Abstract
Fine-tuning plant cell wall properties to render plant biomass more amenable to biofuel conversion is a colossal challenge. A deep knowledge of the biosynthesis and regulation of plant cell wall and a high-precision genome engineering toolset are the two essential pillars of efforts to alter plant cell walls and reduce biomass recalcitrance. The past decade has seen a meteoric rise in use of transcriptomics and high-resolution imaging methods resulting in fresh insights into composition, structure, formation and deconstruction of plant cell walls. Subsequent gene manipulation approaches, however, commonly include ubiquitous mis-expression of a single candidate gene in a host that carries an intact copy of the native gene. The challenges posed by pleiotropic and unintended changes resulting from such an approach are moving the field towards synthetic biology approaches. Synthetic biology builds on a systems biology knowledge base and leverages high-precision tools for high-throughput assembly of multigene constructs and pathways, precision genome editing and site-specific gene stacking, silencing and/or removal. Here, we summarize the recent breakthroughs in biosynthesis and remodelling of major secondary cell wall components, assess the impediments in obtaining a systems-level understanding and explore the potential opportunities in leveraging synthetic biology approaches to reduce biomass recalcitrance.
Keywords: biomass, biofuels, recalcitrance, plant cell wall, synthetic biology, systems biology
Introduction
Plant cell walls, which include primary walls composed largely of cellulose, pectin and hemicellulose and secondary walls composed mainly of cellulose, lignin and hemicellulose, determine the development and morphology, strength properties, water and solute transport mechanisms, and pest resistance, at both cellular and whole plant levels. For lignocellulosic biofuel production, it is known that chemical as well as structural features of the cell walls and biomass are important determinants of feedstock recalcitrance or its resistance to breakdown to sugars. Several cell wall properties, such as content and quality of lignin and cellulose, discernible chemical (i.e. extent of acetylation and methylation, and heteropolysaccharide composition) and structural features of the wall at the angstrom-level (i.e. inter chain covalent, H-bond and van der Waals interactions) and nanometre level (i.e. tonoplastic space and pore density), have a direct impact on recalcitrance and in turn, on the liquid transportation fuel production per unit area of land (Davison et al., 2006; Ding et al., 2012; Himmel et al., 2007; McCann and Carpita, 2008; Mohnen, 2008; Scheller and Ulvskov, 2010). Fine-tuning these cell wall properties to render plant biomass more amenable to biofuel conversion is therefore, a prime goal of the biofuels research community (Davison et al., 2013; Kalluri and Keller, 2010).
Within the context of the present report, the term ‘biomass’ is used to refer to lignocellulosic plant material, ‘systems biology’ is used to refer to an integrative approach combining large amounts of biological data to understand and predict emergent complex biological phenomena, and ‘synthetic biology’ is used to refer to the targeted design and redesign of the biological machinery and living systems using a parts-based approach and engineering principles (Schmidt, 2012). The adoption of synthetic biology approaches to precisely engineer metabolic pathways and generate effective designer enzymes holds tremendous potential in improving biomass properties. Promising and/or or well-studied energy crop plants being explored for lignocellulosic biomass-based biofuels production include switchgrass (cultivars of Panicum virgatum), miscanthus (Miscanthus spp.), poplar (Populus trichocarpa, other Populus spp. and interspecific hybrids of Populus spp.), willow (Salix spp.), pine (Pinus spp.), eucalyptus (Eucalyptus spp.), sorghum (Sorghum spp.) and energycane (Saccharum complex). Additionally, plant models such as Arabidopsis (Arabidopsis thaliana), rice (Ozyza sativa), brachypodium (Brachypodium distachyon), foxtail millet (Setaria italica), tobacco (Nicotiana tabacum) and medicago (Medicago truncatula) that have shorter life-cycles and greater amenability to genomics and biotechnology methods continue to be adopted as references for attaining translational breakthroughs in fundamental science.
The rapid progress in our fundamental understanding and improvement of bioenergy crops in the past decade has included gene-centric as well as pathway- and genome-wide studies. The general approach of undertaking a transcriptomics characterization followed by manipulation of a single gene at a time, has recognized limitations and is being addressed by the community, which is now embracing a systems-wide view. In this review, we summarize the recent progress made in our understanding of plant cell wall biosynthesis and remodelling, assess the major impediments in obtaining a systems-level understanding of cell wall formation and explore the potential opportunities in leveraging systems and synthetic biology approaches to reducing biomass recalcitrance.
Cellulose biosynthesis pathway and recent breakthroughs
Cellulose biosynthesis occurs at the plasma membrane, and various soluble and membrane proteins have been reported to play a role in this pathway (Endler and Persson, 2011). Cellulose synthases, designated as CesAs or CESAs, belonging to the Family 2 glycosyltransferases, are the catalytic enzymes that build beta-glucan chains from UDP-glucose substrate (Delmer, 1999). The CesA-containing functional complexes, CSC, or terminal complexes (localized at nonreducing ends of a growing microfibril) from which a cellulose microfibril extends, have been observed to occur as rosette structures (∼30 nm in diameter) in freeze-fracture electron microscopy of plasma membranes obtained from higher plants (Kimura et al., 1999). Several research reports and reviews have that suggested that factors such as (i) composition, number and arrangement of rosettes, (ii) membrane dynamics, due to action of wall enzymes, changes in lipids in response to developmental cues or environmental stresses, (iii) associated wall proteins KORRIGAN, KOBITO, COBRA (whose molecular functions are still unknown) and (iv) pH [H+ acid growth]/ion [Ca++ levels] changes are determinants of cellulose properties in plant cell walls (Endler and Persson, 2011; Mizrachi et al., 2012). The regulatory and signalling pathways critical to rapid turnover of CesAs and CSCs and cellulose production are not well understood.
The recent discovery of small microtubule-tethered CesA containing compartments (SmaCCs/MASCs) that move by depolymerization of microtubule ends (Crowell et al., 2009; Gutierrez et al., 2009) has provided a working hypothesis on distribution of the Golgi-formed CSC in the plasma membrane (Crowell et al., 2010). A role for other cytoskeleton-associated proteins in cellulose synthesis is also beginning to emerge. For example, disruption of a dynamin-related protein (Collings et al., 2008; Xiong et al., 2010) and a kinesin (Zhong et al., 2002) has been shown to result in altered cellulose synthesis and microfibril orientation. It has also been proposed that cysteine proteases interact with Zn finger domain of CesA and play a role in the turnover and degradation of rosette complexes (Jacob-Wilk et al., 2006).
Based on mutant phenotypic analysis, mutant complementation and domain-swapping experiments, it is proposed that distinct sets of at least three primary or secondary wall-specific CesAs isoforms are essential CSC components (Somerville, 2006; Taylor et al., 2003), where the CesA composition may be nonredundant and the tail region of CesA is important in multisubunit interactions within CSC (Burn et al., 2002; Wang et al., 2006). On the basis of high-resolution live-cell fluorescence microscopy, it was shown that CesA moves along microtubules, thereby possibly affecting the direction of cellulose deposition (Paredez et al., 2006). In a protein interaction screen to identify CSC components, Gu et al. (2010) identified an Armadillo repeat containing protein. Further examination confirmed colocalization with CesAs and direct interaction with microtubule and CesA. The precise constitution of functional CSCs is unclarified. The challenge of extending single subunit visualization techniques (e.g. EM-based immunolocalization or live tracking of protein movement) to all CesA proteins, as well as to enable easy visualization of rosettes multisubunits, is a limiting concern in the field (Itoh et al., 2004).
One of the salient recent developments in the cellulose research field has been the discovery of the structure of the first CesA (Morgan et al., 2013). This pioneering work included crystallographic analysis of a bacterial cellulose synthesis and transport (channelling) protein subunit pair, BcsA-BcsB, with or without substrate addition. The structure of the more complex plant CesA remains elusive although there are several efforts to experimentally or computationally clarify the structure, such as the 3D atomistic simulation of the catalytic domain of cotton CesA (Sethaphong et al., 2013).
The natural evolution of the cellulose research field has steadily yielded valuable insights, yet the pace has been slow relative to the abbreviated timelines, we face in understanding and improvement biomass properties, specifically increasing the content of cellulose. A systems approach offers the potential to identify the molecular and genetic underpinning of the linkages between properties of cellulose synthase and cell wall such as processivity, glucan chain length, degree of polymerization, crystallinity, interactions with other wall components, and sugar release efficiency to facilitate biosystems design of plant cell walls for bioenergy.
Lignin biosynthesis pathway and recent breakthroughs
Lignin, a polyphenolic heteropolymer of monolignols found mainly in secondary walled cells, is to date the plant feature most correlated with recalcitrance of biomass. The abundance, distribution and composition of lignin [i.e. ratio of syringyl (S), guaiacyl (G) and 4-hydroxyphenyl (H) units] is variable across plant species, individuals of a population and even within tissue types of an individual plant. The cross-linkages between lignin and wall polysaccharides confer strength properties to cell wall and plant as a whole, and present a challenge in the form of recalcitrance of biomass to biofuels conversion processes.
Several review articles have provided a detailed view of the lignin biosynthesis pathway in the past (Boerjan et al., 2003; Bonawitz and Chapple, 2010; Zhao and Dixon, 2011). Additionally, the effect of manipulating lignin pathway genes on sugar release efficiency of biomass was first demonstrated in alfalfa (Chen and Dixon, 2007). The vast amount of knowledge gathered for lignin biosynthesis pathway from multiple plant species over the years has generated a consensus model for the pathway (Vanholme et al., 2012b). The present article elaborates on selected recent works (within past 5 years) pertinent to lignin pathway engineering strategies to reduce biomass recalcitrance.
Among the cellulosic feedstock crops, switchgrass, a fast growing perennial grass, has received the considerable attention in the past 5 years. While a switchgrass genome sequencing effort is underway, the polyploid outcrossing nature of the species is a current impediment to precisely identifying genetic basis of biomass traits such as saccharification or sugar release efficiency. Nonetheless, researchers have made tremendous strides by finding creative solutions in the form of homology driven analyses of RNAseq and EST profiles, design of microarrays, developing a cell culture system and achieving impressive transformation efficacy for switchgrass plants (Shen et al., 2013). Candidate genes obtained on the basis of the aforementioned approaches are beginning to be validated as genetically controlling lignin content and S : G ratios. A significant breakthrough with switchgrass transgenic studies of cell wall recalcitrance was the discovery that down-regulation of a single caffeic acid O-methyltransferase, COMT, resulted in changes in lignin properties and a 38% increase in ethanol yield (Fu et al., 2011a). There have been several other reports of transgenic switchgrass characterization involving mis-expression of lignin pathway genes such as cinnamoyl CoA reductase, CCR, (Escamilla-Trevino et al., 2010); cinnamyl alcohol dehydrogenase, CAD (Fu et al., 2011b; Saathoff et al., 2011) and 4-coumarate CoA ligase, 4CL, (Xu et al., 2011).
Lignin engineering efforts are primarily focused on modifications of lignin composition, often with introduction of novel lignin forms in plants (Vanholme et al., 2012a), to reduce cross-linking with wall components resulting in easier extractability, digestibility and recalcitrance of usable wall polysaccharides. Vanholme et al. (2010) and Weng et al. (2010) showed that a combinatorial down-regulation of COMT and overexpression of F5H lead to engineering of cell walls with unusual lignin (>90% benzodiance). Eudes et al. (2012) also successfully engineered novel lignin into Arabidopsis stems by expressing a bacterial hydroxycinnamoyl-CoA hydratase-lyase (HCHL) gene that synthesizes and enhances C6C1 monomers, considered key in reducing lignin polymer length, in cell walls resulting in enhanced saccharification.
Recent studies directed towards a deeper understanding of transcriptome level differences among the collection of Arabidopsis lignin biosynthesis mutants has given rise to an insightful ‘systems biology’ view of impacts of gene perturbation in Arabidopsis (Vanholme et al., 2012b). Such level of systems analysis together with validation of transcriptional nodes and hubs is critical to realizing the potential of synthetic biology in enabling high-precision engineering of biomass properties.
In a seminal effort in higher precision engineering of cell wall properties, C4H gene was expressed under a xylem vessel cell-specific VND6 promoter and concomitantly the NST1 was expressed under the IRX8 promoter in Arabidopsis. Yang et al. (2013) called this an ‘artificial positive feedback loop system’ as the IRX8 promoter is also induced by NST1. This successful demonstration of simultaneous increase in polysaccharide and decrease in lignin in stems of Arabidopsis plants without gross modifications in plant growth is an example of the potential precision engineering holds. The possibilities of extending this gene modification principle to other combinations of genes are exciting.
A new development in the lignin field is the rapid emergence of interest in maximizing the economic value of lignin as a coproduct (Ragauskas et al., 2014). For many biomaterial applications, such as carbon fibre production, unbranched lignin with a S/G ratio >2 is a preferred substrate. However, typical lignin polymers are rather random in orientation. System-wide genetic modification of plant biomass to create lignins that are more linear or restricted in their monomer composition, giving them better properties, appears possible. A recent study reported the natural occurrence of a new type of a lignin polymer, catechyl or C-lignin, in high concentrations (>80%) in the seed coat of vanilla orchid plants (Chen et al., 2012). Furthermore, certain species of cacti exclusively contain C-lignin in seeds (Chen et al. 2013). The C-lignin is interesting as it is an unbranched, homopolymer of catechyl units with abundant benzodioxane units. Similar benzodioxanes have been reported to occur in ccomt and comt mutant plants. Although the genetic underpinnings of C-lignin production are not specifically worked out, it is believed that well-characterized enzymes in the consensus lignin biosynthesis pathway may be important in production of C-lignin.
Hemicellulose biosynthesis and recent breakthroughs
Noncellulosic polysaccharides such as hemicelluloses (xylan, mannan, glucomannan and xyloglucan) and pectin, composed of mixed sugars with branching and potentially crossed with wall proteins, are variable between primary and secondary walls and between plant groups. The subunit components or in some cases, full-length backbones are synthesized in ER–Golgi and secreted via trans-Golgi network to plasma membrane, the site of integration, and ultimately deposited in the cell walls. There are comprehensive reviews of the biosynthesis of noncellulosic wall polysaccharides recently published (Hao and Mohnen, 2014; Pauly et al., 2013; Rennie and Scheller, 2014), and hence this topic is dealt in brevity in the present review.
Briefly, several members of cellulose synthase-like gene family (Csl) have been reported to synthesize mixed-linked sugar backbones (Dhugga, 2012). More recently, progress made through characterization of Arabidopsis mutants for stem structural (e.g. irregular xylem) and chemical features (e.g. NMR analyses of signature hemicellulose linkages), and in vitro enzyme assays have revealed a suit of genes coding for enzymes responsible for synthesis of backbone [e.g. (1-4)-beta xylan oligomer], side chain (e.g. 4-O-methyl GlcA) and moieties (e.g. acetyl). These include genes such as IRX8 (GAUT12), IRX9 (GT43), IRX10 (GT47), IRX14 (GT43), IRX15 (DUF579), PARVUS (GATL1), IRX7/FRA8 (GT47), RWA (REDUCED WALL ACETYLATION) and O-acetyl transferases (Carpita, 2012; Hao and Mohnen, 2014; Pauly et al., 2013). Classic forward and reverse genetics approaches clearly resulted in fundamental insights into the biosynthesis of hemicellulose polymers in the past decade. The diversity of sugars and complexity of biosynthesis processes involved, however, poses a practical challenge in undertaking an immediate systems-level understanding of biogenesis and remodelling of these cell wall components.
Transcription factor-mediated control on biomass recalcitrance-related properties
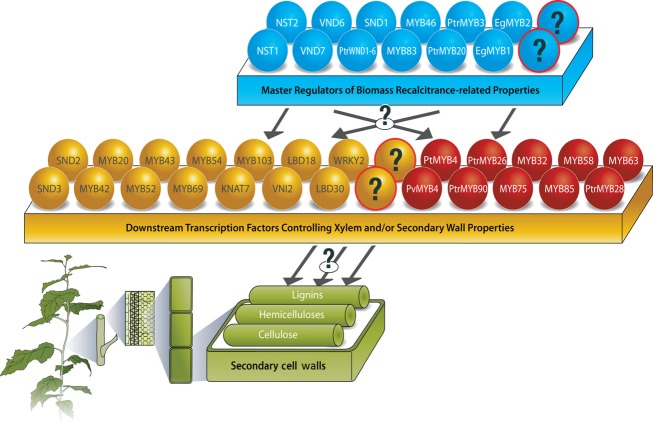
As secondary cell walls of xylem fibres and vessels are the prime components of biomass, alterations in xylem development and secondary cell wall properties will have a significant effect on biomass quality. Transcript profiling experiments that target secondary cell wall biosynthesis across various species have indicated the involvement of specific transcriptional networks. Mutant and transgenic analysis of many transcription factors from these network studies have confirmed their functional role in secondary wall formation (Figure1).
Figure 1.
A conceptual model of transcription factor-mediated control on biomass recalcitrance-related properties such as xylem development, secondary cell wall formation, lignin and other cell wall components. The model is based on current knowledge from bioenergy crop types such as Populus, Pinus, Eucalyptus and Panicum spp. as well as the plant model Arabidopsis thaliana presented in several recent publications (Hussey et al., 2013; Ko et al., 2014; Shen et al., 2012; Wang et al., 2011; Zhong et al., 2010, 2011). Transcription factors (activators or repressors) reported to be control lignin biosynthesis pathway are indicted in red circles. Additional unidentified transcription factors relating to each of these properties are indicated by a ‘?’ symbol embedded within circles of respective colours. The bottom panel of the figure reflects the phenotypic properties of lignocellulosic biomass such as stem development, secondary wall formation under the influence of reported transcription factors.
It has been shown in Arabidopsis that SND1 and its close homologs, NST1 (fibres), NST2, VND6, and VND7 (fibres) (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Yamaguchi et al., 2008; Zhong et al., 2006, 2007), act as ‘master switches’ of secondary wall formation, which in turn regulate genes encoding other transcription factors such as MYB46, MYB83, SND3, MYB103 and KNAT7 (McCarthy et al., 2009; Zhong et al., 2006, 2007) (Figure1). PtrMYB3 and PtrMYB20 have been shown to be involved in secondary cell wall formation in poplar (McCarthy et al., 2010) and MYB85, MYB63, and MYB58 are reported to regulate lignin biosynthesis (Zhong et al., 2008; Zhou et al., 2009). Ko et al. (2009) followed the transcriptome dynamics across a time series of secondary wall formation by generating a dexamethasone-inducible MYB46 secondary wall thickening system in Arabidopsis. Arabidopsis GeneChip microarray and Illumina digital gene expression profiles at 0, 1, 3 and 6 h postinduction and confirmatory in vivo assays showed that AtC3CH14, MYB52 and MYB63 are downstream to MYB46. Furthermore, their work suggests that the C3H-type zinc-finger protein AtC3H14 is possibly another master regulator of genes involved in secondary wall biosynthesis (such as CesA4, CesA7, CesA8, PAL4, CCoAOMT, LAC10). In Arabidopsis, one AP2/EREBP transcriptional factor (SHINE2) has been identified as a key regulator responsible for the increasing of cellulose and decreasing in lignin biosynthesis (Aharoni et al., 2004; Ambavaram et al., 2011). A common set of transcription factors, along with pathway-specific transcription factors, may regulate secondary wall biosynthetic pathways (Ko et al., 2009; Mellerowicz et al., 2001; Mellerowicz and Sundberg 2008).
Mis-expression of these transcription factors has been reported to often result in weak walls, short stature, disease susceptibility, or imbalance in other wall chemicals, suggesting that a robust systems biology model of cell wall changes and a strategy for achieving the desired wall property precisely without compromising other critical attributes is still not available. Detailed knowledge about how cellulose and lignin biosynthetic genes are regulated may provide an opportunity to design the reorganization of plant cell wall compositions.
Bonawitz et al. (2014) recently showed that the dwarf and lignin-deficient phenotype of Arabidopsis ref8 mutant is rescued by disrupting MED5a/REF4 and MED5b/RFR1, which are subunits of a transcriptional co-regulatory Mediator complex. med5a/5b ref8 plants have a wildtype phenotype except that the synthesis of guaiacyl and syringyl is not rescued and a novel p-hydroxyphenyl-enriched lignin is formed, and the saccharification efficiency is enhanced. This work suggests a possible role of Mediator underlying the other dwarf lignin mutant phenotype reports. It also underscores the fact that molecular components of transcriptional machinery and signalling pathways are important to evaluate as potential higher precision targets in biomass improvement efforts.
MyB4, a secondary cell wall transcription factor, has previously been shown to repress lignin biosynthesis in Pinus spp. (Patzlaff et al., 2003) (Figure1). Overexpression of the transcription factor gene, PvMYB4, in switchgrass dramatically improved the yield of cellulosic ethanol yield from switchgrass when compared to nontreated controls or COMT transgenic plants (Shen et al., 2013). These results extend the proven utility of transcriptional regulators into perennials and potential biofeedstocks. PvMYB4-OX switchgrass is a complementary model system for understanding recalcitrance, and provides new germplasm for developing switchgrass cultivars as biomass feedstocks for biofuel production.
Status of collective goal to reduce biomass recalcitrance
As noted in the previous sections, the number of research efforts attempting to improve biomass properties have increased dramatically. In general, the targeting points include the following: key transcriptional factors that control expression of several enzymes, enzymes or transporters involved in nucleotide sugar conversion, and enzymes that generate or modify polysaccharides (Doblin et al., 2014) (Table1).
Table 1.
Recent (within past 5 years) examples of engineering plant cell walls to improve biofuel production
| Species | Engineering approach | Outcome | References |
|---|---|---|---|
| Panicum virgatum | RNAi of Caffeic acid O-methyltransferase | Increase ethanol yield by up to 38% | Fu et al. (2011a) |
| P. virgatum | Down-regulation of Cinnamyl Alcohol Dehydrogenase (CAD) | Improved saccharification efficiency | Fu et al. (2011b), Saathoff et al. (2011) |
| P. virgatum | Overexpression of Myb transcriptional factor PvMYB4 | Increase ethanol yield by 2.6-fold | Shen et al. (2013) |
| Saccharum spp. hybrids | RNAi of Caffeic acid O-methyltransferase | Reduction in total lignin by 6% improved saccharification efficiency by 19%–23% | Jung et al. (2012a) |
| Arabidopsis thaliana | pVND6::C4H+ pIRX8::NST | Increased polysaccharide deposition in the fibre cell | Yang et al. (2013) |
| A. thaliana | Overexpression of bacterial hydroxycinnamoyl-CoA hydratase-lyase | Improve saccharification and reduce lignin polymerization degree | Eudes et al. (2012) |
| P. virgatum | Silencing of 4-coumarate:coenzyme A ligase | Reduce lignin content and increase the efficiency of fermentable sugar | Xu et al. (2011) |
Many emerging technologies in genomics, cell biology and cell wall chemistry have been critical to advances in our understanding of plant cell wall formation. Such techniques include (i) molecular techniques relevant to robust, reliable and rapid single cell genomics, single-molecule detection, protein profiling from very small samples, genetic engineering, and transient and stable transformation, (ii) phenotyping techniques for in situ cell wall measurements include label-free methods for chemical imaging methods such as coherent Raman scattering microscopy (Zeng et al., 2012), 3D time-of-flight secondary-ion mass spectrometry (Jung et al., 2012b), fluorescence tag-based single-molecule tracking of protein and carbohydrate-binding modules for in situ cellulose biosynthesis and cellulose degradation studies (Gu et al., 2010; Liu et al., 2012), and imaging of fluorescent lignin monomers in cell walls (Ding et al., 2012; Tobimatsu et al., 2013). Additional key resources include (iii) appreciably annotated genome sequences from dozens of plant species and hundreds of genotypes; and (iv) standards-adhered data analysis, and reconstructions of transcriptional networks and nodes.
As a general observation, these genomics-based plant improvement efforts typically involve an initial phase of genome-wide ‘omics’ studies (such as transcriptomics, proteomics and metabolomics) and pathway analyses followed by formulation and testing of hypotheses for genetic basis of observed functional feature that simply involves a single candidate gene. The adoption of such an approach of manipulating expression of a single gene under a ubiquitous promoter, while fundamentally informative, has often resulted in pleiotropic and undesirable effects and therefore considered an impediment in the path to rapid and precise improvement of biomass properties.
Despite the extensive efforts, there are few examples shown to be effective in engineering the cell wall components in a predictable manner. This may partly be explained by the (i) failure to identify and target the most effective gene mix or master regulator, (ii) existence of functional endogenous copies of the same or related gene and (iii) lack of knowledge of the best allele- and stoichiometry-related to paralogous or distinct protein family members required in the pathway.
Opportunities, potential and promise of emerging systems and synthetic biology approaches
Precision genome engineering requires advanced knowledge base, informed design approaches and versatile biotechnological tools. With the availability of extensive genome sequence and expression resources, systems biology has emerged as a multidisciplinary research in understanding from interactions of molecules, signal transduction pathways to functional modules of biological systems (Bruggeman and Westerhoff, 2007; Chuang et al., 2010; Sheth and Thaker, 2014).
Systems-level strategies and methodologies will be essential for interpreting and leveraging data/knowledge from different omics platforms (Bruggeman and Westerhoff, 2007; Sheth and Thaker, 2014). In general, top-down and bottom-up methods are two fundamental approaches to coordinate various levels of information in systems biology (Bruggeman and Westerhoff, 2007; Edwards and Thiele, 2013; Klipp et al., 2007; Schneider, 2013). Top-down methods characterize multivariate data as a whole, and perform global analyses, such as principal components analysis (PCA), machine learning and pattern recognition, to fit a priori and post priori models. Bottom-up methods, on the other hand, are designed to build models or test hypotheses by utilizing existing knowledge of network. For example, due to the redundancy of a large laccase gene family in P. trichocarpa, the molecular functions of laccases are difficult to elucidate. Transcriptomics and metabolomics analyses of transgenic P. trichocarpa plants overexpressing Ptr-MIR397a have revealed the post-transcriptional regulation of multiple laccases genes and their roles in the regulation of lignin biosynthesis (Lu et al., 2013). Practically, systems biology, more than simply collecting huge amount of omics data, relies heavily on mathematical modelling and computational simulations to aid the understanding of complex biological systems (Bruggeman and Westerhoff, 2007; Edwards and Thiele, 2013).
Synthetic biology aims to create living systems with designed functions, a distinguishing feature from traditional genetic selection or manipulation. The potential applications of synthetic biology have been extended to many aspects of life sciences from production of new drugs to biofuels (Church et al., 2014). Despite the ambitious scope, recent progress in plants has not reached previous expectations mainly due to a lack of efficient genetic tools (Khalil and Collins, 2010; Lienert et al., 2014). Thus in order to precisely reprogram biological parts to construct new bio-systems with novel functions, building solid foundations of well-defined tools (e.g. DNA synthesis, genome assembly, activation circuits) is the key to the success of synthetic biology (Brophy and Voigt 2014; Khalil and Collins, 2010; Lienert et al., 2014). Many tools have recently been developed for creating multigene constructs, precision genome editing and site-specific gene stacking and removal (Liu et al., 2013).
High-throughput assembly of multigene constructs
To design and optimize the metabolic pathways leading to a desirable production of low recalcitrant biomass feedstock requires transformation of multiple genes. A vector harbouring large DNA molecules with changeable elements provides an opportunity to efficiently deliver multiple genes. For example, the development of multigene vector systems, such as GoldenGate, ePathBricks and BioBrick, allows assembly of several genes in a predictable manner (Appleton et al., 2014; DePaoli et al., 2014; Liu et al., 2013; Sarrion-Perdigones et al., 2013, 2014; Xu and Koffas, 2013). By combining de novo DNA synthesis with seamless assembly methods, it is now feasible to construct multigene vectors (Kosuri and Church, 2014).
Precision genome editing
As an emerging discipline, precise genome editing creates DNA deletion, mutation or integration at a specific site in the host genome, and represents a profound potential for plant biotechnology. The engineering of zinc-finger nucleases or TALE nucleases could also target any genomic sequence specifically, and generate DNA breaks (Sanjana et al., 2012). This technology has been successfully used in many plant species to generate targeted mutants (Osakabe et al., 2010; Shan et al., 2013a; Shukla et al., 2009). Similarly, a recent method called clustered regularly interspaced short palindromic repeats (CRISPRs) generate DNA mutation using precursor RNA guided mechanism to edit genomic sequences (Haurwitz et al., 2010; Liu et al., 2013). The CRISPR-Cas9 system is best characterized between the different types of CRISPRs, and successful examples have been reported in tobacco, Arabidopsis and rice (Chen and Gao, 2014; Li et al., 2013; Shan et al., 2013b).
Site-specific gene stacking and removal
As opposed to generating mutation in targeted regions of a genome, site-specific gene stacking and removal offers the ability of controlling the transgene insertion and removal. Reuse of selectable markers for multiple rounds of transformation and integration of transgene into specific target sites can reduce the risks of unintended consequences (Yau and Stewart, 2013). The application of recombinases systems in plants has been reported. For example, combining the expression of CRE recombinase and ϕC31 integrases in Arabidopsis, De Paepe et al. (2013) have shown about 9% of transformants generated site-specific integration of T-DNA and removal of the resident selectable marker.
RNA-editing
A promising new technology is based on pentatricopeptide repeat (PPR) proteins known for their affinity to bind RNA rather DNA molecules (Yagi et al., 2014). The nucleotide sequence specificity of PPR proteins for RNA binding and editing leads itself amenable to design of customizable enzymes suited to precision RNA engineering.
Here, we present a conceptual scheme of integration of systems biology and synthetic biology for understanding the molecular basis of cell wall biosynthesis and its regulation (Figure2). Firstly, from a top-down view, the various omics data are processed and analysed by common platform, and further integrated to form mathematical models; from a bottom-up view, to test hypotheses or models, individual perturbations are designed to generate new data sets for analysis. Secondly, through the systems biology iterations, meaningful models or genetic circuits would guide the design of synthetic biology to perform high-precision engineering tasks (Figure2). Specifically, systems biology studies could provide biological ‘parts’ (e.g. genes, promoters) and framework (e.g. gene network, spatial and temporal expression, metabolic flux and balance) for synthetic biology approaches such as precision genome editing and multiple gene transfer to reduce biomass recalcitrance.
Figure 2.
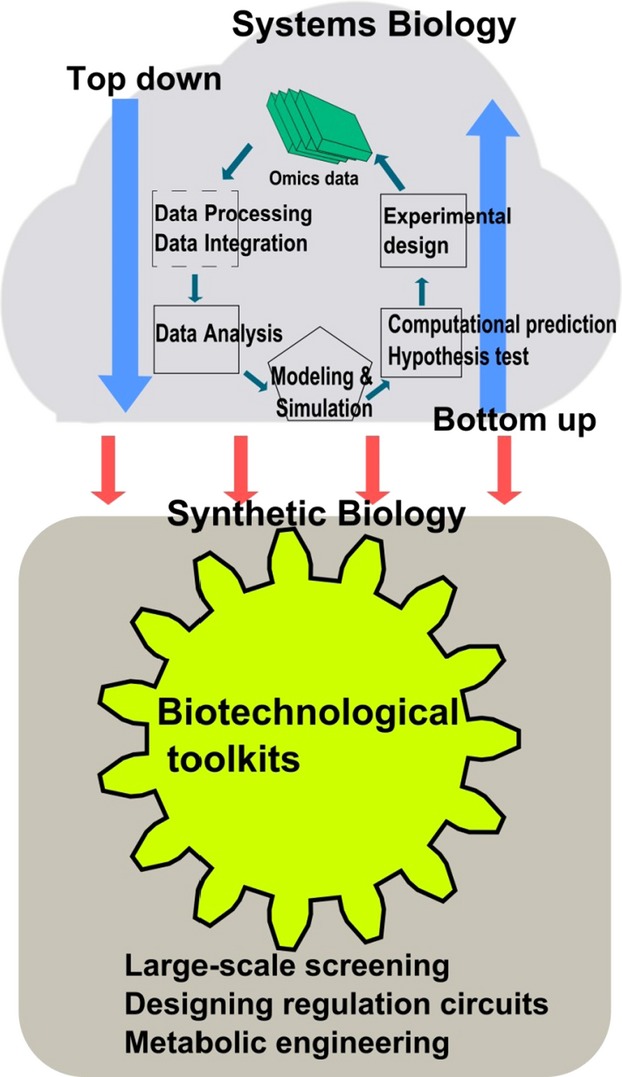
A conceptual scheme describing the integration of systems and synthetic biology. In the upper panel, a ‘cloud’ of systems biology integrates top-down and bottom-up approaches to exploit the omics data. The knowledge from systems biology can guide the design of precision engineering for synthetic biology. The lower panel highlights that the key element of synthetic biology is to develop versatile biotechnological tools for its various applications.
In addition to the emerging tools of genetic engineering mentioned above, methods also exist for ‘rational’ engineering of enzyme structure. There are two main approaches to improving or creating new protein functions, rational design of proteins and directed evolution. Put simply, rational design is the is the generation of new designer proteins based on prior knowledge, and directed evolution is the selection of new functions on the basis of screening of randomly generated variant proteins.
Enzymes of cellulase or glucohydrolase family have been employed in ‘rational design’ strategies to improve the catalytic efficiency during biomass conversion, (Buckeridge and Gustavo, 2011). In general, site-directed mutagenesis and random mutagenesis are two common strategies by which modifications of key residues of proteins of interest. Through three-dimensional structure and molecular dynamics simulation analyses of normal and mutated protein, the knowledge of biochemical importance of key residues would guide the rational design to alter protein structure (McLaughlin et al., 2012). For example, based on the structures of class II cellobiohydrolases, Heinzelman et al. (2009) screened over 6000 of chimeras and identified three thermostable fungal chimeric cellulases.
Conclusion and future perspective
It is an exciting time for plant biologists addressing grand challenge questions. The vast amount of knowledge being garnered on the basis of widely available sequencing, omics profiling, genome engineering and higher resolution phenotyping technologies is moving the field towards the targeted goal of designer plants with improved biomass properties.
It is clear that the recalcitrance of lignocellulosic biomass is determined by plant properties of growth, xylem development and patterning, secondary cell wall formation and cell wall composition. However, the complex sets of biosynthetic and remodelling pathways, and their interactions with transcription factors and signalling effectors underlying recalcitrance properties are not fully understood. Therefore, there is an immediate need to fill the critical knowledge gaps using advanced systems biology studies. While single gene manipulations may have measureable effects on target phenotype, it is also clear that synthetic biology approaches provide an unprecedented opportunity to engineer the cell wall composition, minimize off-target effects and reduce biomass recalcitrance for bioenergy production. The example of ref8 mutant study in A. thaliana has demonstrated a two-step strategy of genetic engineering wherein lignin composition is altered, saccharification efficiency is improved and negative growth effects are restored. A timely convergence of systems and synthetic biology is necessary to address the challenging goal of plant cell wall modification for efficient conversion of biomass to biofuels.
Beyond improved cell wall characteristics, an ideotypic bioenergy crop will also require improved plant performance resulting in high productivity under conditions of suboptimal soil structure, nutrient composition, temperature, moisture and resiliency to various biotic stresses. The systems biology and synthetic biology tools, barriers and opportunities discussed in the context of feedstock improvement for cell wall properties, are expected to be largely universal to research in other areas of plant improvement as well as studies of other plant-derived biofuels and biomaterials.
Acknowledgments
The authors would like to thank Dr. Jerry Tuskan for his valuable comments on this manuscript. This work was funded by the U.S. Department of Energy (DOE) BioEnergy Science Center project. The BioEnergy Science Center is a Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science. ORNL is managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract Number DE–AC05–00OR22725.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambavaram MMR, Krishnan A, Trijatmiko KR, Pereira A. Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 2011;155:916–931. doi: 10.1104/pp.110.168641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleton E, Tao J, Haddock T, Densmore D. Interactive assembly algorithms for molecular cloning. Nat. Methods. 2014;6:657–662. doi: 10.1038/nmeth.2939. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 2010;44:337–363. doi: 10.1146/annurev-genet-102209-163508. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Kim JI, Tobimatsu Y, Ciesielski PN, Anderson NA, Ximenes E, Maeda J, Ralph J, Donohoe BS, Ladisch M, Chapple C. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature. 2014;509:376–380. doi: 10.1038/nature13084. [DOI] [PubMed] [Google Scholar]
- Brophy JA, Voigt CA. Principles of genetic circuit design. Nat. Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman FJ, Westerhoff HV. The nature of systems biology. Trends Microbiol. 2007;15:45–50. doi: 10.1016/j.tim.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Buckeridge MSG, Gustavo H. In: Routes to Cellulosic Ethanol. Buckeridge MSG, Gustavo H, editors. New York, NY: Springer; 2011. [Google Scholar]
- Burn JE, Hocart CH, Birch RJ, Cork AC, Williamson RE. Functional analysis of the cellulose synthase genes CesA1, CesA2, and CesA3 in Arabidopsis. Plant Physiol. 2002;129:797–807. doi: 10.1104/pp.010931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC. Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Curr. Opin. Biotechol. 2012;23:330–337. doi: 10.1016/j.copbio.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Chen F, Dixon RA. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007;25:759–761. doi: 10.1038/nbt1316. [DOI] [PubMed] [Google Scholar]
- Chen K, Gao C. Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 2014;33:575–583. doi: 10.1007/s00299-013-1539-6. [DOI] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Havkin-Frenkel D, Dixon RA, Ralph J. A polymer of caffeyl alcohol in plant seeds. Proc. Natl Acad. Sci. USA. 2012;109:1772–1777. doi: 10.1073/pnas.1120992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tobimatsu Y, Jackson L, Nakashima J, Ralph J, Dixon RA. Novel seed coat lignins in the Cactaceae: structure, distribution and implications for the evolution of lignin diversity. Plant J. 2013;73:201–211. doi: 10.1111/tpj.12012. [DOI] [PubMed] [Google Scholar]
- Chuang HY, Hofree M, Ideker T. A decade of systems biology. Annu. Rev. Cell Dev. Biol. 2010;26:721–744. doi: 10.1146/annurev-cellbio-100109-104122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Elowitz MB, Smolke CD, Voigt CA, Weiss R. Realizing the potential of synthetic biology. Nat. Rev. Mol. Cell Biol. 2014;15:289–294. doi: 10.1038/nrm3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DA, Gebbie LK, Howles PA, Hurley UA, Birch RJ, Cork AH, Hocart CH, Arioli T, Williamson RE. Arabidopsis dynamin-like protein DRP1A: a null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J. Exp. Bot. 2008;59:361–376. doi: 10.1093/jxb/erm324. [DOI] [PubMed] [Google Scholar]
- Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof YD, Schumacher K, Gonneau M, Hofte H, Vernhettes S. Pausing of golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell. 2009;21:1141–1154. doi: 10.1105/tpc.108.065334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell EF, Gonneau M, Stierhof YD, Hofte H, Vernhettes S. Regulated trafficking of cellulose synthases. Curr. Opin. Plant Biol. 2010;13:700–705. doi: 10.1016/j.pbi.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Davison BH, Drescher SR, Tuskan GA, Davis MF, Nghiem NP. Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl. Biochem. Biotechol. 2006;130:427–435. doi: 10.1385/abab:130:1:427. [DOI] [PubMed] [Google Scholar]
- Davison BH, Parks J, Davis MF, Donohoe BS. Plant cell walls: basics of structure, chemistry, accessibility and the influence on conversion. In: Wyman CE, editor. Aqueous Pretreatment of Plant Biomass for Biological and Chemical Conversion to Fuels and Chemicals. Hoboken, NJ: John Wiley & Sons Ltd; 2013. pp. 23–37. [Google Scholar]
- De Paepe A, De Buck S, Nolf J, Van Lerberge E, Depicker A. Site-specific T-DNA integration in Arabidopsis thaliana mediated by the combined action of CRE recombinase and varphiC31 integrase. Plant J. 2013;75:172–184. doi: 10.1111/tpj.12202. [DOI] [PubMed] [Google Scholar]
- Delmer DP. Cellulose biosynthesis: exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:245–276. doi: 10.1146/annurev.arplant.50.1.245. [DOI] [PubMed] [Google Scholar]
- DePaoli HC, Borland AM, Tuskan GA, Cushman JC, Yang X. Synthetic biology as it relates to CAM photosynthesis: challenges and opportunities. J. Exp. Bot. 2014;65:3381–3393. doi: 10.1093/jxb/eru038. [DOI] [PubMed] [Google Scholar]
- Dhugga KS. Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry. 2012;74:8–19. doi: 10.1016/j.phytochem.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Ding SY, Liu YS, Zeng YN, Himmel ME, Baker JO, Bayer EA. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science. 2012;338:1055–1060. doi: 10.1126/science.1227491. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Johnson KL, Humphries J, Newbigin EJ, Bacic A. Are designer plant cell walls a realistic aspiration or will the plasticity of the plant's metabolism win out? Curr. Opin. Biotechol. 2014;26:108–114. doi: 10.1016/j.copbio.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Edwards LM, Thiele I. Applying systems biology methods to the study of human physiology in extreme environments. Extrem. Physiol. Med. 2013;2:8. doi: 10.1186/2046-7648-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Persson S. Cellulose synthases and synthesis in Arabidopsis. Mol. Plant. 2011;4:199–211. doi: 10.1093/mp/ssq079. [DOI] [PubMed] [Google Scholar]
- Escamilla-Trevino LL, Shen H, Uppalapati SR, Ray T, Tang YH, Hernandez T, Yin YB, Xu Y, Dixon RA. Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol. 2010;185:143–155. doi: 10.1111/j.1469-8137.2009.03018.x. [DOI] [PubMed] [Google Scholar]
- Eudes A, George A, Mukerjee P, Kim JS, Pollet B, Benke PI, Yang F, Mitra P, Sun L, Cetinkol OP, Chabout S, Mouille G, Soubigou-Taconnat L, Balzergue S, Singh S, Holmes BM, Mukhopadhyay A, Keasling JD, Simmons BA, Lapierre C, Ralph J, Loque D. Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol. J. 2012;10:609–620. doi: 10.1111/j.1467-7652.2012.00692.x. [DOI] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M, Jr, Chen F, Foston M, Ragauskas A, Bouton J, Dixon RA, Wang ZY. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl Acad. Sci. USA. 2011a;108:3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CX, Xiao XR, Xi YJ, Ge YX, Chen F, Bouton J, Dixon RA, Wang ZY. Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenergy Res. 2011b;4:153–164. [Google Scholar]
- Gu Y, Kaplinsky N, Bringmann M, Cobb A, Carroll A, Sampathkumar A, Baskin TI, Persson S, Somerville CR. Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl Acad. Sci. USA. 2010;107:12866–12871. doi: 10.1073/pnas.1007092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez R, Lindeboom JJ, Paredez AR, Emons AMC, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
- Hao ZY, Mohnen D. A review of xylan and lignin biosynthesis: foundation for studying Arabidopsis irregular xylem mutants with pleiotropic phenotypes. Crit. Rev. Biochem. Mol. Biol. 2014;49:212–241. doi: 10.3109/10409238.2014.889651. [DOI] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzelman P, Snow CD, Wu I, Nguyen C, Villalobos A, Govindarajan S, Minshull J, Arnold FH. A family of thermostable fungal cellulases created by structure-guided recombination. Proc. Natl Acad. Sci. USA. 2009;106:5610–5615. doi: 10.1073/pnas.0901417106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- Hussey SG, Mizrachi E, Creux NM, Myburg AA. Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 2013;4:325. doi: 10.3389/fpls.2013.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Kimura S, Brown RM. Theoretical considerations of immunogold labeling of cellulose synthesizing terminal complexes. Cellulose. 2004;11:385–394. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob-Wilk D, Kurek I, Hogan P, Delmer DP. The cotton fiber zinc-binding domain of cellulose synthase A1 from Gossypium hirsutum displays rapid turnover in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2006;103:12191–12196. doi: 10.1073/pnas.0605098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Fouad WM, Vermerris W, Gallo M, Altpeter F. RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 2012a;10:1067–1076. doi: 10.1111/j.1467-7652.2012.00734.x. [DOI] [PubMed] [Google Scholar]
- Jung S, Foston M, Kalluri UC, Tuskan GA, Ragauskas AJ. 3D chemical image using TOF-SIMS revealing the biopolymer component spatial and lateral distributions in biomass. Angew. Chem. Int. Ed. Engl. 2012b;51:12005–12008. doi: 10.1002/anie.201205243. [DOI] [PubMed] [Google Scholar]
- Kalluri UC, Keller M. Bioenergy research: a new paradigm in multidisciplinary research. J. R. Soc. Interface. 2010;7:1391–1401. doi: 10.1098/rsif.2009.0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Laosinchai W, Itoh T, Cui XJ, Linder CR, Brown RM. Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell. 1999;11:2075–2085. doi: 10.1105/tpc.11.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klipp E, Liebermeister W, Helbig A, Kowald A, Schaber J. Systems biology standards – the community speaks. Nat. Biotechnol. 2007;25:390–391. doi: 10.1038/nbt0407-390. [DOI] [PubMed] [Google Scholar]
- Ko JH, Kim WC, Han KH. Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 2009;60:649–665. doi: 10.1111/j.1365-313X.2009.03989.x. [DOI] [PubMed] [Google Scholar]
- Ko JH, Jeon HW, Kim WC, Kim JY, Han KH. The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 2014 doi: 10.1093/aob/mcu126. doi: 10.1093/aob/mcu126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat. Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Lohmueller JJ, Garg A, Silver PA. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol. 2014;15:95–107. doi: 10.1038/nrm3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YS, Ding SY, Himmel ME. Single-molecule tracking of carbohydrate-binding modules on cellulose using fluorescence microscopy. Methods Mol. Biol. 2012;908:129–140. doi: 10.1007/978-1-61779-956-3_13. [DOI] [PubMed] [Google Scholar]
- Liu W, Yuan JS, Stewart CN., Jr Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 2013;14:781–793. doi: 10.1038/nrg3583. [DOI] [PubMed] [Google Scholar]
- Lu SF, Li QZ, Wei HR, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song JY, Sun YH, Yuan LC, Yeh TF, Peszlen I, Ralph J, Sederoff RR, Chiang VL. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc. Natl Acad. Sci. USA. 2013;110:10848–10853. doi: 10.1073/pnas.1308936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann MC, Carpita NC. Designing the deconstruction of plant cell walls. Curr. Opin. Plant Biol. 2008;11:314–320. doi: 10.1016/j.pbi.2008.04.001. [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Ye ZH. MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 2009;50:1950–1964. doi: 10.1093/pcp/pcp139. [DOI] [PubMed] [Google Scholar]
- McCarthy RL, Zhong RQ, Fowler S, Lyskowski D, Piyasena H, Carleton K, Spicer C, Ye ZH. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 2010;51:1084–1090. doi: 10.1093/pcp/pcq064. [DOI] [PubMed] [Google Scholar]
- McLaughlin RN, Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. The spatial architecture of protein function and adaptation. Nature. 2012;491:138–142. doi: 10.1038/nature11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellerowicz EJ, Sundberg B. Wood cell walls: biosynthesis, developmental dynamics and their implications for wood properties. Curr. Opin. Plant Biol. 2008;11:293–300. doi: 10.1016/j.pbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Baucher M, Sundberg B, Boerjan W. Unravelling cell wall formation in the woody dicot stem. Plant Mol. Biol. 2001;47:239–274. [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M. The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 2005;17:2993–3006. doi: 10.1105/tpc.105.036004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M. NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell. 2007;19:270–280. doi: 10.1105/tpc.106.047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrachi E, Mansfield SD, Myburg AA. Cellulose factories: advancing bioenergy production from forest trees. New Phytol. 2012;194:54–62. doi: 10.1111/j.1469-8137.2011.03971.x. [DOI] [PubMed] [Google Scholar]
- Mohnen D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008;11:266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Morgan JLW, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493:181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe K, Osakabe Y, Toki S. Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2010;107:12034–12039. doi: 10.1073/pnas.1000234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science. 2006;312:1491–1495. doi: 10.1126/science.1126551. [DOI] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, Campbell MM. Characterisation of a pine MYB that regulates lignification. Plant J. 2003;36:743–754. doi: 10.1046/j.1365-313x.2003.01916.x. [DOI] [PubMed] [Google Scholar]
- Pauly M, Gille S, Liu LF, Mansoori N, de Souza A, Schultink A, Xiong GY. Hemicellulose biosynthesis. Planta. 2013;238:627–642. doi: 10.1007/s00425-013-1921-1. [DOI] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, Langan P, Naskar AK, Saddler JN, Tschaplinski TJ, Tuskan GA, Wyman CE. Lignin valorization: improving lignin processing in the biorefinery. Science. 2014;344:709. doi: 10.1126/science.1246843. DOI: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- Rennie EA, Scheller HV. Xylan biosynthesis. Curr. Opin. Biotechol. 2014;26:100–107. doi: 10.1016/j.copbio.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Saathoff AJ, Sarath G, Chow EK, Dien BS, Tobias CM. Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE. 2011;6:e16416. doi: 10.1371/journal.pone.0016416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng GP, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Vazquez-Vilar M, Palaci J, Castelijns B, Forment J, Ziarsolo P, Blanca J, Granell A, Orzaez D. GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013;162:1618–1631. doi: 10.1104/pp.113.217661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrion-Perdigones A, Palaci J, Granell A, Orzaez D. Design and construction of multigenic constructs for plant biotechnology using the GoldenBraid cloning strategy. Methods Mol. Biol. 2014;1116:133–151. doi: 10.1007/978-1-62703-764-8_10. [DOI] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. Hemicelluloses. Annu. Rev. Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Schmidt M. Synthetic Biology: Industrial and Environmental Applications. Weinheim, Germany: Wiley-Blackwell; 2012. pp. 1–67. 3rd edn. [Google Scholar]
- Schneider MV. Defining systems biology: a brief overview of the term and field. Methods Mol. Biol. 2013;1021:1–11. doi: 10.1007/978-1-62703-450-0_1. [DOI] [PubMed] [Google Scholar]
- Sethaphong L, Haigler CH, Kubicki JD, Zimmer J, Bonetta D, DeBolt S, Yingling YG. Tertiary model of a plant cellulose synthase. Proc. Natl Acad. Sci. USA. 2013;110:7512–7517. doi: 10.1073/pnas.1301027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, Zhang K, Liu J, Voytas DF, Zheng X, Zhang Y, Gao C. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant. 2013a;6:1365–1368. doi: 10.1093/mp/sss162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013b;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shen H, He XZ, Poovaiah CR, Wuddineh WA, Ma JY, Mann DGJ, Wang HZ, Jackson L, Tang YH, Stewart CN, Chen F, Dixon RA. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 2012;193:121–136. doi: 10.1111/j.1469-8137.2011.03922.x. [DOI] [PubMed] [Google Scholar]
- Shen H, Poovaiah CR, Ziebell A, Tschaplinski TJ, Pattathil S, Gjersing E, Engle NL, Katahira R, Pu Y, Sykes R, Chen F, Ragauskas AJ, Mielenz JR, Hahn MG, Davis M, Stewart CN, Jr, Dixon RA. Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol. Biofuels. 2013;6:71. doi: 10.1186/1754-6834-6-71. doi: 10.1186/1754-6834-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth BP, Thaker VS. Plant systems biology: insights, advances and challenges. Planta. 2014;240:33–54. doi: 10.1007/s00425-014-2059-5. [DOI] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, Choi VM, Rock JM, Wu YY, Katibah GE, Zhifang G, McCaskill D, Simpson MA, Blakeslee B, Greenwalt SA, Butler HJ, Hinkley SJ, Zhang L, Rebar EJ, Gregory PD, Urnov FD. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- Somerville C. Cellulose synthesis in higher plants. Annu. Rev. Cell Dev. Biol. 2006;22:53–78. doi: 10.1146/annurev.cellbio.22.022206.160206. [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl Acad. Sci. USA. 2003;100:1450–1455. doi: 10.1073/pnas.0337628100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W, Chapple C, Ralph J. Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J. 2013;76:357–366. doi: 10.1111/tpj.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu FC, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, Rohde A, Morreel K, Boerjan W. Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J. 2010;64:885–897. doi: 10.1111/j.1365-313X.2010.04353.x. [DOI] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W. Metabolic engineering of novel lignin in biomass crops. New Phytol. 2012a;196:978–1000. doi: 10.1111/j.1469-8137.2012.04337.x. [DOI] [PubMed] [Google Scholar]
- Vanholme R, Storme V, Vanholme B, Sundin L, Christensen JH, Goeminne G, Halpin C, Rohde A, Morreel K, Boerjan W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell. 2012b;24:3506–3529. doi: 10.1105/tpc.112.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Howles PA, Cork AH, Birch RJ, Williamson RE. Chimeric proteins suggest that the catalytic and/or C-terminal domains give CesA1 and CesA3 access to their specific sites in the cellulose synthase of primary walls. Plant Physiol. 2006;142:685–695. doi: 10.1104/pp.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HZ, Zhao Q, Chen F, Wang MY, Dixon RA. NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 2011;68:1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x. [DOI] [PubMed] [Google Scholar]
- Weng JK, Mo HP, Chapple C. Over-expression of F5H in COMT-deficient Arabidopsis leads to enrichment of an unusual lignin and disruption of pollen wall formation. Plant J. 2010;64:898–911. doi: 10.1111/j.1365-313X.2010.04391.x. [DOI] [PubMed] [Google Scholar]
- Xiong GY, Li R, Qian QA, Song XQ, Liu XL, Yu YC, Zeng DL, Wan JM, Li JY, Zhou YH. The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J. 2010;64:56–70. doi: 10.1111/j.1365-313X.2010.04308.x. [DOI] [PubMed] [Google Scholar]
- Xu P, Koffas MA. Assembly of multi-gene pathways and combinatorial pathway libraries through ePathBrick vectors. Methods Mol. Biol. 2013;1073:107–129. doi: 10.1007/978-1-62703-625-2_10. [DOI] [PubMed] [Google Scholar]
- Xu B, Escamilla-Trevino LL, Sathitsuksanoh N, Shen ZX, Shen H, Zhang YHP, Dixon RA, Zhao BY. Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 2011;192:611–625. doi: 10.1111/j.1469-8137.2011.03830.x. [DOI] [PubMed] [Google Scholar]
- Yagi Y, Nakamura T, Small I. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 2014;78:772–782. doi: 10.1111/tpj.12377. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Kubo M, Fukuda H, Demura T. Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 2008;55:652–664. doi: 10.1111/j.1365-313X.2008.03533.x. [DOI] [PubMed] [Google Scholar]
- Yang F, Mitra P, Zhang L, Prak L, Verhertbruggen Y, Kim JS, Sun L, Zheng K, Tang K, Auer M, Scheller HV, Loque D. Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 2013;11:325–335. doi: 10.1111/pbi.12016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau YY, Stewart CN. Less is more: strategies to remove marker genes from transgenic plants. BMC Biotechnol. 2013;13:36. doi: 10.1186/1472-6750-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Himmel ME, Ding SY. Coherent Raman microscopy analysis of plant cell walls. Methods Mol. Biol. 2012;908:49–60. doi: 10.1007/978-1-61779-956-3_5. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Dixon RA. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 2011;16:227–233. doi: 10.1016/j.tplants.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Zhong RQ, Burk DH, Morrison WH, Ye ZH. A kinesin-like protein is essential for oriented deposition of cellulose microfibrils and cell wall strength. Plant Cell. 2002;14:3101–3117. doi: 10.1105/tpc.005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell. 2006;18:3158–3170. doi: 10.1105/tpc.106.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH. The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell. 2007;19:2776–2792. doi: 10.1105/tpc.107.053678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH. Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 2010;15:625–632. doi: 10.1016/j.tplants.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Lee C, Ye ZH. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011;157:1452–1468. doi: 10.1104/pp.111.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lee C, Zhong R, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21:248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]