of these six independent QTLs, four of them contributed to seed size. Further, all five of the seed size QTLs had moderate effects, each explaining only 7–11% of the parental divergence in this trait. Further, the seed size QTLs all had additive effects in the same direction, with the FIL2 allele contributing to smaller seeds at all loci. Finally, it should be noted that seed size did not have a QTL in the vicinity of the large effect colocalizing QTLs on linkage group 5.
Synteny mapping demonstrated that the clustering of large effect QTLs on linkage group 5 in P. hallii colocalized with a flowering time QTL discovered in a recent study of foxtail millet (Mauro-Herrera et al., 2013). The foxtail millet study identified a number of flowering time candidate genes in this region of synteny, including homologues of FVE/OsFVE (Ausin et al., 2004; Baek et al., 2008), OsLFL1 (Peng et al., 2007, 2008), SPA1 (Laubinger et al., 2006), and OsFTL9/ZCN12 (Danilevskaya et al., 2008). The linkage group 5 QTL of P. hallii is also in the vicinity of a homologue of developmental gene MORE AXILLARY BRANCHES1 (MAX1).
Stepwise QTL model fitting revealed a significant (LOD = 17.274) pairwise epistatic interaction for hybrid sterility established by seed set (Fig.5). This pairwise interaction appears to be a classic Dobzhansky–Muller incompatibility (Coyne & Orr, 2004; Sweigart & Willis, 2012; Bomblies, 2013). Here, individual plants were sterile when homozygous for the HAL2 allele at position 83 cM on linkage group 7 and homozygous for the FIL2 allele at position 65 cM on linkage group 2.
Fig 5.
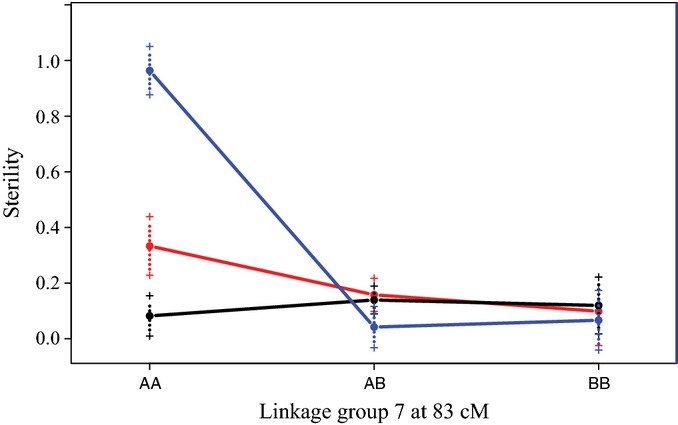
A two-locus Dobzhansky–Muller hybrid sterility incompatibility system between the HAL2 and FIL2 lines of Panicum hallii. Mean genotypic effects for the homozygous HAL2 individuals (AA), homozygous FIL2 individuals (BB), and heterozygous individuals (AB) are indicated for the locus on linkage group 7 on the x-axis and by colored lines (AA, red line; BB, blue line; AB, black line) for the locus on linkage group 2 at 65 cM. A high level of sterility occurs in individuals that are homozygous for the HAL2 allele on linkage group 7 and homozygous for the FIL2 allele on linkage group 2. cM, centimorgan.
Discussion
Determining the genetic architecture of traits involved in the adaptive divergence between ecotypes is crucial to understanding the process of speciation. In this study, we used genetic mapping to evaluate whether or not QTLs for traits involved in the divergence between xeric and mesic ecotypes of P. hallii colocalize to particular genomic regions. In the process of conducting this study, we assembled the first genetic map for P. hallii, which revealed a high level of synteny with foxtail millet. We mapped at least one QTL for every morphological trait, as well as QTL for five out of seven physiological traits. We found large effect colocalizing QTLs on linkage group 5, which controlled multiple classic ecotype-differentiating traits including tiller and leaf size and shape as well as flowering time. We also identified a two-locus epistatic hybrid incompatibility system that causes sterility in hybrids. Overall, our study suggests that some ecotype-differentiating traits evolve independently while others may evolve in tandem due to genetic colocalization resulting from linkage or pleiotropy.
The genetics of divergence between ecotypes
The evolution of plant ecotypes adapted to xeric vs mesic habitats is a common phenomenon that serves as a model of how ecology can contribute to the process of speciation (Clausen, 1951; Clausen & Heisey, 1958; Grant, 1981; Kruckeberg, 1986; Rajakaruna, 2004; Lowry, 2012). One of the key discoveries of our study was the colocalization of a major QTL for five traits to the same region of linkage group 5. The effects of this region on multiple traits could be due to tight linkage of multiple genes with effects on individual traits, or could be caused by pleiotropic effects of a single gene on multiple traits. Regardless, it is interesting that so much of the phenotypic divergence between the ecotypes maps to one genomic location and that the additive effects of these QTLs are all in the same direction as the ecotype trait divergences. The linkage group 5 QTLs contrasts with those that clustered on linkage group 3, where the additive effects of three of the QTLs were in the opposite direction of the ecotype divergence.
The finding that multiple major-effect ecotype-differentiating QTLs colocalize to linkage group 5 suggests that the evolution of these traits is nonindependent. This could be the result of fundamental trade-offs between above ground growth and the demand for water imposed by transpiration (Geber & Dawson, 1990; Dudley, 1996; Ackerly et al., 2000). There may be structural developmental reasons for those trade-offs, especially if the same genetic loci are contributing to evolution of many of these traits. One possible reason why multiple ecotype-differentiating traits have colocalizing QTLs could be that those traits can be simultaneously altered by mutations in major developmental genes and hormone pathways. In fact, mutations in hormone pathways result in phenotypic effects that mimic the developmental divergence between xeric and mesic ecotypes (Fleet & Sun, 2005; Domagalska et al., 2007; Zhao, 2010). Further, recent studies in rice (Xue et al., 2008; Yan et al., 2011) have cloned the QTLs that pleiotropically control flowering time, plant height, and the number of seeds per panicle in the same additive directions as the colocalizing QTLs on linkage group 5 in P. hallii. Thus, there is precedent that the set of traits involved in xeric/mesic ecotype divergence can be controlled pleiotropically in other grass species. We identified multiple potential candidate genes underlying the pleiotropic QTL on linkage group five through synteny analysis between P. hallii and foxtail millet. The gene MAX1, which is found in this region, is known to repress axillary bud primordia and control branching in A. thaliana (Stirnberg et al., 2002; Waters et al., 2012) and is thus a candidate gene underlying the QTLs that colocalize in this region.
Another possible reason why the QTLs for so many traits colocalize to the same genomic region could be that haplotypes of multiple alleles are held together by a chromosomal inversion (Kirkpatrick, 2010; Lowry & Willis, 2010). A set of ecotype-differentiating traits, similar to those found in P. hallii, all mapped to a single chromosomal inversion polymorphism in Mimulus guttatus (Hall et al., 2006, 2010; Lowry & Willis, 2010). While we found no evidence that a chromosomal rearrangement is in the vicinity of the colocalizing QTLs on linkage group 5, we cannot yet rule out this possibility without further fine genetic mapping.
Four of the five QTLs for seed size did not have overlapping confidence intervals with any other QTLs. Thus, this trait may be able to evolve more independently of many of the other ecotype-differentiating morphological traits, especially those that colocalize on linkage group 5. Classic studies have identified associations between seed size and many environmental factors (Baker, 1972; Salisbury, 1974). Baker (1972) surveyed seed size in nearly 2500 plant taxa in California and found plants typically had larger seed sizes in locations where there was a greater likelihood of exposure to drought after germination. Provisioning in larger seeds can provide a seedling with greater potential to establish a large root system before the onset of periods of low water availability (Kneebone & Cremer, 1955; Glewen & Vogel, 1984; Venable & Brown, 1988). P. hallii follows this classic pattern with larger seeds in the xeric var. hallii and smaller seeds in the mesic var. filipes. Our study suggests a highly polygenic genetic architecture involved in seed size evolution in this system, with the five significant moderate effect QTLs only accounting for c. 51% of the divergence between the two varieties. This finding may also be relevant to the closely related bioenergy crop Panicum virgatum (switchgrass), where seed size is a focal trait for breeding because it is positively correlated with seedling establishment (Aiken & Springer, 1995; Smart & Moser, 1999; Boe, 2003). Just like P. hallii, P. virgatum has a wet lowland habitat ecotype with smaller seeds than its dry habitat upland ecotype (Seepaul, 2010; Lowry et al., 2014).
Genetics and physiology
Regulation of photosynthesis and stomatal gas exchange is crucial for plant response to drought (Hiesey & Milner, 1965; Ackerly et al., 2000; Chaves et al., 2009; Pinheiro & Chaves, 2011; Taylor et al., 2011, 2014). We found that FIL2 showed greater capacity to draw down CO2 concentrations inside the leaf, supported by consistent but nonsignificant differences in chlorophyll fluorescence traits, and indicating greater intrinsic water-use efficiency than HAL2. Given the greater canopy size of FIL2 individuals, and the expectation that transpiration will scale with the canopy size, lower gs may help to reduce the possibility of hydraulic failure in FIL2 (Taylor et al., 2014). Equally, as HAL2 flowers significantly more quickly than FIL2, greater gs in HAL2 may help to improve the return on investments in shorter-lived leaves (Wright et al., 2003).
Although we were able to identify a colocalizing QTL for four physiologically inter-linked traits, we do not yet know how the two varieties respond to decreased levels of available soil moisture, or have data on leaf lifespan and structural investment, which will be crucial in understanding the evolution of divergence between P. hallii ecotypes. The morphological and physiological traits underlying photosynthetic performance and water-use efficiency are notoriously plastic (Flood et al., 2011). Our physiological measurements were made on flowering plants in a glasshouse under well-watered conditions, but genetic differences contributing to divergence between P. hallii ecotypes may also manifest at particular ontogenetic stages and/or as plastic responses to decreasing soil water availability (Teng et al., 2004; Van Kleunen & Fischer, 2005; Picotte et al., 2007; Maherali et al., 2009; Pinheiro & Chaves, 2011).
One of the most striking results of our study was that while most of the physiological traits did not strongly differ between HAL2 and FIL2, many of these traits were highly transgressive in their segregation in the F2 hybrids. Transgressive segregation is often caused by alleles with opposite additive effects across loci being shuffled into different combinations through hybridization (Rieseberg et al., 1999; Lexer et al., 2005). Morphological traits were less transgressive than the physiological traits. Even so, we did identify morphological QTLs (leaf width and number of flowers) that had opposite additive effects consistent with transgressive segregation.
Reproductive isolation between the ecotypes
Understanding what reproductive isolating barriers are important for reducing gene flow between ecotypes is key to better understanding the process of speciation (Coyne & Orr, 2004; Nosil et al., 2005; Sobel et al., 2010). P. hallii var. hallii and var. filipes have an overlapping geographic range in southern Texas and northern Mexico and occur in sympatry in areas where both dry upland and wet lowland habitats exist in close proximity (Waller, 1976). We have now identified multiple potential reproductive isolating barriers that could explain the maintenance of the ecotypes in sympatry.
Approximately one-fifth of the F2 hybrids had a high level of sterility. This hybrid sterility is partially caused by a two-locus nuclear Dobzhansky–Muller incompatibility between linkage groups 2 and 7. Dobzhansky–Muller incompatibilities are thought to play a major role in the evolution of new species by causing postzygotic reproductive isolation (Coyne & Orr, 2004) and are frequently found in genetic mapping studies between diverging plant varieties and species (Moyle & Graham, 2005; Sweigart et al., 2006; Mizuta et al., 2010; Kubo et al., 2011). In some cases, the genes underlying these epistatic incompatibility systems in plants have even been identified (reviewed in Rieseberg & Blackman, 2010; Sweigart & Willis, 2012; Bomblies, 2013). The incompatibility system we identified here required homozygote genotypes for both varieties at each of two loci. The relatively low level of hybrid sterility, and no evidence of hybrid inviability, suggests that postzygotic isolation may not be the main factor maintaining the distinctness of the P. hallii varieties.
Ecological isolating barriers often form as a byproduct of divergent adaptations between ecotypes and species (Coyne & Orr, 2004; Nosil, 2012). The differences in flowering time between the ecotypes suggest that temporal isolation may be a significant barrier to gene flow. Further, the fact that var. hallii and var. filipes are found in different microhabitats suggests a potential role for habitat isolation and extrinsic postzygotic isolation as barriers to gene flow. Extrinsic postzygotic isolation can also result from transgressive segregation, where increased phenotypic variability leads to the production of maladapted hybrids (Rogers & Bernatchez, 2006; Renaut et al., 2009). Hybrids between the two ecotypes were highly transgressive in physiological traits, which could lead to decreased fitness in nature.
The most important barrier for P. hallii may be mating system isolation. Our recent population genetic survey of P. hallii revealed that it has a near obligate self-fertilization mating system (Lowry et al., 2013). We have yet to identify a population of P. hallii with an appreciable level of outcrossing. The low level of polymorphism in the genome sequence of the FIL2 accession produced for this study further supports the idea that P. hallii plants typically occur in nature as highly inbred nearly-homozygous lines as a result of their selfing mating system. With such low levels of outcrossing, the possibility of hybridization between the varieties is very low. Martin & Willis (2007) recently showed that mating system can be a very strong barrier to gene flow, even when only one out of a pair of hybridizing species has a high rate of self-fertilization.
Conclusions
It has long been known that a common set of traits often differentiates xeric and mesic ecotypes across plant species (Clausen & Heisey, 1958). Our study reinforces the pattern found recently in other systems (Latta & Gardner, 2009; Lowry & Willis, 2010; Lovell et al., 2013) that QTLs for ecotype-differentiating traits frequently colocalize to the same genomic position. The same set of ecotype-differentiating traits found between xeric and mesic varieties of P. hallii are also involved in the divergence of upland and lowland ecotypes of the closely related bioenergy crop switchgrass (P. virgatum; Lowry et al., 2014). Gaining insight into the molecular details of colocalizing QTL in these systems will be an important next step in understanding the factors constraining or facilitating adaptation and ecotypic differentiation to different habitats in nature.
Acknowledgments
We would like to thank A. Asmus, C. Purmal, T. Liu, L. Villareal, and A. Hiers for help in the glasshouse and with phenotyping. The Lady Bird Johnson Wildflower Center and J. L. Reilly provided us with seeds for HAL2 and FIL2. S. Merrell was vital to working out glasshouse logistics. The Texas Advanced Computing Center provided computing time on the Lonestar cluster that made our analyses possible. Thanks to K. Bomblies, L. Delph, B. Logan, R. Shaw and six anonymous reviewers for discussion and comments that greatly improved this manuscript. The University of Texas Freshman Research Initiative provided funding for training of undergraduate students that worked on this project. The National Science Foundation provided funding through a Plant Genome Research Program Award (IOS-0922457) to T.E.J. and a Postdoctoral Research Fellowship in Biology (DBI 1103668) to K.H. A United States Department of Agriculture NIFA-AFRI Postdoctoral Fellowship (2011-67012-30696) supported D.B.L. The work conducted by the US Department of Energy Joint Genome Institute was supported by the Office of Science of the US Department of Energy under Contract no. DE-AC02-05CH11231.
Supporting Information
Additional supporting information may be found in the online version of this article.
Flowchart of the pipeline for mapping and SNP calling of RAD-tag markers for the FIL2 × HAL2 hybrid F2 mapping population of Panicum hallii.
Fig. S2 Flowchart of the pipeline for synteny analysis between the Panicum hallii linkage map and the genome of foxtail millet (Setaria italica).
Fig. S3 Histograms of phenotypic variation for morphological traits in the F2 hybrid population of Panicum hallii.
Fig. S4 Histograms of phenotypic variation for physiological traits in the F2 hybrid population of Panicum hallii.
Fig. S5 Plot of recombination fractions across the Panicum hallii FIL2 × HAL2 genetic map.
Methods S1 Supplementary methods.
Summary of Illumina libraries used in the preliminary Meraculous Panicum hallii assembly
Table S2 R/qtl input file for QTL mapping of morphological traits
Table S3 R/qtl input file for QTL mapping of physiological traits
References
- Ackerly DD, Dudley SA, Sultan SE, Schmitt J, Coleman JS, Linder CR, Sandquist DR, Geber MA, Evans AS, Dawson TE, et al. The evolution of plant ecophysiological traits: recent advances and future directions. BioScience. 2000;50:979–995. [Google Scholar]
- Aiken GE, Springer TL. Seed size distribution, germination, and emergence of 6 switchgrass cultivars. Journal of Range Management. 1995;48:455–458. [Google Scholar]
- Andrew RL, Kane NC, Baute GJ, Grassa CJ, Rieseberg LH. Recent nonhybrid origin of sunflower ecotypes in a novel habitat. Molecular Ecology. 2013;22:799–813. doi: 10.1111/mec.12038. [DOI] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Regulation of flowering time by FVE, a retinoblastoma–associated protein. Nature Genetics. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- Baek IS, Park HY, You MK, Lee JH, Kim JK. Functional conservation and divergence of FVE genes that control flowering time and cold response in rice and Arabidopsis. Molecules and Cells. 2008;26:368–372. [PubMed] [Google Scholar]
- Baker HG. Seed weight in relation to environmental conditions in California. Ecology. 1972;53:997–1010. [Google Scholar]
- Bates D, Maechler M, Bolker B. Lme4: Linear mixed-effects models using S4 classes. 2013. WWW document] URL http://CRAN.R-project.org/package=lme4 [R package version 0.999999-2] [accessed 15 January 2014]
- Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, Pontaroli AC, Estep M, Feng L, Vaughn JN, Grimwood J, et al. Reference genome sequence of the model plant Setaria. Nature Biotechnology. 2012;30:555–561. doi: 10.1038/nbt.2196. [DOI] [PubMed] [Google Scholar]
- Boe A. Genetic and environmental effects on seed weight and seed yield in switchgrass. Crop Science. 2003;43:63–67. [Google Scholar]
- Bomblies K. Genes causing postzygotic hybrid incompatibility in plants: a window into co-evolution. In: Chen ZJ, Birchler JA, editors. Polyploid and hybrid genomics. Oxford, UK: John Wiley & Sons Inc; 2013. pp. 225–240. [Google Scholar]
- Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. doi: 10.1038/nature02106. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Chapman JA, Ho I, Sunkara S, Luo S, Schroth GP, Rokhsar DS. Meraculous: de novo genome assembly with short paired-end reads. PLoS ONE. 2011;6:e23501. doi: 10.1371/journal.pone.0023501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Annals of Botany London. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen J. Stages in the evolution of plant species. Ithaca, NY, USA: Cornell University Press; 1951. [Google Scholar]
- Clausen J, Heisey WM. Experimental studies on the nature of species IV. Genetic structure of ecological races. Washington, DC, USA: Carnegie Institute of Washington; 1958. [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, MA, USA: Sinauer Associates; 2004. [Google Scholar]
- Danilevskaya ON, Meng X, Hou ZL, Ananiev EV, Simmons CR. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiology. 2008;146:250–264. doi: 10.1104/pp.107.109538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, Ilie L, Brudno M. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics. 2011;27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. New York, NY, USA: Columbia University Press; 1937. [Google Scholar]
- Domagalska MA, Schomburg FM, Amasino RM, Vierstra RD, Nagy F, Davis SJ. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development. 2007;134:2841–2850. doi: 10.1242/dev.02866. [DOI] [PubMed] [Google Scholar]
- Dudley SA. The response to differing selection on plant physiological traits: evidence for local adaptation. Evolution. 1996;50:103–110. doi: 10.1111/j.1558-5646.1996.tb04476.x. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, Richards RA. Isotopic composition of plant carbon correlates with water-use efficiency of wheat genotypes. Functional Plant Biology. 1984;11:539–552. [Google Scholar]
- Fleet CM, Sun T-P. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Current Opinions in Plant Biology. 2005;8:77–85. doi: 10.1016/j.pbi.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Flood PJ, Harbinson J, Aarts MG. Natural genetic variation in plant photosynthesis. Trends in Plant Science. 2011;16:327–335. doi: 10.1016/j.tplants.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Franks SJ. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytologist. 2011;190:249–257. doi: 10.1111/j.1469-8137.2010.03603.x. [DOI] [PubMed] [Google Scholar]
- Geber MA, Dawson TE. Genetic variation in and covariation between leaf gas exchange, morphology, and development in Polygonum arenastrum, an annual plant. Oecologia. 1990;85:153–158. doi: 10.1007/BF00319396. [DOI] [PubMed] [Google Scholar]
- Glewen K, Vogel K. Partitioning the genetic variability for seedling growth in sand bluestem into its seed size and seedling vigor components. Crop Science. 1984;24:137–141. [Google Scholar]
- Gould FW. Chromosome numbers in southwestern grasses. American Journal of Botany. 1958;45:757–767. [Google Scholar]
- Gould FW. The grasses of Texas. College Station, TX, USA: Texas A&M University Press; 1975. [Google Scholar]
- Grant V. Plant speciation. New York, NY, USA: Columbia University Press; 1981. 2nd edn. [Google Scholar]
- Hall MC, Basten C, Willis JH. Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics. 2006;172:1829–1844. doi: 10.1534/genetics.105.051227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Lowry DB, Willis JH. Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Molecular Ecology. 2010;19:2739–2753. doi: 10.1111/j.1365-294X.2010.04680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch SL, Schuster JL, Drawe DL. Grasses of the Texas Gulf prairies and marshes. College Station, TX, USA: Texas A&M University Press; 2003. [Google Scholar]
- Hiesey WM, Milner HW. Physiology of ecological races and species. Annual Reviews in Plant Physiology. 1965;16:203–213. [Google Scholar]
- Juenger TE. Natural variation and genetic constraints on drought tolerance. Current Opinions in Plant Biology. 2013;16:274–281. doi: 10.1016/j.pbi.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. How and why chromosome inversions evolve. PLoS Biology. 2010;8:e1000501. doi: 10.1371/journal.pbio.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneebone W, Cremer C. The relationship of seed size to seedling vigor in some native grass species. Agronomy Journal. 1955;47:472–477. [Google Scholar]
- Kruckeberg AR. An essay: the stimulus of unusual geologies for plant speciation. Systematic Botany. 1986;11:455–463. [Google Scholar]
- Kubo T, Yoshimura A, Kurata N. Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics. 2011;189:1083–1092. doi: 10.1534/genetics.111.132035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta RG, Gardner KM. Natural selection on pleiotropic quantitative trait loci a life-history trade-off in Avena barbata. Evolution. 2009;63:2153–2163. doi: 10.1111/j.1558-5646.2009.00701.x. [DOI] [PubMed] [Google Scholar]
- Laubinger S, Marchal V, Gentilhomme J, Wenkel S, Adrian J, Jang S, Kulajta C, Braun H, Coupland G, Hoeckler U. Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development. 2006;133:3213–3222. doi: 10.1242/dev.02481. [DOI] [PubMed] [Google Scholar]
- Lexer C, Rosenthal DM, Raymond O, Donovan LA, Rieseberg LH. Genetics of species differences in the wild annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2005;169:2225–2239. doi: 10.1534/genetics.104.031195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JT, Juenger TE, Michaels SD, Lasky JR, Platt A, Richards JH, Yu X, Easlon HM, Sen S, McKay JK. Pleiotropy of FRIGIDA enhances the potential for multivariate adaptation. Proceedings of the Royal Society of London. Series B, Biological Sciences. 2013;280:20131043. doi: 10.1098/rspb.2013.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB. Ecotypes and the controversy over stages in the formation of new species. Biological Journal of the Linnean Society. 2012;106:241–257. [Google Scholar]
- Lowry DB, Behrman KD, Grabowski P, Morris GP, Kiniry JR, Juenger TE. Adaptation between ecotypes and along environmental gradients in Panicum virgatum. American Naturalist. 2014;183:682–692. doi: 10.1086/675760. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Purmal CT, Juenger TE. A population genetic transect of Panicum hallii (Poaceae) American Journal of Botany. 2013;100:592–601. doi: 10.3732/ajb.1200379. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Purmal CT, Meyer E, Juenger TE. Microsatellite markers for the native Texas perennial grass, Panicum hallii (Poaceae) American Journal of Botany. 2012;99:e114–e116. doi: 10.3732/ajb.1100430. [DOI] [PubMed] [Google Scholar]
- Lowry DB, Rockwood RC, Willis JH. Ecological reproductive isolation of coast and inland races of Mimulus guttatus. Evolution. 2008;62:2196–2214. doi: 10.1111/j.1558-5646.2008.00457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry DB, Willis JH. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biology. 2010;8:e1000500. doi: 10.1371/journal.pbio.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow MM. Strategies of response to water stress. In: Kreeb KH, Richter H, Minckley TM, editors. Structural and functional responses to environmental stress. The Hague, the Netherlands: SPB Academic; 1989. pp. 269–281. [Google Scholar]
- Maherali H, Caruso CM, Sherrard ME. The adaptive significance of ontogenetic changes in leaf physiology: a test with Avena barbata. New Phytologist. 2009;183:908–918. doi: 10.1111/j.1469-8137.2009.02845.x. [DOI] [PubMed] [Google Scholar]
- Markwell J, Osterman JC, Mitchell JL. Calibration of the Minolta SPAD-502 leaf chlorophyll meter. Photosynthesis Research. 1995;46:467–472. doi: 10.1007/BF00032301. [DOI] [PubMed] [Google Scholar]
- Martin NH, Willis JH. Ecological divergence associated with mating system causes nearly complete reproductive isolation between sympatric Mimulus species. Evolution. 2007;61:68–82. doi: 10.1111/j.1558-5646.2007.00006.x. [DOI] [PubMed] [Google Scholar]
- Mauro-Herrera M, Wang X, Barbier H, Brutnell TP, Devos KM, Doust AN. Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae) Genes, Genomes, and Genetics. 2013;3:283–295. doi: 10.1534/g3.112.005207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. Chlorophyll fluorescence – a practical guide. Journal of Experimental Botany. 2000;51:659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana. I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology. 2003;12:1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Nemali KS, Ervin O, Rice KJ, Sen S, Mitchell-Olds T, Stahl E, Wayne T, Boles S, et al. Genetics of drought adaptation in Arabidopsis thaliana II. QTL analysis of a new mapping population, Kas-1 × Tsu-1. Evolution. 2008;62:3014–3026. doi: 10.1111/j.1558-5646.2008.00474.x. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E, Logan TL, Juenger TE. Transcriptome analysis and gene expression atlas for Panicum hallii var. filipes, a diploid model for biofuel research. Plant Journal. 2012;70:879–890. doi: 10.1111/j.1365-313X.2012.04938.x. [DOI] [PubMed] [Google Scholar]
- Mizuta Y, Harushima Y, Kurata N. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proceedings of the National Academy of Science, USA. 2010;107:20417–20422. doi: 10.1073/pnas.1003124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC, Graham EB. Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics. 2005;169:355–373. doi: 10.1534/genetics.104.029546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P. Ecological speciation. Oxford, UK: Oxford University Press; 2012. [Google Scholar]
- Nosil P, Vines TH, Funk DJ. Reproductive isolation caused by natural selection against immigrants from divergent habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- Osnas JLD, Lichstein JW, Reich PB, Pacala SW. Global leaf trait relationships: mass, area, and the leaf economics spectrum. Science. 2013;340:741–744. doi: 10.1126/science.1231574. [DOI] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochemical and Biophysical Research Communications. 2007;360:251–256. doi: 10.1016/j.bbrc.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Peng LT, Shi ZY, Li L, Shen GZ, Zhang JL. Overexpression of transcription factor OsLFL1 delays flowering time in Oryza sativa. Journal of Plant Physiology. 2008;165:876–885. doi: 10.1016/j.jplph.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Picotte JJ, Rosenthal DM, Rhode JM, Cruzan MB. Plastic responses to temporal variation in moisture availability: consequences for water use efficiency and plant performance. Oecologia. 2007;153:821–832. doi: 10.1007/s00442-007-0794-z. [DOI] [PubMed] [Google Scholar]
- Pinheiro C, Chaves MM. Photosynthesis and drought: can we make metabolic connections from available data? Journal of Experimental Botany. 2011;62:869–882. doi: 10.1093/jxb/erq340. [DOI] [PubMed] [Google Scholar]
- Porter CL. An analysis of variation between upland and lowland Switchgrass Panicum virgatum L in Central Oklahoma. Ecology. 1966;47:980–992. [Google Scholar]
- Rajakaruna N. The edaphic factor in the origin of plant species. International Geology Review. 2004;46:471–478. [Google Scholar]
- Renaut S, Nolte AW, Bernatchez L. Gene expression divergence and hybrid misexpression between lake whitefish species pairs (Coregonus spp. Salmonidae) Molecular Biology and Evolution. 2009;26:925–936. doi: 10.1093/molbev/msp017. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Blackman BK. Speciation genes in plants. Annals of Botany London. 2010;106:439–455. doi: 10.1093/aob/mcq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SM, Bernatchez L. The genetic basis of intrinsic and extrinsic post-zygotic reproductive isolation jointly promoting speciation in the lake whitefish species complex (Coregonus clupeaformis. Journal of Evolutionary Biology. 2007;19:1979–1994. doi: 10.1111/j.1420-9101.2006.01150.x. [DOI] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Le Corre V. How to be early flowering: an evolutionary perspective. Trends in Plant Science. 2006;11:375–381. doi: 10.1016/j.tplants.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Salisbury E. Seed size and mass in relation to environment. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1974;186:83–88. [Google Scholar]
- Schemske DW. Adaptation and the origin of species. American Naturalist. 2010;176:S4–S25. doi: 10.1086/657060. [DOI] [PubMed] [Google Scholar]
- Seepaul R. Switchgrass (Panicum virgatum L.) intraspecific variation and temperature tolerance classification using in vitro seed germination assay. Mississippi State, MS, USA: Mississippi State University; 2010. MS Thesis. [Google Scholar]
- Smart AJ, Moser LE. Switchgrass seedling development as affected by seed size. Agronomy Journal. 1999;91:335–338. [Google Scholar]
- Sobel JM, Chen GF, Watt LR, Schemske DW. The biology of speciation. Evolution. 2010;64:295–315. doi: 10.1111/j.1558-5646.2009.00877.x. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Aridity as a stimulus to plant evolution. American Naturalist. 1952;86:33–44. [Google Scholar]
- Stirnberg P, van De Sande K, Leyser HO. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129:1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- Sweigart AL, Fishman L, Willis JH. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics. 2006;172:2465–2479. doi: 10.1534/genetics.105.053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart AL, Willis JH. Molecular evolution and genetics of postzygotic reproductive isolation in plants. F1000 Biology Reports. 2012;4:23. doi: 10.3410/B4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SH, Ripley BS, Martin T, De-Wet LA, Woodward FI, Osborne CP. Physiological advantages of C4 grasses in the field: a comparative experiment demonstrating the importance of drought. Global Change Biology. 2014;20:1992–2003. doi: 10.1111/gcb.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SH, Ripley BS, Woodward FI, Osborne CP. Drought limitation of photosynthesis differs between C3 and C4 grass species in a comparative experiment. Plant, Cell & Environment. 2011;34:65–75. doi: 10.1111/j.1365-3040.2010.02226.x. [DOI] [PubMed] [Google Scholar]
- Teng S, Qian Q, Zeng D, Kunihiro Y, Fujimoto K, Huang D, Zhu L. QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.) Euphytica. 2004;135:1–7. [Google Scholar]
- Turesson G. The species and variety as ecological units. Hereditas. 1922;3:100–113. [Google Scholar]
- Van Kleunen M, Fischer M. Constraints on the evolution of adaptive phenotypic plasticity in plants. New Phytologist. 2005;166:49–60. doi: 10.1111/j.1469-8137.2004.01296.x. [DOI] [PubMed] [Google Scholar]
- Van Ooijen JW. JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations. Wageningen, the Netherlands: Kyazma BV; 2006. [Google Scholar]
- Venable DL, Brown JS. The selective interactions of dispersal, dormancy, and seed size as adaptations for reducing risk in variable environments. American Naturalist. 1988;131:360–384. [Google Scholar]
- Waller FR. A biosystematic study of Panicum section Diffusa (Poaceae) in North America. College Station, TX, USA: Texas A&M University; 1976. PhD thesis. [Google Scholar]
- Wang S, Meyer E, McKay JK, Matz MV. 2b-RAD: a simple and flexible method for genome-wide genotyping. Nature Methods. 2012;9:808–810. doi: 10.1038/nmeth.2023. [DOI] [PubMed] [Google Scholar]
- Waters MT, Brewer PB, Bussell JD, Smith SM, Beveridge CA. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiology. 2012;159:1073–1085. doi: 10.1104/pp.112.196253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker RH. Communities and ecosystems. New York, NY, USA: Mcmillan; 1975. [Google Scholar]
- Woodward FI, Lomas MR, Kelly CK. Global climate and the distribution of plant biomes. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 2004;359:1465–1476. doi: 10.1098/rstb.2004.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright IJ, Reich PB, Westoby M. Least-cost input mixtures of water and nitrogen for photosynthesis. American Naturalist. 2003;161:98–111. doi: 10.1086/344920. [DOI] [PubMed] [Google Scholar]
- Wu CI. The genic view of the process of speciation. Journal of Evolutionary Biology. 2001;14:851–865. [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, Zhang Q. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yan WH, Wang P, Chen HX, Zhou HJ, Li QP, Wang CR, Ding ZH, Zhang YS, Yu SB, Xing YZ, Zhang QF. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Molecular Plant. 2011;4:319–330. doi: 10.1093/mp/ssq070. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Zalapa JE, Jakubowski AR, Price DL, Acharya A, Wei YL, Brummer EC, Kaeppler SM, Cassler MD. Post-glacial evolution of Panicum virgatum: centers of diversity and gene pools revealed by SSR markers and cpDNA sequences. Genetica. 2011;139:933–948. doi: 10.1007/s10709-011-9597-6. [DOI] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annual Review of Plant Biology. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart of the pipeline for mapping and SNP calling of RAD-tag markers for the FIL2 × HAL2 hybrid F2 mapping population of Panicum hallii.
Fig. S2 Flowchart of the pipeline for synteny analysis between the Panicum hallii linkage map and the genome of foxtail millet (Setaria italica).
Fig. S3 Histograms of phenotypic variation for morphological traits in the F2 hybrid population of Panicum hallii.
Fig. S4 Histograms of phenotypic variation for physiological traits in the F2 hybrid population of Panicum hallii.
Fig. S5 Plot of recombination fractions across the Panicum hallii FIL2 × HAL2 genetic map.
Methods S1 Supplementary methods.
Summary of Illumina libraries used in the preliminary Meraculous Panicum hallii assembly
Table S2 R/qtl input file for QTL mapping of morphological traits
Table S3 R/qtl input file for QTL mapping of physiological traits


