Abstract
Phosphocitrate (PC) inhibited calcium crystal-associated osteoarthritis (OA) in Hartley guinea pigs. However, the molecular mechanisms remain elusive. This study sought to determine PC targeted genes and the expression of select PC targeted genes in OA menisci to test hypothesis that PC exerts its disease modifying activity in part by reversing abnormal expressions of genes involved in OA. We found that PC downregulated the expression of numerous genes classified in immune response, inflammatory response, and angiogenesis, including chemokine (C-C motif) ligand 5, Fc fragment of IgG, low affinity IIIb receptor (FCGR3B), and leukocyte immunoglobulin-like receptor, subfamily B member 3 (LILRB3). In contrast, PC upregulated the expression of many genes classified in skeletal development, including collagen type II alpha1, fibroblast growth factor receptor 3 (FGFR3), and SRY- (sex determining region Y-) box 9 (SOX-9). Immunohistochemical examinations revealed higher levels of FCGR3B and LILRB3 and lower level of SOX-9 in OA menisci. These findings indicate that OA is a disease associated with immune system activation and decreased expression of SOX-9 gene in OA menisci. PC exerts its disease modifying activity on OA, at least in part, by targeting immune system activation and the production of extracellular matrix and selecting chondroprotective proteins.
1. Introduction
Osteoarthritis (OA) is one of the most prevalent causes of disability in the aging population and has enormous economic and social consequences. However, existing nonsurgical treatment options only provide symptomatic relief but have no effect on the progression of the disease. The lack of progress in the development of structural disease-modifying drugs for OA therapy is largely due to our limited understanding of the pathogenesis of OA and insufficient knowledge regarding the molecular targets or key OA disease genes for therapeutic intervention.
OA is not merely an articular cartilage disease, but a disease of the whole joint. An important local factor to the health of the knee joints is the structural integrity and biochemical properties of the knee meniscus. Knee meniscus is a specialized tissue that plays a vital role in load transmission, shock absorption, and joint stability. In recent years there has been a dramatic advance in our understanding of the integral role of the meniscus for the knee functions and the consequences of meniscal abnormality in cartilage degeneration. Studies found that meniscal degeneration is a general feature of OA [1, 2]; meniscal lesions at baseline were more common in the knees that developed OA than in the knees that did not develop OA [3] and that OA meniscal cells displayed a distinct gene expression profile different from normal meniscal cells [4]. These findings indicate that meniscal changes or abnormalities are involved in the OA disease process. The involvement of meniscal changes or abnormalities in the OA disease process has also been highlighted by recent findings that meniscal extrusion, vascular penetration (angiogenesis), and calcification are associated with cartilage degeneration and subchondral lesions in OA [5–7]. Meniscal abnormalities such as meniscal degeneration, inflammation, and angiogenesis may represent as new targets for the development of disease-modifying drugs for OA therapy, especially for a subgroup of OA patients who develop severe meniscal lesions before developing severe cartilage degeneration [8, 9].
Phosphocitrate (PC), a potent calcification inhibitor, is a naturally occurring compound originally identified in rat liver mitochondrial extract [10, 11]. PC prevented soft tissue calcification and inhibited calcium crystal-induced mitogenesis, crystal-induced expression of matrix metalloproteinases (MMPs), and crystal-induced cell death [12–15]. In Hartley guinea pig model of crystal-associated OA, PC inhibited meniscal calcification and reduced the severity of cartilage degeneration [16]. These findings provide support for the notion that calcification inhibitors are potentially disease modifying drugs for crystal-associated OA therapy. It is believed that PC exerts its disease modifying activity by inhibiting the formation of articular calcium crystals and the detrimental interaction between the crystals and cells [17]. However, two studies found that bisphosphonates, which are also potent calcification inhibitors, failed to inhibit cartilage degeneration in animal models of OA, including the Hartley guinea pig model of crystal-associated OA [18, 19], raising doubts as to whether calcification inhibitors are potentially disease-modifying drugs for OA therapy. An alternative mechanism underlying the disease modifying activity of PC may be present.
We previously reported that PC downregulated the expression of many genes classified in inflammatory response and angiogenesis in OA fibroblast-like synoviocytes (FLSs) and OA meniscal cells in the absence of calcium crystals [20, 21]. These findings suggest that the molecular mechanism underlying the disease-modifying activity of PC is more complicated than originally thought. In this study, we sought to further investigate the gene expression-modulating activity of PC and determine the expressions of select PC-targeted genes in menisci derived from OA patients. The hypothesis to be tested is that PC exerts its disease modifying activity, at least in part, by modulating the abnormal expressions of genes involved in the OA disease process. The information gained is not only important for a better understanding of the molecular mechanisms underlying the disease modifying activity of PC but may also valuable for the identification of disease candidate genes involved in the OA disease process.
2. Materials and Methods
2.1. Materials
Dulbecco's modified eagle medium (DMEM), fetal bovine serum (FBS), stock antibiotic/antimycotic mixture are products of Invitrogen (Carlsbad, CA, USA). Superfrost-Plus microscope slides and neutral buffered formalin (10%) were obtained from Allegiance Inc. (McGaw Park, IL, USA). PC was prepared as described [22]. Antibodies specific to Fc fragment of IgG, low affinity IIIb receptor (FCGR3B), SRY (sex determining region Y)-box 9 (Sox-9), and fibroblast growth factor receptor 3 (FGFR-3) were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). Antibody specific to leukocyte immunoglobulin-like receptor, subfamily B member 3 (LILRB3) was obtained from Lifespan Biosciences (Seattle, WA, USA).
2.2. Meniscal Explant Culture and RNA Extraction
OA meniscal tissue specimens were minced into small pieces and cultured in a six well-cluster plate (350 mg per well) at 37°C in DMEM containing 1% FBS and 0.5% antibiotic/antimycotic solution. Twenty-four hours later, the medium in the top three wells was replaced with DMEM containing 1% FBS and 1 mM of PC and the medium in the bottom three wells was replaced with DMEM containing 1% FBS without PC as control. Every three days, the medium was changed. On the eighth day, the medium was changed again. Twenty-four hours later, the meniscal explants were collected, snap-freezed, and stored in −70°C freezer until use. Total RNA was extracted from these snap-freezed meniscal explants using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and purified using Oligotex kit (Qiagen, Valencia, CA, USA). These RNA samples were used for microarray analysis.
Meniscal tissue specimens were collected with the approval of the authors' Institutional Review Board from end-stage OA patients undergoing knee joint replacement surgery and osteosarcoma patients undergoing lower limb amputation surgery at Carolinas Medical Center. The need for informed consent was waived because these meniscal specimens were surgical waste and no private patient information was collected. Meniscal specimens were collected in sterilized containers filled with tissue culture medium and transported to the laboratory from operating room using an ice box.
2.3. Microarray
RNA samples extracted from two independent experiments were used for microarray analysis. Briefly, double stranded DNA was synthesized using SuperScript double stranded cDNA synthesis kit using these RNA samples (Invitrogen, San Diego, CA, USA). The DNA product was purified using GeneChip sample cleanup module (Affymetrix, Santa Clara, CA, USA). cRNA was synthesized and biotin labeled using BioArray high yield RNA transcript labeling kit (Enzo Life Sciences, Farmingdale, NY, USA). The cRNA product was purified using GeneChip sample cleanup module and subsequently chemically fragmented. The fragmented and biotinylated cRNA was hybridized to HG-U133_Plus_2 gene chip using Affymetrix Fluidics Station 400 (Affymetrix, Santa Clara, CA, USA). The fluorescent signal was quantified during two scans by Agilent Gene Array Scanner G2500A (Agilent Technologies, Palo Alto, CA) and GeneChip operating Software (Affymetrix, Santa Clara, CA, USA). Genesifter software (VizX Labs, Seattle, WA, USA) was used for the analysis of differential gene expression and gene ontology.
2.4. Real-Time RT-PCR
Briefly, cDNA was synthesized using TaqMan Reverse Transcription Reagents (Applied Biosystems, University Park, IL, USA) using the RNA samples described. Quantification of relative transcript levels for selected genes and the housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was performed using ABI7000 Real Time PCR system (Applied Biosystems, University Park, IL, USA). TaqMan Gene Expression assay (Applied Biosystems, University Park, IL, USA) was used. CDNA samples were amplified with an initial Taq DNA polymerase activation step at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds and annealing at 60°C for one minute. Fold change was calculated and the expression level of the genes to be examined was normalized to the expression level of GAPDH. RT-PCR experiment was performed in triplicates using the same RNA sample for the microarray analysis.
2.5. Immunohistochemistry
Medial meniscal specimens derived from 6 end-stage OA patients and 3 osteosarcoma patients were used for examination. These meniscal specimens were fixed in 10% neutral buffered formalin for twenty-four hours and transferred to 70% ethyl alcohol. A portion of 5 mm wide specimen was transversely excised from the middle part of meniscus, embedded in Paraplast Plus paraffin, and sectioned with a Leica RM2025 microtome (Nussloch, Germany) to obtain 4 μm serial transverse sections [23]. Sections were examined with immunohistochemical staining using specific antibodies. Briefly, paraffin-embedded sections were deparaffinized with xylene and rehydrated with graded ethanol. Endogenous peroxidase activity was blocked by incubation of the sections with freshly prepared 3% H2O2 in deionized water for 5 minutes at room temperature. Nonspecific binding was blocked by incubation of the sections with 100 μL of 10% normal horse serum diluted in base solution (4% BSA and 5% nonfat dry milk in PBS) for 20 minutes. These sections were incubatedwith a specific primary antibody (2 μg/mL) for 1 hour, followed with the secondary reagent specific for each antibody for 30 minutes (Immpress reagent kit, Vector, Inc., Burlingame, CA). Negative control was performed using mouse IgG to replace the primary specific antibody. Slides were rinsed in phosphate buffered saline three times and visualized using 3,3′-diaminobenzidine for 5 minutes. Slides were then counterstained with light green, dehydrated, and mounted with resinous mounting media. These immunostainings were graded on a scale of 0–3, where 0 = very weak staining; 1 = weak staining; 2 = moderate staining; 3 = strong staining.
2.6. Statistical Analysis
The difference between the immunostaining grades of the OA meniscal group and control group was analyzed using the Wilcoxon rank-sum test. P values less than 0.05 were considered significant. Statistical analysis was performed using the statistical analysis tool in the Sigma Plot software, version 12 (Systat software Inc., San Jose, CA).
3. Results
3.1. Effect of PC on Gene Expressions
Microarrayanalysis revealed that of the more than 50,000 transcripts examined, 2561 transcripts displayed significant differential expressions (more than 2.0 fold) in PC-treated OA meniscal explants via untreated OA meniscal explants. A total of 1430 transcripts displayed decreased expressions and 1131 transcripts displayed increased expressions. The genes that fell into specific biological processes previously implicated in OA or suspected to have a role in OA are listed in Tables 1, 2, and 3.
Table 1.
Genes classified in the immune response.
| Biological process | Gene name | Gene ID | Differ expre* | Description |
|---|---|---|---|---|
| Immune response | ||||
| CCL20 | NM_004591 | −103.53 | Chemokine (C-C motif) ligand 20 | |
| CCL5 | NM_002985 | −3.54 | Chemokine (C-C motif) ligand 5 | |
| CCR5 | NM_000579 | −2.03 | Chemokine (C-C motif) receptor 5 | |
| CXCL3 | NM_002090 | −10.03 | Chemokine (C-X-C motif) ligand 3 | |
| CXCL5 | AK026546 | −4.49 | Chemokine (C-X-C motif) ligand 5 | |
| CXCL2 | M57731 | −2.94 | Chemokine (C-X-C motif) ligand 2 | |
| CXCL1 | NM_001511 | −3.01 | Chemokine (C-X-C motif) ligand 1 | |
| CXCL9 | NM_002416 | −2.65 | Chemokine (C-X-C motif) ligand 9 | |
| CXCL13 | NM_006419 | −2.62 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) | |
| CXCR4 | AF348491 | −2.63 | Chemokine (C-X-C motif) receptor 4 | |
| IGKC | BC005332 | −37.44 | Netrin 2-like (chicken) | |
| IL6 | NM_000600 | −32.07 | Interleukin 6 (interferon, beta 2) | |
| IL8 | NM_000584 | −8.17 | Interleukin 8 | |
| IL23A | M15564 | −7.89 | Enhancer of polycomb homolog 1 (Drosophila) | |
| IL24 | NM_006850 | −3.73 | Interleukin 24 | |
| IL15 | NM_000585 | −3.70 | Interleukin 15 | |
| IL7R | BE217880 | −3.31 | Interleukin 7 receptor | |
| IL1B | NM_000576 | −2.28 | Interleukin 1, beta | |
| IL1RN | BE563442 | −2.80 | Interleukin 1 receptor antagonist | |
| MS4A2 | NM_000139 | −28.91 | Membrane-spanning 4-domains, subfamily A, member 2 | |
| FCGR3B | NM_000570 | −22.56 | Fc fragment of IgG, low affinity IIIb, receptor (CD16b) | |
| FCGR2B | U90940 | −7.13 | Fc fragment of IgG, low affinity IIc, receptor for (CD32) | |
| FCGR1B | L03419 | −3.30 | Fc fragment of IgG, high affinity Ib, receptor (CD64) | |
| FCER1A | BC005912 | −5.19 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | |
| HLA-C | AW575927 | −22.41 | Major histocompatibility complex, class I, C | |
| HLA-F | BE138825 | −2.22 | Major histocompatibility complex, class I, F | |
| AQP9 | NM_020980 | −17.49 | Aquaporin 9 | |
| GZMA | NM_006144 | −14.63 | Granzyme A | |
| IGHG1 | M87789 | −14.43 | Immunoglobulin heavy constant gamma 1 | |
| LBP | M35533 | −11.36 | Lipopolysaccharide binding protein | |
| EREG | NM_001432 | −8.60 | Epiregulin | |
| CD86 | NM_006889 | −7.11 | CD86 molecule | |
| CD74 | M28590 | −2.76 | CD74 molecule | |
| CD40 | NM_001250 | −3.68 | CD40 molecule, TNF receptor superfamily member 5 | |
| CD1D | NM_001766 | −5.17 | CD1d molecule | |
| CD8A | AW006735 | −4.99 | CD8a molecule | |
| CD14 | NM_000591 | −3.25 | CD14 molecule | |
| CD209 | AF290886 | −2.50 | CD209 molecule | |
| KYNU | BC000879 | −6.69 | Kynureninase (L-kynurenine hydrolase) | |
| PTPRC | NM_002838 | −5.88 | Protein tyrosine phosphatase, receptor type, C | |
| TLR8 | AW872374 | −5.55 | Toll-like receptor 8 | |
| TLR7 | NM_016562 | −3.34 | Toll-like receptor 7 | |
| TLR5 | AF051151 | −3.00 | Toll-like receptor 5 | |
| TLR4 | NM_003266 | −2.40 | Toll-like receptor 4 | |
| TLR2 | NM_003264 | −2.20 | Toll-like receptor 2 | |
| TLR1 | AL050262 | −2.66 | Toll-like receptor 1 | |
| SLC11A1 | L32185 | −5.41 | Solute carrier family 11, member 1 | |
| CFD | NM_001928 | −4.99 | Complement factor D (adipsin) | |
| CFI | BC020718 | −3.37 | Complement factor I | |
| CFB | NM_001710 | −2.39 | Complement factor B | |
| C3 | NM_000064 | −2.32 | Complement component 3 | |
| CR1 | AI052659 | −2.24 | Complement component (3b/4b) receptor 1 (Knops blood group) | |
| C1QA | NM_015991 | −2.20 | Complement component 1, q subcomponent, A chain | |
| C1QB | NM_000491 | −2.10 | Complement component 1, q subcomponent, B chain | |
| C1RL | NM_016546 | 2.01 | Complement component 1, r subcomponent-like | |
| FYB | BF679849 | −4.64 | FYN binding protein (FYB-120/130) | |
| TREM1 | NM_018643 | −4.52 | Triggering receptor expressed on myeloid cells 1 | |
| BMP6 | NM_001718 | −4.48 | Bone morphogenetic protein 6 | |
| INPP5D | U53470 | −4.47 | Inositol polyphosphate-5-phosphatase, 145 kDa | |
| LILRB1 | NM_006669 | −4.33 | Leukocyte immunoglobulin-like receptor, subfamily B, member 1 | |
| LILRB2 | AF004231 | −4.32 | Leukocyte immunoglobulin-like receptor, subfamily B, member 2 | |
| LILRB3 | AF009634 | −4.23 | leukocyte immunoglobulin-like receptor, subfamily B, member 3 | |
| LILRB4 | U82979 | −2.06 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | |
| LILRB5 | NM_006840 | −2.31 | Leukocyte immunoglobulin-like receptor, subfamily B, member 5 | |
| LILRA2 | NM_006866 | −2.15 | Leukocyte immunoglobulin-like receptor, subfamily A, member 2 | |
| ZEB1 | NM_030751 | −4.12 | Zinc finger E-box binding homeobox 1 | |
| PLA2G7 | M80436 | −3.95 | Platelet-activating factor receptor | |
| MASP1 | AI274095 | −3.67 | Mannan-binding lectin serine peptidase 1 | |
| EBI2 | NM_004951 | −3.59 | Epstein-Barr virus induced gene 2 | |
| NOD2 | NM_022162 | −3.57 | Nucleotide-binding oligomerization domain containing 2 | |
| LAIR1 | NM_021708 | −3.54 | Leukocyte-associated immunoglobulin-like receptor 1 | |
| HLA-DQA1 | BG397856 | −3.38 | Major histocompatibility complex, class II, DQ alpha 1 | |
| HLA-DQB1 | M17955 | −2.99 | Major histocompatibility complex, class II, DQ beta 1 | |
| HLA-DRA | M60333 | −3.22 | Major histocompatibility complex, class II, DR alpha | |
| HLA-DPA1 | M27487 | −2.83 | Major histocompatibility complex, class II, DP alpha 1 | |
| HLA-DPB1 | NM_002121 | −2.15 | Major histocompatibility complex, class II, DP beta 1 | |
| HLA-DRB4 | BC005312 | −2.81 | Major histocompatibility complex, class II, DR beta 4 | |
| HLA-DRB1 | AJ297586 | −2.72 | Major histocompatibility complex, class II, DR beta 3 | |
| HLA-DMB | NM_002118 | −2.68 | Major histocompatibility complex, class II, DM beta | |
| HLA-DMA | X76775 | −2.19 | major histocompatibility complex, class II, DM alpha | |
| LIF | NM_002309 | −3.16 | Leukemia inhibitory factor (cholinergic differentiation factor) | |
| NCF4 | NM_000631 | −2.88 | Neutrophil cytosolic factor 4, 40 kDa | |
| PAG1 | BF589359 | −2.86 | Phosphoprotein associated with glycosphingolipid microdomains 1 | |
| FYN | AK090692 | −2.85 | FYN oncogene related to SRC, FGR, YES | |
| TREM2 | NM_018965 | −2.84 | Triggering receptor expressed on myeloid cells 2 | |
| BST2 | NM_004335 | −2.84 | Bone marrow stromal cell antigen 2 | |
| CTSG | NM_001911 | −2.78 | Cathepsin G | |
| CTSS | AK024855 | −2.60 | Cathepsin S | |
| MS4A1 | AW474852 | −2.74 | Membrane-spanning 4-domains, subfamily A, member 1 | |
| NCF2 | BC001606 | −2.70 | Neutrophil cytosolic factor 2 | |
| GPR65 | NM_003608 | −2.68 | G protein-coupled receptor 65 | |
| GBP4 | BG260886 | −2.59 | Guanylate binding protein 4 | |
| VAV1 | NM_005428 | −2.53 | Vav 1 guanine nucleotide exchange factor | |
| LCK | NM_005356 | −2.49 | Lymphocyte-specific protein tyrosine kinase | |
| SYK | NM_003177 | −2.42 | Spleen tyrosine kinase | |
| LY86 | NM_004271 | −2.36 | Lymphocyte antigen 86 | |
| TNFSF10 | U57059 | −2.36 | Tumor necrosis factor (ligand) superfamily, member 10 | |
| TNFSF13B | AF134715 | −2.29 | Tumor necrosis factor (ligand) superfamily, member 13b | |
| IRF8 | AI073984 | −2.36 | Interferon regulatory factor 8 | |
| RELB | NM_006509 | −2.33 | V-rel reticuloendotheliosis viral oncogene homolog B | |
| SMAD6 | AI628464 | −2.25 | SMAD family member 6 | |
| MBP | N37023 | −2.24 | Myelin basic protein | |
| BCL6 | S67779 | −2.17 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | |
| IGKC | BG485135 | −2.12 | Netrin 2-like (chicken) | |
| CLEC7A | AF313468 | −2.12 | C-type lectin domain family 7, member A | |
| LCP2 | AI123251 | −2.05 | Lymphocyte cytosolic protein 2 | |
| IL31RA | AI123586 | 6.61 | Interleukin 31 receptor A | |
| C4BPA | NM_000715 | 3.34 | Complement component 4 binding protein, alpha | |
| ULBP2 | AA831769 | 3.07 | UL16 binding protein 2 | |
| CLEC4E | NM_014358 | 2.61 | C-type lectin domain family 4, member E | |
| TNFSF9 | NM_003811 | 2.43 | Tumor necrosis factor (ligand) superfamily, member 9 | |
| C7 | NM_000587 | 2.36 | Complement component 7 | |
| TGFB2 | NM_003238 | 2.36 | Transforming growth factor, beta 2 | |
| FAS | X83493 | 2.36 | Fas (TNF receptor superfamily, member 6) | |
| LAG3 | NM_002286 | 2.26 | Lymphocyte-activation gene 3 | |
| IL27RA | NM_004843 | 2.20 | Interleukin 27 receptor, alpha | |
| CD276 | NM_025240 | 2.19 | CD276 molecule | |
| IL26 | NM_018402 | 2.03 | Interleukin 26 | |
| OAS1 | NM_016816 | 2.02 | 2,5-oligoadenylate synthetase 1, 40/46 kDa |
*Negative number indicates decreased expression and positive number indicates increased expression (fold change) in PC-treated OA meniscal explants compared with untreated OA meniscal explants.
Table 2.
Genes classified in inflammatory response and angiogenesis.
| Biological process | Gene name | Gene ID | Differ expre (fold)* | Description |
|---|---|---|---|---|
| Inflammatory response | ||||
| CCL20 | NM_004591 | −103.53 | Chemokine (C-C motif) ligand 20 | |
| CCL5 | NM_002985 | −3.54 | Chemokine (C-C motif) ligand 5 | |
| CCR5 | NM_000579 | −2.03 | Chemokine (C-C motif) receptor 5 | |
| CXCL3 | NM_002090 | −10.03 | Chemokine (C-X-C motif) ligand 3 | |
| CXCL2 | M57731 | −2.94 | Chemokine (C-X-C motif) ligand 2 | |
| CXCL1 | NM_001511 | −3.01 | Chemokine (C-X-C motif) ligand 1 | |
| CXCL9 | NM_002416 | −2.65 | Chemokine (C-X-C motif) ligand 9 | |
| CXCL13 | NM_006419 | −2.62 | Chemokine (C-X-C motif) ligand 13 (B-cell chemoattractant) | |
| CXCR4 | AF348491 | −2.63 | Chemokine (C-X-C motif) receptor 4 | |
| PTGS2 | AY151286 | −34.01 | Prostaglandin-endoperoxide synthase 2 | |
| IL6 | NM_000600 | −32.07 | Interleukin 6 (interferon, beta 2) | |
| IL23A | AF043179 | −16.44 | Enhancer of polycomb homolog 1 (Drosophila) | |
| IL8 | NM_000584 | −8.17 | Interleukin 8 | |
| IL1B | NM_000576 | −2.28 | Interleukin 1, beta | |
| IL1RN | BE563442 | −2.80 | Interleukin 1 receptor antagonist | |
| S100A8 | NM_002964 | −16.53 | S100 calcium binding protein A8 | |
| S100A9 | NM_002965 | −2.79 | S100 calcium binding protein A9 | |
| LBP | M35533 | −11.36 | Lipopolysaccharide binding protein | |
| APOE | NM_000041 | −9.23 | Apolipoprotein E | |
| FABP4 | AI766029 | −6.98 | Fatty acid binding protein 4, adipocyte | |
| LYZ | U25677 | −6.37 | Lysozyme (renal amyloidosis) | |
| ITIH4 | AI004137 | −6.33 | Inter-alpha (globulin) inhibitor H4 | |
| BDKRB2 | NM_000623 | −6.01 | Bradykinin receptor B2 | |
| TLR8 | AW872374 | −5.55 | Toll-like receptor 8 | |
| TLR7 | NM_016562 | −3.34 | Toll-like receptor 7 | |
| TLR5 | AF051151 | −3.00 | Toll-like receptor 5 | |
| TLR4 | NM_003266 | −2.40 | Toll-like receptor 4 | |
| TLR2 | NM_003264 | −2.20 | Toll-like receptor 2 | |
| TLR1 | AL050262 | −2.66 | Toll-like receptor 1 | |
| AOX1 | AB046692 | −5.34 | Aldehyde oxidase 1 | |
| FCER1A | BC005912 | −5.19 | Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide | |
| CFD | NM_001928 | −4.99 | Complement factor D (adipsin) | |
| CFI | BC020718 | −3.37 | Complement factor I | |
| CFB | NM_001710 | −2.39 | Complement factor B | |
| C3 | NM_000064 | −2.32 | Complement component 3 | |
| CR1 | AI052659 | −2.24 | Complement component (3b/4b) receptor 1 (Knops blood group) | |
| C1RL | NM_016546 | −2.01 | Complement component 1, r subcomponent-like | |
| C1QA | NM_015991 | −2.20 | Complement component 1, q subcomponent, A chain | |
| C1QB | NM_000491 | −2.10 | Complement component 1, q subcomponent, B chain | |
| AOAH | NM_001637 | −4.89 | Acyloxyacyl hydrolase (neutrophil) | |
| AGT | NM_000029 | −4.66 | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | |
| BMP6 | NM_001718 | −4.48 | Bone morphogenetic protein 6 | |
| PLA2G7 | M80436 | −3.95 | Platelet-activating factor receptor | |
| FOS | BC004490 | −3.95 | V-fos FBJ murine osteosarcoma viral oncogene homolog | |
| CD40 | NM_001250 | −3.68 | CD40 molecule, TNF receptor superfamily member 5 | |
| CD163 | NM_004244 | −3.68 | CD163 molecule | |
| CD14 | NM_000591 | −3.25 | CD14 molecule | |
| MASP1 | AI274095 | −3.67 | Mannan-binding lectin serine peptidase 1 | |
| F11R | AF191495 | −3.16 | F11 receptor | |
| ITGB2 | L78790 | −2.98 | Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | |
| NLRC4 | NM_021209 | −2.92 | NLR family, CARD domain containing 4 | |
| AIF1 | U19713 | −2.76 | Allograft inflammatory factor 1 | |
| ALOX5 | NM_000698 | −2.71 | Arachidonate 5-lipoxygenase | |
| NOD1 | AF126484 | −2.66 | Nucleotide-binding oligomerization domain containing 1 | |
| JMJD3 | AI830331 | −2.46 | Jumonji domain containing 3, histone lysine demethylase | |
| SIGLEC1 | NM_023068 | −2.46 | Sialic acid binding Ig-like lectin 1, sialoadhesin | |
| TNFAIP6 | AW188198 | −2.37 | Tumor necrosis factor, alpha-induced protein 6 | |
| LY86 | NM_004271 | −2.36 | Lymphocyte antigen 86 | |
| AOC3 | NM_003734 | −2.31 | Amine oxidase, copper containing 3 (vascular adhesion protein 1) | |
| TNFRSF1B | NM_001066 | −2.21 | Tumor necrosis factor receptor superfamily, member 1B | |
| BCL6 | S67779 | −2.17 | B-cell CLL/lymphoma 6 (zinc finger protein 51) | |
| CLEC7A | AF313468 | −2.12 | C-type lectin domain family 7, member A | |
| CDO1 | NM_001801 | −2.11 | Cysteine dioxygenase, type I | |
| NFATC4 | AI806528 | −2.04 | NF of activated T-cells, cytoplasmic, calcineurin-dependent 4 | |
| SERPINA3 | NM_001085 | 5.20 | Serpin peptidase inhibitor, clade A, member 3 | |
| SERPINA1 | AF119873 | 3.58 | Serpin peptidase inhibitor, clade A, member 1 | |
| C4BPA | NM_000715 | 3.34 | Complement component 4 binding protein, alpha | |
| FN1 | AJ276395 | 3.03 | Fibronectin 1 | |
| B4GALT1 | D29805 | 2.99 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 1 | |
| GPR68 | AI805006 | 2.40 | G protein-coupled receptor 68 | |
| C7 | NM_000587 | 2.36 | Complement component 7 | |
| ANXA1 | AU155094 | 2.23 | Annexin A1 | |
| KLKB1 | NM_000892 | 2.21 | Cytochrome P450, family 4, subfamily V, polypeptide 2 | |
| Angiogenesis | ||||
| PTGS2 | AY151286 | −34.00 | Prostaglandin-endoperoxide synthase 2 | |
| BAI3 | NM_001704 | −33.48 | Brain-specific angiogenesis inhibitor 3 | |
| IL6 | NM_000600 | −32.08 | Interleukin 6 (interferon, beta 2) | |
| IL8 | NM_000584 | −8.18 | Interleukin 8 | |
| IL1B | NM_000576 | −2.28 | Interleukin 1, beta | |
| SFRP1 | AF017987 | −17.55 | Secreted frizzled-related protein 1 | |
| ANGPTL4 | AF169312 | −11.43 | Angiopoietin-like 4 | |
| ANGPT2 | BE501356 | −2.56 | Angiopoietin 2 | |
| EREG | NM_001432 | −8.60 | Epiregulin | |
| VEGFA | M27281 | −7.91 | Vascular endothelial growth factor A | |
| VASH1 | AU152507 | −4.24 | Vasohibin 1 | |
| FLT1 | U01134 | −3.62 | Fms-related tyrosine kinase 1 | |
| PTPRB | AL080103 | −3.48 | Protein tyrosine phosphatase, receptor type, B | |
| FGF10 | NM_004465 | −3.34 | Fibroblast growth factor 10 | |
| LIF | NM_002309 | −3.16 | Leukemia inhibitory factor (cholinergic differentiation factor) | |
| BTG1 | BC009050 | −3.11 | B-cell translocation gene 1, anti-proliferative | |
| DLL4 | AB036931 | −2.85 | Delta-like 4 (Drosophila) | |
| SOX17 | NM_022454 | −2.45 | SRY (sex determining region Y)-box 17 | |
| EPAS1 | NM_001430 | −2.43 | Endothelial PAS domain protein 1 | |
| CTNNB1 | AB062292 | −2.35 | Catenin (cadherin-associated protein), beta 1, 88 kDa | |
| TGFBR2 | NM_003242 | −2.35 | Transforming growth factor, beta receptor II (70/80 kDa) | |
| TYMP | NM_001953 | −2.34 | Thymidine phosphorylase | |
| C3 | NM_000064 | −2.32 | Complement component 3 | |
| NRP1 | AF280547 | −2.19 | Neuropilin 1 | |
| NOTCH4 | AI743713 | −2.17 | Notch homolog 4 (Drosophila) | |
| TSPAN12 | AI056699 | −2.16 | Tetraspanin 12 | |
| ADAM8 | NM_001109 | −2.15 | ADAM metallopeptidase domain 8 | |
| RHOB | AI263909 | −2.12 | Ras homolog gene family, member B | |
| HHEX | NM_001529 | −2.12 | Hematopoietically expressed homeobox | |
| THBS1 | BF055462 | −2.05 | Thrombospondin 1 | |
| KDR | NM_002253 | −2.03 | Kinase insert domain receptor (a type III receptor tyrosine kinase) | |
| GREM1 | NM_013372 | 15.86 | Gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) | |
| FGFR2 | AB030078 | 4.49 | Fibroblast growth factor receptor 2 | |
| FGF1 | X59065 | 2.39 | fibroblast growth factor 1 (acidic) | |
| FGF18 | AI798863 | 2.33 | Fibroblast growth factor 18 | |
| COL8A2 | AI806793 | 4.46 | Collagen, type VIII, alpha 2 | |
| COL8A1 | BE877796 | 3.30 | Collagen, type VIII, alpha 1 | |
| ARHGAP22 | NM_021226 | 4.45 | Rho GTPase activating protein 22 | |
| TNFRSF12A | NM_016639 | 3.23 | Tumor necrosis factor receptor superfamily, member 12A | |
| CSPG4 | BE857703 | 3.14 | Chondroitin sulfate proteoglycan 4 | |
| WNT5B | NM_030775 | 3.10 | Wingless-type MMTV integration site family, member 5B | |
| FN1 | AJ276395 | 3.03 | Fibronectin 1 | |
| SFRP2 | AF311912 | 3.00 | Secreted frizzled-related protein 2 | |
| B4GALT1 | D29805 | 2.99 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 1 | |
| ANGPT1 | NM_001146 | 2.62 | Angiopoietin 1 | |
| MFGE8 | BC003610 | 2.39 | Milk fat globule-EGF factor 8 protein | |
| TGFB2 | NM_003238 | 2.36 | Transforming growth factor, beta 2 | |
| CYR61 | NM_001554 | 2.30 | Cysteine-rich, angiogenic inducer, 61 | |
| MEOX2 | NM_005924 | 2.15 | Mesenchyme homeobox 2 | |
| PLXDC1 | NM_020405 | 2.09 | Plexin domain containing 1 | |
| TBXA2R | NM_001060 | 2.09 | Thromboxane A2 receptor |
*Negative number indicates decreased expression and positive number indicates increased expression (fold change).
Table 3.
Genes classified in skeletal development, steroid biosynthetic process, and DNA repair.
| Biological process | Gene name | Gene ID | Differ expre (fold)* | Description |
|---|---|---|---|---|
| Skeletal development | ||||
| COL2A1 | X06268 | 21.29 | Collagen, type II, alpha 1 | |
| COL11A1 | J04177 | 9.86 | Collagen, type XI, alpha 1 | |
| COL1A1 | AI743621 | 2.59 | Collagen, type I, alpha 1 | |
| ACAN | NM_001135 | 9.17 | Aggrecan | |
| POSTN | AW137148 | 5.57 | Periostin, osteoblast specific factor | |
| PAX1 | AA725078 | 4.73 | Paired box 1 | |
| FRZB | U91903 | 3.22 | Frizzled-related protein | |
| MMP9 | NM_004994 | 2.58 | Matrix metallopeptidase 9 | |
| GLI2 | NM_030379 | 2.54 | GLI-Kruppel family member GLI2 | |
| FGFR3 | NM_000142 | 2.40 | Fibroblast growth factor receptor 3 | |
| FGF18 | AI798863 | 2.33 | Fibroblast growth factor 18 | |
| TGFB2 | NM_003238 | 2.36 | Transforming growth factor, beta 2 | |
| TRPS1 | AK000948 | 2.32 | Trichorhinophalangeal syndrome I | |
| RUNX2 | AW469546 | 2.24 | Runt-related transcription factor 2 | |
| PRELP | U41344 | 2.15 | Proline/arginine-rich end leucine-rich repeat protein | |
| PAPSS2 | AW299958 | 2.03 | 3-phosphoadenosine 5-phosphosulfate synthase 2 | |
| SOX9 | NM_000346 | 2.00 | SRY (sex determining region Y)-box 9 | |
| MEPE | NM_020203 | −13.5 | Matrix, extracellular phosphoglycoprotein with ASARM motif | |
| PTHLH | BC005961 | −6.79 | Parathyroid hormone-like hormone | |
| CHRDL2 | AF332891 | −6.20 | Chordin-like 2 | |
| COL10A | X98568 | −5.40 | collagen, type X, alpha 1 (Schmid metaphyseal chondrodysplasia) | |
| BMP6 | NM_001718 | −4.48 | Bone morphogenetic protein 6 | |
| GDF10 | NM_004962 | −2.99 | Growth differentiation factor 10 | |
| BMP8B | AA610122 | −2.70 | Bone morphogenetic protein 8b | |
| TGFBR2 | NM_003242 | −2.35 | Transforming growth factor, beta receptor II (70/80 kDa) | |
| IGFBP4 | NM_001552 | −2.30 | Insulin-like growth factor binding protein 4 | |
| Steroid biosynthetic process | ||||
| SQLE | AA639705 | 3.81 | Squalene epoxidase | |
| SRD5A1 | NM_001047 | 3.75 | Steroid-5-alpha-reductase, alpha polypeptide 1 | |
| FDXR | NM_004110 | 3.26 | Ferredoxin reductase | |
| HMGCS1 | NM_002130 | 3.12 | 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) | |
| HMGCR | AL518627 | 2.15 | 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | |
| LSS | D63807 | 3.04 | Lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) | |
| DHCR24 | NM_014762 | 2.66 | 24-dehydrocholesterol reductase | |
| DHCR7 | NM_001360 | 2.57 | 7-dehydrocholesterol reductase | |
| OPRS1 | NM_005866 | 2.62 | Opioid receptor, sigma 1 | |
| CYB5R2 | NM_016229 | 2.37 | Cytochrome b5 reductase 2 | |
| SC5D | D85181 | 2.06 | Sterol-C5-desaturase | |
| BMP6 | NM_001718 | −4.48 | Bone morphogenetic protein 6 | |
| CYP39A1 | NM_016593 | −2.21 | Cytochrome P450, family 39, subfamily A, polypeptide 1 | |
| HSD3B7 | BC004929 | −2.10 | 3 beta-hydroxysteroid dehydrogenase type 7 | |
| ADM | NM_001124 | −2.06 | Adrenomedullin | |
| ABCG1 | NM_004915 | −2.04 | ATP-binding cassette, sub-family G (WHITE), member 1 | |
| DNA repair | ||||
| TOP2A | AU159942 | 3.75 | Topoisomerase (DNA) II alpha 170 kDa | |
| RAD52B | AF125949 | 2.83 | RAD52 homolog (S. cerevisiae) | |
| RAD54B | NM_012415 | 2.05 | RAD54 homolog B (S. cerevisiae) | |
| TYMS | NM_001071 | 2.40 | Thymidylate synthetase | |
| RFC5 | BG260658 | 2.39 | Replication factor C (activator 1) 5, 36.5 kDa | |
| SOD2 | AL050388 | −9.58 | Superoxide dismutase 2, mitochondrial | |
| REV3L | NM_002912 | −3.08 | REV3-like, catalytic subunit of DNA polymerase zeta (yeast) |
*Negative number indicates decreased expression and positive number indicates increased expression (fold change).
As shown in Table 1, the expressions of numerous genes classified in the immune response were downregulated by PC. Of the 120 differentially-expressed genes classified in the immune response, the expressions of 106 genes, including many genes encoding chemokines and cytokines, such as chemokine (C-C motif) ligand 20 (CCL20, −103.53 fold change), chemokine (C-C motif) ligand 5 (CCL5, −3.54 fold change), chemokine (C-C motif) receptor 5 (CCR5, −2.03 fold change), chemokine (C-X-C motif) ligand 3 (CXCL3, −10.03 fold), interleukin 6 (IL-6, −32.07 fold change), interleukin 7 receptor (IL-7R. −3.31 fold change), IL-8 (−8.17 fold change), IL-23, alpha subunit (IL23A, −5.46 fold change), and IL-1 beta (IL-1β, −2.28 fold change) genes, were downregulated by PC.
The genes downregulated by PC also included many Fc fragments of IgG receptors (FCGRs), leukocyte immunoglobulin-like receptors (LILRs), toll-like receptors (TLRs), and major histocompatibility complex (MHC) class II molecules genes, such as FCGR3B (−22.56 fold change), FCGR2B (−7.13 fold change), LILRB3 (−4.23 fold change), LILRB2 (−4.32 fold change), LILRB1 (−4.33 fold change), TLR8 (−5.55 fold change), TLR7 (−3.34 fold change), MHC class II, DP alpha 1 (HLA-DPA1, −2.83 fold change), MHC class II, DQ alpha 1 (HLA-DQA1, −3.39 fold), MHC class II, DR beta 1 (HLA-DRB1, −2.72 fold change), and MHC class II, DR beta 4 genes (HLA-DRB4, −2.81 fold change).
The expressions of many genes classified in inflammatory response and angiogenesis were also downregulated by PC. As shown in Table 2, of the 73 differentially-expressed genes classified in inflammatory response, the expressions of 64 genes, including prostaglandin-endoperoxide synthase 2 (PTGS2/Cox-2, −34.01 fold change), S100 calcium binding protein A8 (S100A8, −16.53 fold change), complement factor D (CFD, −4.99 fold change), and allograft inflammatory factor 1 (AIF1, −2.76 fold change) genes, were downregulated by PC. Of the 51 differentially-expressed genes classified in angiogenesis, the expressions of 31 genes, including brain-specific angiogenesis inhibitor 3 (BAI3, −33.48 fold change), angiopoietin-like 4 (ANGPTL4, −11.43 fold change), and vascular endothelial growth factor A genes (VEGFA, −7.91 fold change), were downregulated by PC.
In contrast, the expressions of many genes classified in skeletal development, steroid biosynthetic process, and DNA repair were upregulated by PC. As shown in Table 3, of the 26 differentially-expressed genes classified in skeletal development, the expressions of 17 genes, including collagen type II, alpha 1 (COL2A1, 21.29 fold change), collagen type XI, alpha 1 (COL11A1, 9.86 fold change), aggrecan (ACAN, 9.17 fold change), FGFR3 (2.40 fold change), FGF18 (2.33 fold change), and SOX-9 (2.00 fold change) genes, were upregulated by PC. Of the 16 differentially expressed genes classified in steroid biosynthetic process, the expressions of 11 genes, including squalene epoxidase (SQLE, 3.81 fold change) and steroid-5-alpha-reductase and alpha polypeptide 1 (SRD5A1, 3.75 fold change) genes, were upregulated by PC. Of the 7 differentially expressed genes classified in DNA repair, the expressions of 5 genes, including topoisomerase (DNA) II alpha 170 kDa (TOP2A, 3.75 fold change) and RAD52 homolog (RAD52B, 2.83 fold change) genes, were upregulated by PC.
We performed another independent microarray analysis using RNA samples extracted from meniscal explants derived from a different OA patient. Results from the two microarray analyses for select genes were listed in Table 4. As shown, the results from the two microarray analyses were consistent. We also examined the expressions of selected genes using real-time quantitative RT-PCR. The results from the real-time RT-PCR for the genes examined were consistent with the results from the microarray analyses (Table 4).
Table 4.
Differential expression of selected genes.
| Gene name | Gene ID | Differential expression* microarray | Differential expression** microarray | Differential expression*** real-time RT-PCR |
|---|---|---|---|---|
| CCL5 | NM_002985 | −3.54 | −6.28 | −3.69 |
| CCR5 | NM_000579 | −2.03 | −2.95 | |
| FCGR3B | NM_000570 | −6.49 | −2.09 | |
| FCGR2B | M31933 | −3.45 | −3.72 | |
| IL6 | NM_000600 | −32.07 | −1.85 | |
| IL7R | BE217880 | −3.31 | −11.0 | −2.85 |
| IL8 | NM_000584 | −8.17 | −2.25 | |
| IL23A | M15564 | −7.89 | −8.98 | |
| LILRB1 | NM_006669 | −4.33 | −3.06 | |
| LILRB3 | AF009634 | −4.23 | −3.38 | |
| TLR8 | AW872374 | −5.55 | −3.55 | |
| TLR7 | NM_016562 | −3.34 | −2.54 | |
| HLA-DRB1 | AJ297586 | −2.72 | −3.41 | −1.97 |
| S100A8 | NM_002964 | −16.53 | −9.95 | |
| S100A9 | NM_002965 | −2.79 | −4.70 | |
| PTPRC | NM_002838 | −5.88 | −2.85 | |
| SYK | NM_003177 | −2.42 | −2.99 | |
| BMP6 | NM_001718 | −4.48 | −3.00 | |
| CPA3 | NM_001870 | −15.23 | −9.37 | |
| CPM | BE878495 | −3.17 | −2.74 | |
| ADAMTS5 | BI254089 | −2.84 | −1.98 | −2.65 |
| ADAM28 | NM_021778 | −14.27 | −3.59 | |
| MMP10 | NM_002425 | −2.41 | −1.56 | |
| MMP1 | NM_002421 | −2.15 | −1.83 | −2.01 |
| FGFR3 | NM_000142 | 2.40 | 1.52 | |
| SOX-9 | NM_000346 | 2.00 | 1.70 | |
| POSTN | AW137148 | 5.57 | 2.76 | |
| COL2A1 | X06268 | 21.29 | 1.76 | 3.11 |
| COL11A1 | J04177 | 9.86 | 2.29 | |
| COL1A1 | AI743621 | 2.59 | 1.79 | |
| ACAN | NM_001135 | 9.17 | 2.02 | 2.65 |
*Negative number indicates decreased expression and positive number indicates increased expression (fold change).
**Second microarray using different RNA samples extracted from meniscal explants derived from a different OA patient.
***Results of real-time RC-PCR.
3.2. Immunohistochemical Staining
To investigate whether PC-targeted genes were associated with OA, we decided to examine the expressions of selected PC-targeted genes in OA and normal menisci. Medial menisci derived from 6 end-stage OA patients and 3 osteosarcoma patients were used for the examinations. Representative images of the normal meniscus and OA menisci are shown in Figure 1.
Figure 1.
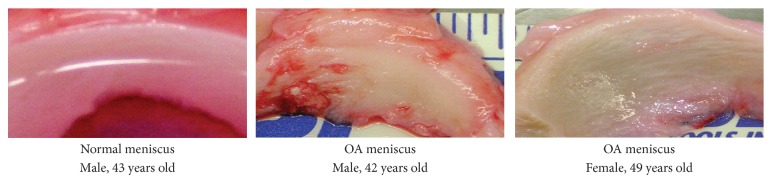
Images of normal and OA menisci. Meniscus derived from a male osteosarcoma patient, a male OA patient and a female OA patient. Menisci derived from OA patients exhibited discoloration and rough surface, with clear signs of degeneration whereas the normal control menisci had a smooth and glistening surface, with no signs of degeneration.
The expressions of genes we selected for immunohistochemical examinations included FCGR3B, LILRB3, FGFR3 and SOX-9. FCGR3B and LILRB3 are two genes classified in the immune and inflammatory responses. FGFR3 and SOX-9 were two genes classified in the skeletal development. We first tested the antibodies specific to these proteins using human tissues known for their expression before using these antibodies to examine the meniscal tissues.
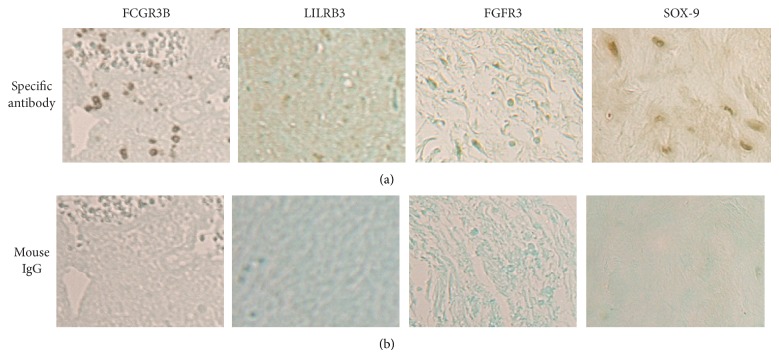
As shown in Figure 2, positive immunostainings were observed when these primary antibodies specific to these proteins were used whereas no immunostainings were observed when these primary antibodies were replaced by mouse IgG, confirming that these antibodies could be used to examine the expressions of FCGR3B, LILRB3, FGFR3, and SOX-9 in human tissues. We then used these antibodies to examine medial meniscal specimens derived from 6 end-stage OA patients (diseased tissues) and 3 osteosarcoma patients (control tissues). Representative images of FCGR3B immunostaining are provided in Figure 2(a).
Figure 2.
(a) Positive FCGR3B immunostaining in human tonsil tissue, positive LILRB3 immunostaining in human tonsil tissue, positive FGFR3 immunostaining in human skin tissue, and positive SOX-9 immunostaining in human cartilage tissue. (b) Negative immunostaining of these proteins when primary antibodies were replaced by mouse IgG.
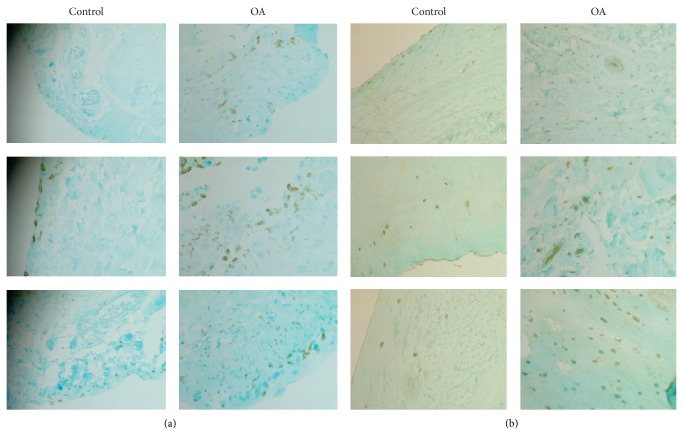
As shown, FCGR3B protein was detected in the surface zone of normal menisci. In contrast, FCGR3B was detected in both the surface and middle zones of OA menisci. It was clear that OA menisci contained more FCGR3B immunostaining-positive cells than the normal menisci, suggesting infiltration of inflammatory cells within the OA menisci. Representative images of LILRB3 immunostaining are provided in Figure 3(b). As shown, LILRB3 protein was detected in all 3 zones (surface, middle, and deep zones) of the normal and OA menisci. OA menisci not only contained more LILRB3 immunostaining-positive cells but also displayed more intensified LILRB3 immunostaining compared to the normal menisci.
Figure 3.
Representative images of FCGR3B and LILR3B immunostaining (magnification 10x). (a) FCGR3B immunostaining of normal menisci and OA menisci. (b) LILRB3 immunostaining of normal menisci and OA menisci.
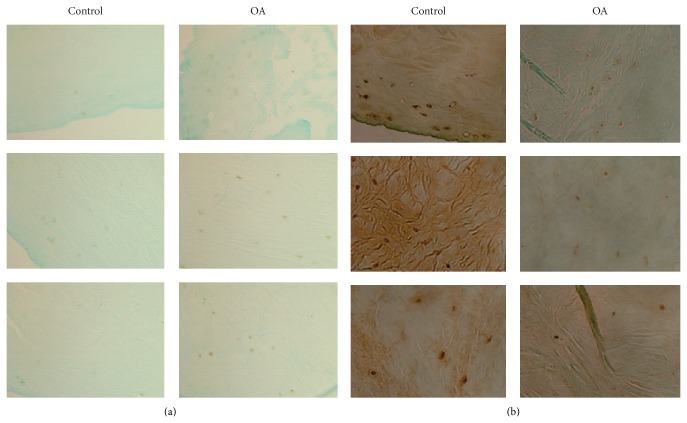
Representative images of FGFR3 and SOX-9 immunostaining are provided in Figure 4. As shown in Figure 4(a), few FGFR3 immunostaining-positive cells were detected in the normal menisci and only a small number of FGFR3 immunostaining-positive cells were detected in the OA menisci. In contrast, strong SOX-9 immunostaining was detected in the normal menisci and the intensity of SOX-9 immunostaining and number of SOX-9 immunostaining-positive cells were decreased significantly in the OA menisci compared to the normal menisci (Figure 4(b)).
Figure 4.
Representative images of FGFR3 and SOX-9 immunostaining (magnification 10x). (a) FGFR3 immunostaining of normal menisci and OA menisci. (b) SOX-9 immunostaining of normal menisci and OA menisci.
The immunostainings of FCGR3B, LILRB3, FGFR3, and SOX-9 were graded according to the scale described in Methods. The results along with the demographic patient information are listed in Table 5. As shown, the mean grades of FCGR3B, LILRB3, FGFR3, and SOX-9 immunostainings for the normal menisci were 0.33, 1.33, 0.00, and 3.00, respectively, and the mean grades of FCGR3B, LILRB3, FGFR3, and SOX-9 immunostaining for the OA menisci were 2.00, 2.67, 1.17, and 1.50, respectively. The difference between the mean grades of FCGR3B, LILRB3, FGFR3, and SOX-9 immunostaining of the OA menisci and the normal menisci were statistically significant (P = 0.023, 0.010, 0.002, and 0.024, resp.).
Table 5.
Grades of immunostaining of FCGR3B, LILRB3, FGFR3, and SOX-9.
| Control 12 F |
Control 43 M |
Control 39 F |
OA 77 M |
OA 49 F |
OA 66 F |
OA 70 F |
OA 57 M |
OA 65 F |
|
|---|---|---|---|---|---|---|---|---|---|
| FCGR3B | 0 | 1 | 0 | 3 | 3 | 2 | 2 | 1 | 1 |
| Mean (FCGR3B) | 0.33 | 2.00 | |||||||
|
| |||||||||
| LILRB3 | 1 | 2 | 1 | 2 | 3 | 3 | 2 | 3 | 3 |
| Mean (LILRB3) | 1.33 | 2.67 | |||||||
|
| |||||||||
| FGFR3 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 2 | 1 |
| Mean (FGFR3) | 0.00 | 1.17 | |||||||
|
| |||||||||
| SOX-9 | 3 | 3 | 3 | 2 | 1 | 1 | 1 | 2 | 2 |
| Mean (SOX-9) | 3.00 | 1.50 | |||||||
Ages of the patients are listed in years; M = male; F = female.
4. Discussion
There is increasing evidence indicating the involvement of immune system in OA. The expressions of several TLRs genes, which play a key role in innate immune system, were increased in OA cartilage and correlated with the severity of cartilage degeneration [24, 25]. The expressions of several MHC class II genes were increased in degenerative menisci of older patients and OA meniscal cells compared to younger patients and normal meniscal cells [4, 26]. In this study, we demonstrated that PC, which inhibited cartilage degeneration in Hartley guinea pigs [16], downregulated the expression of numerous genes classified in the immune response, including many TLRs genes (Table 1, such as TLR-4 and TLR-8), MHC class II genes (such as HLA-DPA1, HLA-DRA, and HLA-DRB1), FCGRs genes (such as FCGR2B and FCGR3B), and LILRs genes (such as LILRA2, LILRB1, and LILRB3). These findings suggest that PC may be capable of reversing the abnormal expressions of many genes involved in immune system activation in OA menisci and cartilage.
Studies found that the expression of FCGRs, which help to bridge the adaptive and innate immune responses, and the expression of LILRs, which exert influence on signaling pathways of both innate and adaptive immune systems, were increased in inflammatory arthritis such as rheumatoid arthritis (RA) [27–30]. In addition, studies found that the numbers of FCGRs- and LILRs-positive immune cells were decreased in RA patients who responded to treatment with anti-rheumatic drugs [31, 32]. These previous findings indicate that abnormal expressions of FCGRs and LILRs are associated with inflammatory arthritis. In this study, we demonstrated that the expressions of FCGR3B and LILRB3 genes were increased in OA menisci and that their expressions in OA meniscal explant culture were inhibited by PC. These findings indicate that abnormal expressions of FCGRs and LILRs are also associated with OA. PC exerts its disease-modifying activity on OA, at least in part, by targeting abnormal immune system activation in OA.
Inflammation and angiogenesis are closely integrated processes in OA [33, 34]. A study demonstrated that inhibition of inflammation and angiogenesis reduced pain and retard joint damage in a rat model of OA [35]. In our study, we demonstrated that PC downregulated the expression of numerous genes classified in the inflammatory response and angiogenesis, including CCL5, CCR5, IL-8, IL-7R, IL-6, IL-1β, PTGS2/Cox-2, S100A8, ANGPTL4, and VEGFA. It is worth noting that abnormal expressions of these genes are associated with either OA or RA. For example, the protein levels of CCL5, IL-6, IL-8, IL-1β, S100A8, ANGPTL4, and VEGFA were increased in chondrocytes, cartilage, synovium, or synovial fluid derived from OA patients, which in turn stimulated the expression of MMPs [36–44]; the expression of IL-7R was elevated in RA FLSs and blockade of IL-7R reduced joint inflammation and cartilage destruction [45, 46]; PTGS2/Cox-2 is a key molecular target for the management of arthritis pain [47]. These findings together suggest that PC exerts its disease-modifying activity on OA, at least in part, by targeting abnormal inflammatory response and angiogenesis in OA. These findings also suggest that abnormal inflammatory response and angiogenesis in the menisci may be new target for OA intervention.
PC upregulated the expressions of many genes classified in skeletal development, including putative chondroprotective proteins FGFR3 and SOX-9 [48, 49]. To identify which proteins might be involved in the OA disease process, we examined the protein levels of FGFR3 and SOX-9 in normal and OA menisci. We found that FGFR3 protein was barely detected in the normal menisci and was slightly increased in OA menisci, suggesting that FGFR3 gene is unlikely a key OA disease candidate gene. In contrast, the protein level of SOX-9 was very high in the normal menisci but was significantly decreased in the OA menisci. This finding is consistent with the previous findings that the expression of SOX-9 gene was significantly decreased in OA articular cartilage and chondrocytes [50–52]. Taken together, it suggests that SOX-9 may be an OA disease candidate gene and that PC exerts its disease modifying activity on OA in part by reversing the abnormal expression of SOX-9 in OA menisci or cartilage. Study with SOX-9 knok-in or knock-out using an animal model of OA may provide more clues about the role of SOX-9 in OA.
A recent study reported that hundreds of genes were differentially expressed in degenerative menisci derived from older patients and younger patients [26]. The genes displayed higher expressions in the degenerative menisci derived from older patients compared to younger patients included HLA-DRB1 (15.01 fold change), FCER1A (4.15 fold change), and IL-7R (2.83 fold change) [26]. These findings are consistent with our findings and indicate that immune system activation occurs in the degenerative menisci. These findings, together with our findings, also suggest that increased expressions of HLA-DRB and IL-7R is likely a phenomenon associated with both the normal meniscal aging process and OA disease process whereas the increased expression of FCER1A is a phenomenon only associated with the normal meniscal aging process.
The genes displayed lower expression in the degenerative menisci derived from older patients included COL2A1 (−10.38 fold change) and FGFR3 (−4.65 fold change) [26]. These findings, together with our findings [23], indicate that the decreased expression of COL2A1 is likely a phenomenon associated with both the normal meniscal aging process and OA disease process whereas the decreased expression of FGFR3 is a phenomenon associated with the meniscal aging process. Our findings presented in this study demonstrate that PC affects the expressions of many genes involved in both OA disease process and meniscal aging process in the absence of calcium crystals. This suggests that PC is not only potentially a disease-modifying drug for calcification-induced OA therapy but also potentially a disease-modifying drug for noncalcification-induced arthritis therapy such as posttraumatic OA.
5. Conclusions
OA is a disease associated with immune system activation and decreased expression of chondroprotective protein SOX-9. PC exerts its disease-modifying activity on OA, at least in part, by suppressing immune system activation and stimulating the production of extracellular cellular matrix proteins and chondroprotective proteins. PC is potentially a disease-modifying drug for noncalcification-induced arthritis therapy.
Acknowledgments
This study is supported in part by a Charlotte-Mecklenburg Education and Research Foundation grant and a Mecklenburg County Medical Society Smith Arthritis Fund grant (to Yubo Sun). This study was performed at Carolinas Medical Center, Charlotte, NC, USA.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Bennett L. D., Buckland-Wright J. C. Meniscal and articular cartilage changes in knee osteoarthritis: a cross-sectional double-contrast macroradiograpahic study. Rheumatology. 2002;41(8):917–923. doi: 10.1093/rheumatology/41.8.917. [DOI] [PubMed] [Google Scholar]
- 2.Hunter D. J., Zhang Y. Q., Tu X., LaValley M., Niu J. B., Amin S., Guermazi A., Genant H., Gale D., Felson D. T. Change in joint space width: hyaline articular cartilage loss or alteration in meniscus? Arthritis & Rheumatism. 2006;54(8):2488–2495. doi: 10.1002/art.22016. [DOI] [PubMed] [Google Scholar]
- 3.Englund M., Guermazi A., Roemer F. W., Aliabadi P., Yang M., Lewis C. E., Torner J., Nevitt M. C., Sack B., Felson D. T. Meniscal tear in knees without surgery and the development of radiographic osteoarthritis among middle-aged and elderly persons: the multicenter osteoarthritis study. Arthritis and Rheumatism. 2009;60(3):831–839. doi: 10.1002/art.24383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y., Mauerhan D. R., Honeycutt P. R., Kneisl J. S., Norton J. H., Hanley E. N., Gruber H. E. Analysis of meniscal degeneration and meniscal gene expression. BMC Musculoskeletal Disorders. 2010;11, article 19 doi: 10.1186/1471-2474-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Wluka A. E., Pelletier J.-P., Martel-Pelletier J., Abram F., Ding C., Cicuttini F. M. Meniscal extrusion predicts increases in subchondral bone marrow lesions and bone cysts and expansion of subchondral bone in osteoarthritic knees. Rheumatology (Oxford, England) 2010;49(5):997–1004. doi: 10.1093/rheumatology/keq034. [DOI] [PubMed] [Google Scholar]
- 6.Ashraf S., Wibberley H., Mapp P. I., Hill R., Wilson D., Walsh D. A. Increased vascular penetration and nerve growth in the meniscus: a potential source of pain in osteoarthritis. Annals of the Rheumatic Diseases. 2011;70(3):523–529. doi: 10.1136/ard.2010.137844. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y., Mauerhan D. R., Honeycutt P. R., Kneisl J. S., Norton H. J., Zinchenko N., Hanley E. N., Jr., Gruber H. E. Calcium deposition in osteoarthritic meniscus and meniscal cell culture. Arthritis Research and Therapy. 2010;12(2, article R56) doi: 10.1186/ar2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y., Mauerhan D. R. Meniscal calcification, pathogenesis and implications. Current Opinion in Rheumatology. 2012;24(2):152–157. doi: 10.1097/BOR.0b013e32834e90c1. [DOI] [PubMed] [Google Scholar]
- 9.Englund M., Guermazi A., Lohmander L. S. The meniscus in knee osteoarthritis. Rheumatic Disease Clinics of North America. 2009;35(3):579–590. doi: 10.1016/j.rdc.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Tew W. P., Malis C. D., Howard J. E., Lehninger A. L. Phosphocitrate inhibits mitochondrial and cytosolic accumulation of calcium in kidney cells in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(9):5528–5532. doi: 10.1073/pnas.78.9.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehninger A. L. Mitochondria and biological mineralization processes: an exploration. Horizons in Biochemistry and Biophysics. 1977;4:1–30. [PubMed] [Google Scholar]
- 12.Shankar R., Crowden S., Sallis J. D. Phosphocitrate and its analogue N-sulpho-2-amino tricarballylate inhibit aortic calcification. Atherosclerosis. 1984;52(2):191–198. doi: 10.1016/0021-9150(84)90117-5. [DOI] [PubMed] [Google Scholar]
- 13.Nair D., Misra R. P., Sallis J. D., Cheung H. S. Phosphocitrate inhibits a basic calcium phosphate and calcium pyrophosphate dihydrate crystal-induced mitogen-activated protein kinase cascade signal transduction pathway. Journal of Biological Chemistry. 1997;272(30):18920–18925. doi: 10.1074/jbc.272.30.18920. [DOI] [PubMed] [Google Scholar]
- 14.Cheung H. S., Sallis J. D., Struve J. A. Specific inhibition of basic calcium phosphate and calcium pyrophosphate crystal-induction of metalloproteinase synthesis by phosphocitrate. Biochimica et Biophysica Acta: Molecular Basis of Disease. 1996;1315(2):105–111. doi: 10.1016/0925-4439(95)00106-9. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y., Reuben P., Wenger L., Sallis J. D., Demadis K. D., Cheung H. S. Inhibition of calcium phosphate-DNA coprecipitates induced cell death by phosphocitrates. Frontiers in Bioscience. 2005;10:803–808. doi: 10.2741/1574. [DOI] [PubMed] [Google Scholar]
- 16.Cheung H. S., Sallis J. D., Demadis K. D., Wierzbicki A. Phosphocitrate blocks calcification-induced articular joint degeneration in a guinea pig model. Arthritis and Rheumatism. 2006;54(8):2452–2461. doi: 10.1002/art.22017. [DOI] [PubMed] [Google Scholar]
- 17.Cheung H. S. Phosphocitrate as a potential therapeutic strategy for crystal deposition disease. Current Rheumatology Reports. 2001;3(1):24–28. doi: 10.1007/s11926-001-0047-9. [DOI] [PubMed] [Google Scholar]
- 18.Ding M., Danielsen C. C., Hvid I. The effects of bone remodeling inhibition by alendronate on three-dimensional microarchitecture of subchondral bone tissues in guinea pig primary osteoarthrosis. Calcified Tissue International. 2008;82(1):77–86. doi: 10.1007/s00223-007-9093-2. [DOI] [PubMed] [Google Scholar]
- 19.Walton M. The effects of long-term administration of ethane-1-hydroxy-1, 1-diphosphonate on osteoarthrosis and heterotopic ossification in the mouse knee joint. Clinical Orthopaedics and Related Research. 1981;155:218–223. [PubMed] [Google Scholar]
- 20.Sun Y., Mauerhan D. R., Franklin A. M., Norton J., Hanley E. N., Gruber H. E. Phosphocitrate is potentially a disease-modifying drug for noncrystal-associated osteoarthritis. BioMed Research International. 2013;2013 doi: 10.1155/2013/326267.326267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y., Roberts A., Mauerhan D. R., Sun A. R., Norton H. J., Hanley E. N., Jr. Biological activities of phosphocitrate: a potential meniscal protective agent. BioMed Research International. 2013;2013 doi: 10.1155/2013/726581.726581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turhanen P. A., Demadis K. D., Peräniemi S., Vepsäläinen J. J. A novel strategy for the preparation of naturally occurring phosphocitrate and its partially esterified derivatives. Journal of Organic Chemistry. 2007;72(4):1468–1471. doi: 10.1021/jo061709c. [DOI] [PubMed] [Google Scholar]
- 23.Sun Y., Mauerhan D. R., Kneisl J. S., James Norton H., Zinchenko N., Ingram J., Hanley E. N., Jr., Gruber H. E. Histological examination of collagen and proteoglycan changes in osteoarthritic menisci. The Open Rheumatology Journal. 2012;6(1):24–32. doi: 10.2174/1874312901206010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H. A., Cho M.-L., Choi H. Y., Yoon C. S., Jhun J. Y., Oh H. J., Kim H.-Y. The catabolic pathway mediated by Toll-like receptors in human osteoarthritic chondrocytes. Arthritis & Rheumatism. 2006;54(7):2152–2163. doi: 10.1002/art.21951. [DOI] [PubMed] [Google Scholar]
- 25.Barreto G., Sillat T., Soininen A., Ylinen P., Salem A., Konttinen Y. T., Al-Samadi A., Nordström D. C. E. Do changing toll-like receptor profiles in different layers and grades of osteoarthritis cartilage reflect disease severity? The Journal of Rheumatology. 2013;40(5):695–702. doi: 10.3899/jrheum.121159. [DOI] [PubMed] [Google Scholar]
- 26.Rai M. F., Patra D., Sandell L. J., Brophy R. H. Transcriptome analysis of injured human meniscus reveals a distinct phenotype of meniscus degeneration with aging. Arthritis and Rheumatism. 2013;65(8):2090–2101. doi: 10.1002/art.37984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masuda M., Morimoto T., Kobatake S., Nishimura N., Nakamoto K., Dong X. H., Komiyama Y., Ogawa R., Takahashi H. Measurement of soluble Fcγ receptor type IIIa derived from macrophages in plasma: increase in patients with rheumatoid arthritis. Clinical and Experimental Immunology. 2003;132(3):477–484. doi: 10.1046/j.1365-2249.2003.02168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijngaarden S., van Roon J. A. G., Bijlsma J. W. J., van de Winkel J. G. J., Lafeber F. P. J. G. Fcγ receptor expression levels on monocytes are elevated in rheumatoid arthritis patients with high erythrocyte sedimentation rate who do not use anti-rheumatic drugs. Rheumatology. 2003;42(5):681–688. doi: 10.1093/rheumatology/keg174. [DOI] [PubMed] [Google Scholar]
- 29.Tedla N., Gibson K., McNeil H. P., Cosman D., Borges L., Arm J. P. The co-expression of activating and inhibitory leukocyte immunoglobulin-like receptors in rheumatoid synovium. The American Journal of Pathology. 2002;160(2):425–431. doi: 10.1016/S0002-9440(10)64861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tedla N., An H., Borges L., Vollmer-Conna U., Bryant K., Geczy C., Patrick Mcneil H. Expression of activating and inhibitory leukocyte immunoglobulin-like receptors in rheumatoid synovium: correlations to disease activity. Tissue Antigens. 2011;77(4):305–316. doi: 10.1111/j.1399-0039.2011.01633.x. [DOI] [PubMed] [Google Scholar]
- 31.Coulthard L. R., Geiler J., Mathews R. J., Church L. D., Dickie L. J., Cooper D. L., Wong C., Savic S., Bryer D., Buch M. H., Emery P., Morgan A. W., Mcdermott M. F. Differential effects of infliximab on absolute circulating blood leucocyte counts of innate immune cells in early and late rheumatoid arthritis patients. Clinical and Experimental Immunology. 2012;170(1):36–46. doi: 10.1111/j.1365-2249.2012.04626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huynh O. A., Hampartzoumian T., Arm J. P., Hunt J., Borges L., Ahern M., Smith M., Geczy C. L., McNeil H. P., Tedla N. Down-regulation of leucocyte immunoglobulin-like receptor expression in the synovium of rheumatoid arthritis patients after treatment with disease-modifying anti-rheumatic drugs. Rheumatology. 2007;46(5):742–751. doi: 10.1093/rheumatology/kel405. [DOI] [PubMed] [Google Scholar]
- 33.Haywood L., McWilliams D. F., Pearson C. I., Gill S. E., Ganesan A., Wilson D., Walsh D. A. Inflammation and angiogenesis in osteoarthritis. Arthritis & Rheumatism. 2003;48(8):2173–2177. doi: 10.1002/art.11094. [DOI] [PubMed] [Google Scholar]
- 34.Bonnet C. S., Walsh D. A. Osteoarthritis, angiogenesis and inflammation. Rheumatology. 2005;44(1):7–16. doi: 10.1093/rheumatology/keh344. [DOI] [PubMed] [Google Scholar]
- 35.Ashraf S., Mapp P. I., Walsh D. A. Contributions of angiogenesis to inflammation, joint damage, and pain in a rat model of osteoarthritis. Arthritis and Rheumatism. 2011;63(9):2700–2710. doi: 10.1002/art.30422. [DOI] [PubMed] [Google Scholar]
- 36.Tang C.-H., Hsu C.-J., Fong Y.-C. The CCL5/CCR5 axis promotes interleukin-6 production in human synovial fibroblasts. Arthritis Care and Research. 2010;62(12):3615–3624. doi: 10.1002/art.27755. [DOI] [PubMed] [Google Scholar]
- 37.Alaaeddine N., Olee T., Hashimoto S., Creighton-Achermann L., Lotz M. Production of the chemokine RANTES by articular chondrocytes and role in cartilage degradation. Arthritis & Rheumatology. 2001;44(7):1633–1643. doi: 10.1002/1529-0131(200107)44:7<1633::AID-ART286>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko S., Satoh T., Chiba J., Ju C., Inoue K., Kagawa J. Interleukin-6 and interleukin-8 levels in serum and synovial fluid of patients with osteoarthritis. Cytokines, Cellular and Molecular Therapy. 2000;6(2):71–79. doi: 10.1080/13684730050515796. [DOI] [PubMed] [Google Scholar]
- 39.Tsuchida A. I., Beekhuizen M., Rutgers M., van Osch G. J. V. M., Bekkers J. E. J., Bot A. G. J., Geurts B., Dhert W. J. A., Saris D. B. F., Creemers L. B. Interleukin-6 is elevated in synovial fluid of patients with focal cartilage defects and stimulates cartilage matrix production in an in vitro regeneration model. Arthritis Research and Therapy. 2012;14(6, article R262) doi: 10.1186/ar4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetlow L. C., Adlam D. J., Woolley D. E. Matrix metalloproteinase and proinflammatory cytokine production by chondrocytes of human osteoarthritic cartilage: associations with degenerative changes. Arthritis & Rheumatology. 2001;44:585–594. doi: 10.1002/1529-0131(200103)44:3<585::AID-ANR107>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 41.van Lent P. L., Blom A. B., Schelbergen R. F., Slöetjes A., Lafeber F. P. J. G., Lems W. F., Cats H., Vogl T., Roth J., van den Berg W. B. Active involvement of alarmins S100A8 and S100A9 in the regulation of synovial activation and joint destruction during mouse and human osteoarthritis. Arthritis & Rheumatism. 2012;64(5):1466–1476. doi: 10.1002/art.34315. [DOI] [PubMed] [Google Scholar]
- 42.Enomoto H., Inoki I., Komiya K., Shiomi T., Ikeda E., Obata K.-I., Matsumoto H., Toyama Y., Okada Y. Vascular endothelial growth factor isoforms and their receptors are expressed in human osteoarthritic cartilage. The American Journal of Pathology. 2003;162(1):171–181. doi: 10.1016/S0002-9440(10)63808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermann L. M., Pinkerton M., Jennings K., Yang L., Grom A., Sowders D., Kersten S., Witte D. P., Hirsch R., Thornton S. Angiopoietin-like-4 is a potential angiogenic mediator in arthritis. Clinical Immunology. 2005;115(1):93–101. doi: 10.1016/j.clim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Murata M., Yudo K., Nakamura H., Chiba J., Okamoto K., Suematsu N., Nishioka K., Beppu M., Inoue K., Kato T., Masuko K. Hypoxia upregulates the expression of angiopoietin-like-4 in human articular chondrocytes: role of angiopoietin-like-4 in the expression of matrix metalloproteinases and cartilage degradation. Journal of Orthopaedic Research. 2009;27(1):50–57. doi: 10.1002/jor.20703. [DOI] [PubMed] [Google Scholar]
- 45.Pickens S. R., Chamberlain N. D., Volin M. V., Pope R. M., Talarico N. E., Mandelin A. M., II, Shahrara S. Characterization of interleukin-7 and interleukin-7 receptor in the pathogenesis of rheumatoid arthritis. Arthritis and Rheumatism. 2011;63(10):2884–2893. doi: 10.1002/art.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartgring S. A. Y., Willis C. R., Alcorn D., Nelson L. J., Bijlsma J. W. J., Lafeber F. P. J. G., Van Roon J. A. G. Blockade of the interleukin-7 receptor inhibits collagen-induced arthritis and is associated with reduction of T cell activity and proinflammatory mediators. Arthritis and Rheumatism. 2010;62(9):2716–2725. doi: 10.1002/art.27578. [DOI] [PubMed] [Google Scholar]
- 47.Lane N. E. Pain management in osteoarthritis: the role of COX-2 inhibitors. Journal of Rheumatology. 1997;24(49):20–24. [PubMed] [Google Scholar]
- 48.Valverde-Franco G., Binette J. S., Li W., Wang H., Chai S., Laflamme F., Tran-Khanh N., Quenneville E., Meijers T., Poole A. R., Mort J. S., Buschmann M. D., Henderson J. E. Defects in articular cartilage metabolism and early arthritis in fibroblast growth factor receptor 3 deficient mice. Human Molecular Genetics. 2006;15(11):1783–1792. doi: 10.1093/hmg/ddl100. [DOI] [PubMed] [Google Scholar]
- 49.Cucchiarini M., Thurn T., Weimer A., Kohn D., Terwilliger E. F., Madry H. Restoration of the extracellular matrix in human osteoarthritic articular cartilage by overexpression of the transcription factor SOX9. Arthritis and Rheumatism. 2007;56(1):158–167. doi: 10.1002/art.22299. [DOI] [PubMed] [Google Scholar]
- 50.Tchetina E. V., Squires G., Poole A. R. Increased type II collagen degradation and very early focal cartilage degeneration is associated with upregulation of chondrocyte differentiation related genes in early human articular cartilage lesions. Journal of Rheumatology. 2005;32(5):876–886. [PubMed] [Google Scholar]
- 51.Brew C. J., Clegg P. D., Boot-Handford R. P., Andrew J. G., Hardingham T. Gene expression in human chondrocytes in late osteoarthritis is changed in both fibrillated and intact cartilage without evidence of generalised chondrocyte hypertrophy. Annals of the Rheumatic Diseases. 2010;69(1):234–240. doi: 10.1136/ard.2008.097139. [DOI] [PubMed] [Google Scholar]
- 52.Lee J. S., Im G. I. SOX trio decrease in the articular cartilage with the advancement of osteoarthritis. Connective Tissue Research. 2011;52(6):496–502. doi: 10.3109/03008207.2011.585409. [DOI] [PubMed] [Google Scholar]