SUMMARY
CRISPR-Cas9 is a versatile genome editing technology for studying the function of genetic elements. To broadly enable the application of Cas9 in vivo, we established a Cre-dependent Cas9 knockin mouse. We demonstrated in vivo as well as ex vivo genome editing using adeno-associated virus (AAV)-, lenti-virus-, or particle-mediated delivery of guide RNA in neurons, immune cells, and endothelial cells. Using these mice, we simultaneously modeled the dynamics of KRAS, p53, and LKB1, the top three significantly mutated genes in lung adenocarcinoma. Delivery of a single AAV vector in the lung generated loss-of-function mutations in p53 and LKB1, as well as homology-directed repair-mediated KRASG12D mutations, leading to macroscopic tumors of adeno-carcinoma pathology. Together, these results suggest that Cas9 mice empower a wide range of biological and disease modeling applications.
INTRODUCTION
Molecular genetic studies are continuously transforming our knowledge in biology and medicine. Forward and reverse genetics in cells and animal models is key to discovering causal mechanisms relating molecular events to phenotypes (Griffiths, 2012). Traditionally, genetic manipulations in mammalian spe cies are often made in the germline of an organism, which can then be used to create a stable transgenic strain for experimentation (Griffiths, 2012; Nagy, 2003). Recently, the RNA-guided endonuclease Cas9 from microbial type II CRISPR (clustered regularly interspaced short palindromic repeat) systems (previously referred to as Cas5 or Csn1) (Barrangou et al., 2007; Bolotin et al., 2005; Deltcheva et al., 2011; Garneau et al., 2010; Gasiunas et al., 2012; Jinek et al., 2012; Sapranauskas et al., 2011) has been harnessed to facilitate facile genetic manipulations in a variety of cell types and organisms (reviewed in Hsu et al., 2014).
Cas9 can be easily reprogrammed using RNA guides to generate targeted DNA double-strand breaks (DSBs), which can stimulate genome editing via one of the two DNA damage repair pathways: nonhomologous end-joining (NHEJ), resulting in insertions and deletions (indels) (Bibikova et al., 2002), or homology-directed repair (HDR) (Bibikova et al., 2003; Bibikova et al., 2001; Jasin et al., 1985; Rudin et al., 1989), resulting in precise sequence substitution in the presence of a repair template. Unlike other programmable nuclease systems used for genome editing, a unique advantage of the Cas9 system is that Cas9 can be combined with multiple single-guide RNAs (sgRNAs) (Jinek et al., 2012) to achieve efficient multiplexed genome editing in mammalian cells (Cong et al., 2013; Mali et al., 2013). This accelerates the study of multigenic processes, such as the role of mutation combinations in tumor evolution.
Though Cas9 has been broadly applied in a variety of cell-line- and embryo-based experiments, in vivo applications of Cas9 in somatic tissue remain challenging, owing to a combination of factors such as its large transgene size. Commonly used delivery systems based on lentiviral and adeno-associated viral (AAV) vectors have limited packaging capacity (Kumar et al., 2001; Wu et al., 2010), which renders it challenging for incorporation of Cas9 along with sgRNA expression cassettes and necessary genetic elements (i.e., promoters, fluorescent proteins, and polyadenlyation sequences). Higher-capacity viral vectors, such as adenovirus, can be used to deliver large transgenes (Ding et al., 2014), but generally have higher immunogenicity, limited cell type specificity, and tissue tropism (Schirmbeck et al., 2008). Most recently, hydrodynamic injection, which can accommodate the large transgene size of Cas9, has been employed to achieve Cas9-mediated genome editing in the mouse liver in vivo (Xue et al., 2014; Yin et al., 2014), albeit with low editing efficiencies, and is primarily applicable to hepatocytes within the mouse liver. Given these challenges, there is an urgent need for a more versatile system to enable efficient Cas9-mediated genome editing for in vivo applications.
To facilitate broader applications of CRISPR-Cas9, we generated a Cre-dependent Rosa26 Cas9 knockin mouse to overcome the delivery challenges associated with Cas9. This mouse can be used in conjunction with a variety of guide RNA delivery reagents, providing an attractive model for studying the interplay of mutations in biological processes and disease. Here, we demonstrate that both viral and nonviral sgRNA delivery methods are efficient to facilitate genome editing in multiple tissues in vivo. In particular, we used this Cas9 mouse to generate loss-of-function mutations in tumor suppressor genes as well as gain-of-function mutations in a proto-oncogene. Successful genome editing in lung tissue led to formation of adenocarcinoma, allowing us to rapidly model the dynamics of multiple mutations in tumorigenesis. These applications broadly demonstrate the potential of the Cas9 mouse in facilitating rapid screening of causal genetic mutations in a variety of biological and disease processes.
RESULTS
Generation of a Cre-Dependent Cas9 Knockin Transgenic Mouse
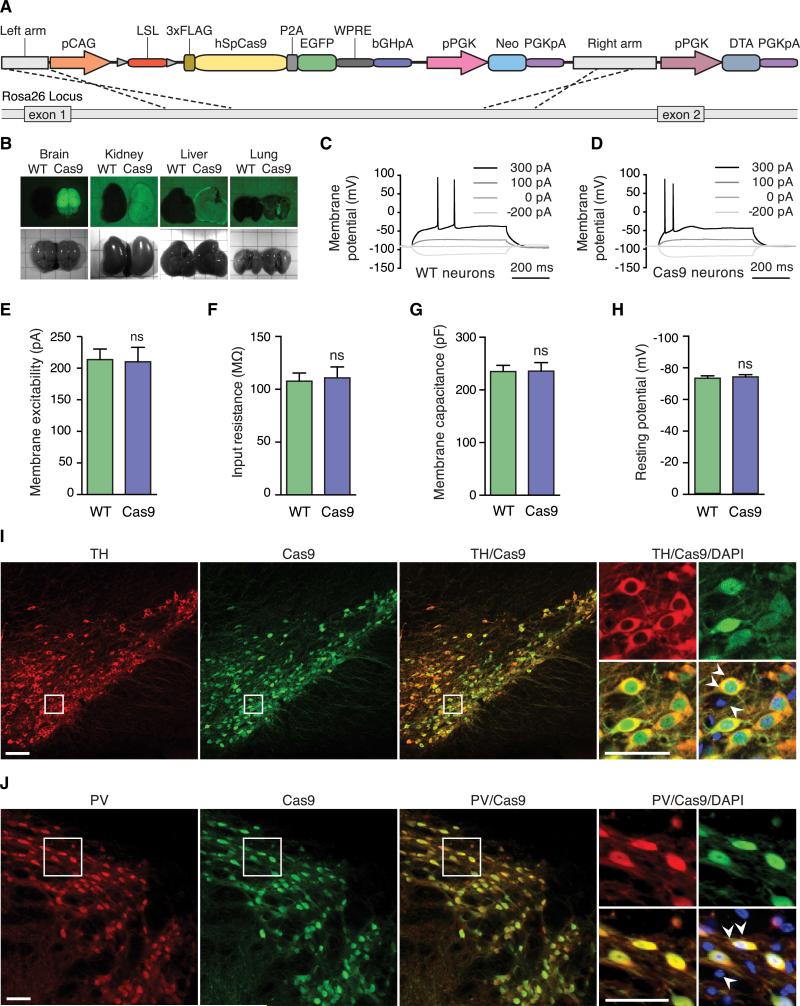
We generated a Cre-dependent Cas9 mouse by inserting a Cas9 transgene expression cassette into the Rosa26 locus (Figure 1A). The transgene consists of a 33 FLAG-tagged Streptococcus pyogenes Cas9 linked via a self-cleaving P2A peptide to an enhanced green fluorescent protein (EGFP) to facilitate visualization of Cas9-expressing cells. The transgene is driven by the ubiquitous CAG promoter and is interrupted by a loxP-stop (33 polyA signal)-loxP (LSL) cassette to render Cas9 expression inducible by the Cre recombinase (Figure 1A).
Figure 1. Generation and Characterization of Cre-Dependent and Constitutive Cas9-Expressing Mice.
(A) Schematic of the Cre-dependent Cas9 Rosa26 targeting vector.
(B) Bright-field and fluorescence stereomicroscope images of tissues dissected from constitutive Cas9-expressing (left) and wild-type (right) mice, showing Cas9-P2A-EGFP expression only in Cas9 mice.
(C and D) Representative current-clamp recordings and evoked action potentials from wild-type (C) and constitutive Cas9-expressing (D) neurons, showing no difference.
(E–H) Electrophysiological characterization of hippocampal neurons in acute slices from constitutive Cas9-expressing and wild-type neurons, showing no significant difference in: membrane excitability (E), input resistance (F), membrane capacitance (G), and resting potential (H). Data are plotted as mean ± SEM; n = 12 neurons from two wild-type mice and n = 15 neurons from two constitutive Cas9-expressing mice. n.s., not significant. See also Table S1.
(I) Representative immunofluorescence images of the substantia nigra in progenies from a Cre-dependent Cas9 mouse crossed with a TH-IRES-Cre driver mouse, showing Cas9 expression is restricted to TH-positive cells. Double arrowheads indicate a cell coexpressing TH and Cas9-P2A-EGFP. Single arrowhead indicates a cell expressing neither TH nor Cas9-P2A-EGFP. Scale bar, 100 μm.
(J) Representative immunofluorescence images of the reticular thalamus in progenies from a Cre-dependent Cas9 mouse crossed with a PV-Cre driver mouse, showing that Cas9 expression is restricted to PV-positive cells. Double arrowheads indicate a cell expressing PV and Cas9-P2A-EGFP. Single arrowhead indicates a cell expressing neither PV nor Cas9-P2A-EGFP. Scale bar, 50 μm.
Characterization of Constitutive- and Tissue-Specific Cas9 Expression in Mice
To evaluate the effect of prolonged Cas9 expression, we generated a constitutive Cas9-expressing mouse line by crossing the Cre-dependent Cas9 mouse to a β-actin Cre driver (Lewandoski et al., 1997). Resulting progenies of this cross were viable, and Cas9-P2A-EGFP expression was observed throughout the body (Figure 1B). The constitutive Cas9-expressing mice were fertile, had normal litter sizes, presented no morphological abnormalities, and were able to breed to homozygosity. At the cellular level, we also found no morphological abnormalities or upregulation in DNA damage and apoptosis markers (Figure S1 available online).
To further gauge whether constitutive Cas9 expression had adverse effects in cellular physiology, we used a panel of electrophysiological measurements to evaluate the health of Cas9-expressing neurons, a cell type particularly sensitive to perturbations. Therefore, we performed whole-cell patch clamp recordings in CA1 pyramidal neurons from acute hippo-campal slices to examine firing threshold (Figures 1C and 1D) and membrane properties (membrane excitability, input resistance, membrane capacitance, resting potential; Figures 1E– 1H and Table S1) and found no significant differences between wild-type and Cas9-expressing neurons.
With the conditional Cas9 mouse, tissue- and cell-type-specific promoters (Lewandoski, 2001) can facilitate defined spatio-temporal expression of Cas9. To demonstrate this, we crossed the Cre-dependent Cas9 mouse with two Cre driver strains, namely the tyrosine hydroxylase (TH-IRES-Cre) driver for dopaminergic neurons and the parvalbumin (PV-Cre) driver for a subtype of inhibitory interneurons (Hippenmeyer et al., 2005; Lindeberg et al., 2004). As predicted, Cas9 expression was restricted to TH- or PV-positive cells in the F1 progenies of these two crosses (Figure 1I–1J).
Ex Vivo Genome Editing in Primary Dendritic Cells by Lentiviral-Mediated sgRNA Expression
To determine whether the Rosa26 knockin construct provided functional levels of Cas9 expression, we set out to test whether a previously described U6-sgRNA lentiviral vector (Sanjana et al., 2014) could mediate indel formation ex vivo in primary immune cells. Several types of immune cells, such as innate immune dendritic cells (DCs), are often not accessible for genetic manipulation due to delivery challenges, short viability terms in culture, or both. Moreover, because existing cell lines do not mimic DC biology well, many studies are performed with primary cells derived ex vivo from precursors isolated from the bone marrow (BMDCs) (Figure 2A), which retain many critical characteristics of DCs in vivo (Amit et al., 2009; Chevrier et al., 2011; Garber et al., 2012; Shalek et al., 2013). We thus reasoned that Cas9-expressing cells derived from the constitutive Cas9-expressing mice may facilitate such applications, as genome editing would only require introduction of sgRNAs, which can be efficiently delivered using lentiviral vectors.
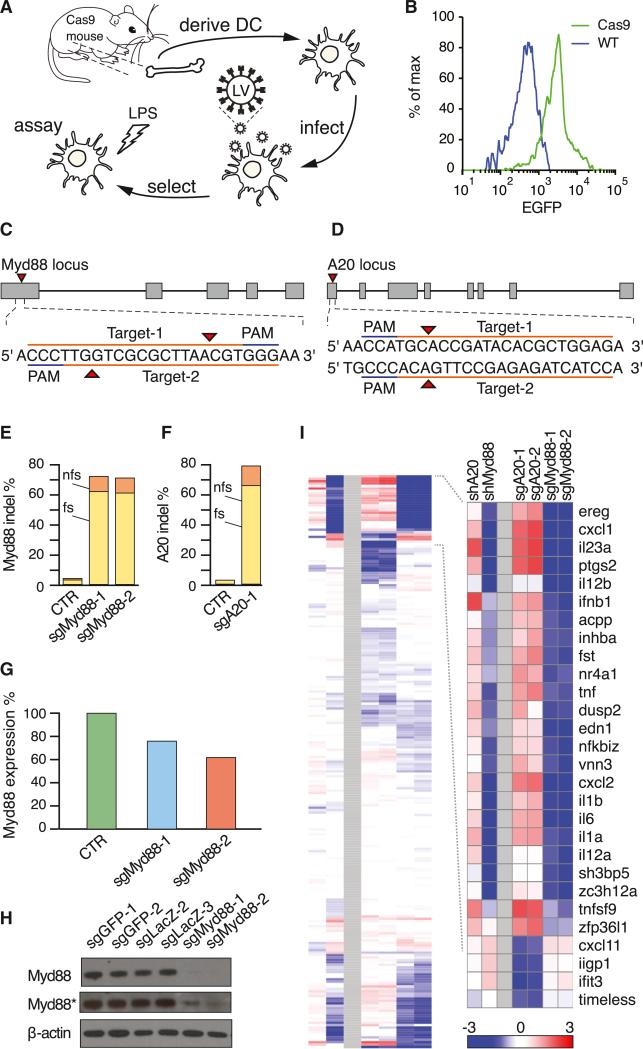
Figure 2. Ex Vivo Genome Editing of Primary Immune Cells Derived from Constitutive Cas9-Expressing Mice.
(A) Schematic of ex vivo genome editing experimental flow.
(B) Flow cytometry histogram of bone marrow cells from constitutive Cas9-expressing (green) and wild-type (blue) mice, showing Cas9-P2A-EGFP expression only in Cas9 mice. Data are plotted as a percentage of the total number of cells.
(C) sgRNA design for targeting the mouse Myd88 locus.
(D) sgRNA design for targeting the mouse A20 locus.
(E) Myd88 indel analysis of constitutive Cas9-expressing DCs transduced with either a Myd88-targeting sgRNA (sgMyd88-1 and sgMyd88-2) or controls (CTR, average of four control sgRNAs), showing indel formation only in Myd88-targeted cells. Data are plotted as the percent of Illumina sequencing reads containing indels at the target site. Mutations are categorized as frameshift (fs, yellow bar) or non-frameshift (nfs, orange bar).
(F) A20 indel analysis of constitutive Cas9-expressing DCs transduced with either an A20-targeting sgRNA (sgA20-1) or controls (CTR, average of four control sgRNAs), showing indel formation only in A20-targeted cells. Data are plotted as the percent of Illumina sequencing reads containing indels at the target site. Mutations are categorized as frameshift (fs, yellow bar) or non-frameshift (nfs, orange bar).
(G) Myd88 mRNA quantification of constitutive Cas9-expressing DCs transduced with either Myd88-targeting sgRNA (sgMyd88-1 or sgMyd88-2) or controls (CTR, average of six control sgRNAs), showing reduced expression only in Myd88-targeted cells. Data are plotted as Myd88 mRNA levels from Nanostring nCounter analysis.
(H) Immunoblot of constitutive Cas9-expressing DCs transduced with either Myd88-targeting sgRNA (sgMyd88-1 or sgMyd88-2) or controls (four control sgRNAs), showing depletion of Myd88 protein only in Myd88-targeted cells. β-actin was used as a loading control. (Asterisk) Overexposed, repeated-measurement.
(I) Nanostring nCounter analysis of constitutive Cas9-expressing DCs transduced with either Myd88-targeting sgRNA (sgMyd88-1 or sgMyd88-2) or shRNA (shMyd88), A20-targeting sgRNA (sgA20-1 or sgA20-2), or shRNA (shA20), showing an altered LPS response. (Inset) The cluster showing the highest difference between Myd88- and A20-targeting sgRNAs, including key inflammatory genes (IL1a, IL1b, Cxcl1, Tnf, etc.). (Red) High; (blue) low; (white) unchanged; based on fold change relative to measurements with six control sgRNAs. See also Figure S2.
We first verified the expression of Cas9 in bone marrow from constitutive Cas9-expressing mice (Figure 2B). Similarly, we validated Cas9 expression in many other immune cell types (Figure S2). Two days after culturing bone marrow cells from the constitutive Cas9-expressing mice, we infected BMDCs with lentivirus encoding two different sgRNAs targeting early exons of either MyD88 (Figure 2C) or A20 (Figure 2D), two well-characterized positive and negative regulators of Toll-like receptor 4 (TLR4) signaling, respectively. At 7 days posttransduction, we activated cells with lipopolysaccharide (LPS) and performed functional analysis (Figure 2A). We found indels in 67%–78% of sequencing reads (Figures 2E and 2F), leading to reduction in mRNA (Figure 2G), and protein (Figure 2H).
DCs specialize in pathogen detection and initiation of appropriate immune responses (Mellman and Steinman, 2001). Therefore, we measured the expression of 276 representative genes of the LPS response, using the Nanostring nCounter, in cells targeted for Myd88 or A20 as compared to controls (Figure 2I). As predicted, depletion of MyD88 resulted in a reduction of inflammatory response genes, whereas depletion of A20 resulted in an increase of inflammatory response genes. These effects were comparable to those observed with shRNA-mediated knockdown in independent experiments (Figure 2I). Taken together, our results demonstrate the potential of constitutive Cas9-expressing mice for efficient perturbation in primary cells.
In Vivo Genome Editing in the Brain
To demonstrate direct genome editing in vivo in the Cre-dependent Cas9 mouse, we applied AAV-mediated expression of Cre and sgRNA in the brain (Figure 3A). We constructed an AAV-U6-sgRNA-Cre vector (Figure 3B) containing an sgRNA targeting a highly expressed neuronal-specific RNA-splicing factor, NeuN (RbFox3) (sgNeuN) (Figure 3C). A chimeric AAV1/2 vector was packaged using an equal mixture of AAV1 and two packaging plasmids, which was previously shown to efficiently infect neurons (Konermann et al., 2013). The packaged virus was delivered via stereotactic injection into the prefrontal cortex of Cre-dependent Cas9 mice (Figure 3A). Three weeks postinjection, we dissected the injected brain region. Deep sequencing of the NeuN locus showed indel formation near the predicted cleavage site (Figure 3C), suggesting that Cas9 was functional and facilitated on-target indel formation. Furthermore, we observed NeuN protein depletion only in the injected region (Figures 3D– 3F), but not in controls (noninjected and LacZ-targeted sgRNA, termed sgLacZ) (Figures 3D–3F). To quantify this effect, we performed immunoblot analysis on tissue samples from three mice and found that NeuN protein expression was reduced by 80% (Figure 3G).
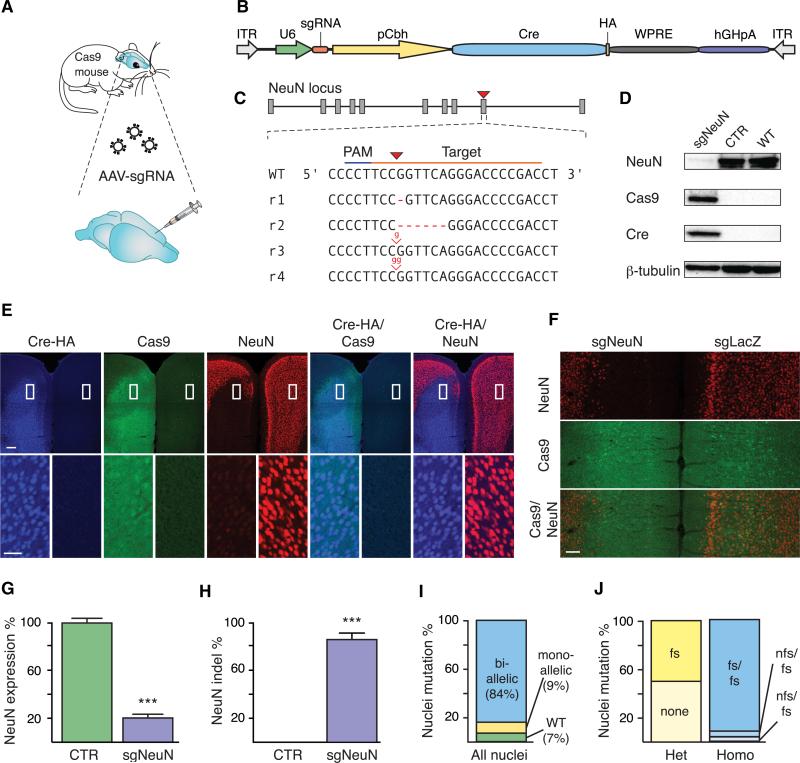
Figure 3. In Vivo Genome Editing in the Brain of Cre-Dependent Cas9 Mice.
(A) Schematic showing experimental procedure for stereotactic delivery of sgNeuN-expressing AAV into the prefrontal cortex of Cre-dependent Cas9 mice.
(B) Schematic of AAV vector for sgRNA expression.
(C) sgRNA design for targeting the mouse NeuN locus and representative Illumina sequencing reads (rn) from Cre-dependent Cas9 mice injected with AAV1/2-NeuN, showing indel formation at the target site (red arrow).
(D) Representative immunoblot of brain tissue dissected from Cre-dependent Cas9 mice injected with either AAV1/2-sgNeuN or AAV1/2-sgLacZ or not injected, showing NeuN depletion only in NeuN-targeted mice. β-tubulin was used as a loading control.
(E) Representative immunofluorescence images of the prefrontal cortex of Cre-dependent Cas9 mice injected with AAV1/2-sgNeuN 3 weeks posttransduction, showing Cre-mediated activation of Cas9-P2A-EGFP and corresponding NeuN depletion (NeuN). Bottom row shows magnified view of the boxed regions. Scale bars, 200 μm (top) and 50 μm (bottom).
(F) Representative immunofluorescence images of the prefrontal cortex of Cre-dependent Cas9 mice injected bilaterally with AAV1/2-sgNeuN (left hemisphere) or AAV1/2-sgLacZ (right hemisphere) 3 weeks posttransduction, showing NeuN depletion only in the NeuN-targeted hemisphere. Scale bar, 200 μm.
(G) Quantification of immunoblots of brain tissues dissected from Cre-dependent Cas9 mice injected with either AAV1/2-sgNeuN or AAV1/2-sgLacZ 3 weeks posttransduction, showing significant NeuN depletion only in NeuN-targeted mice. Data are plotted as mean ± SEM (n = 3 mice). ***p < 0.0005.
(H) NeuN indel analysis of populations of neuronal nuclei from Cre-dependent Cas9 mice injected with an AAV1/2-EGFP-KASH vector expressing either sgNeuN or sgLacZ 3 weeks posttransduction, showing significant indel formation only in NeuN-targeted cells. Data are plotted as the mean ± SEM (n = 3 mice). ***p < 0.0005.
(I) NeuN indel analysis of single neuronal nuclei from Cre-dependent Cas9 mice injected with an AAV1/2-EGFP-KASH vector expressing sgNeuN, showing that 84% of transduced neurons are mutated on both alleles. Individual nuclei are categorized as bi-allelic, mono-allelic, or wild-type. Data are plotted as a percent of nuclei (n = 167).
(J) NeuN indel analysis of single neuronal nuclei from Cre-dependent Cas9 mice injected with an AAV1/2-EGFP-KASH vector expressing sgNeuN, showing that most mutations are frameshift. Individual allele mutations from homozygous (n = 141) or heterozygous (n = 15) nuclei are categorized as either frameshift (fs) or non-frameshift (nfs). Data are plotted as a percent of homozygous (n = 141) or heterozygous (n = 15) nuclei.
To study the mutagenic effect of Cas9-mediated genome editing at the single-cell level, we set out to isolate individual neuronal nuclei according to a previously established method (Okada et al., 2011). To enable this, we generated a single vector (AAV-sgRNA-hSyn-Cre-P2A-EGFP-KASH) that expresses sgRNA, Cre, and EGFP fused to the KASH (Klarsicht, ANC-1, Syne Homology) nuclear transmembrane domain in neurons (L.S. and M. Heidenreich, unpublished data). After harvesting injected brain tissues, we used fluorescence-activated cell sorting (FACS) to isolate single EGFP-positive neuronal nuclei. Targeted amplicon sequencing of the target site from populations of EGFP-positive nuclei revealed an average indel rate of 85% (Figure 3H). Sequencing of 167 single nuclei showed that 84% (n = 141) of transduced cells had biallelic modifications, leaving 9% (n = 15) with monoallelic modification and 7% (n = 11) unmodified (Figure 3I). The 141 biallelic mutant nuclei contained both frame-shift (phase of 3n+1 or 3n+2) and non-frame-shift mutations (phase of 3n), and the 15 monoallelic mutant nuclei all contained frame-shift mutations in the modified alleles (Figure 3J).
In Vivo Modeling of Multigenic Cancer Mutations
The multiplexability of Cas9-mediated genome editing enables modeling of multigenic diseases processes such as tumorigenesis. The genomes of cancer cells possess complex combinations of genetic lesions (Weinberg, 2007), which usually include gain-of-function mutations in proto-oncogenes as well as loss-of-function mutations in tumor suppressor genes (Garraway and Lander, 2013; Lawrence et al., 2013). It is challenging to elucidate the roles of these complex combinations of genetic alterations in tumor evolution largely due to the difficulty of creating transgenic animals with multiple perturbed alleles. We therefore set out to model multi-lesion lung cancer in the Cre-dependent Cas9 mouse.
We chose to model lung cancer mutations involving the onco-gene KRAS and the tumor suppressor genes p53 and LKB1 (Stk11), which are the top three most frequently mutated genes in lung adenocarcinoma in human (46% for p53, 33% for KRAS and 17% for LKB1) (TCGA-Network, 2014). To model the dynamics of mutations in these three genes, we built a single vector capable of generating KRASG12D mutations while simultaneously knocking out p53 and LKB1 in the Cre-dependent Cas9 mouse.
Multiple sgRNAs were designed targeting the first exon near the G12D site in KRAS and early exons in p53 and LKB1 using a previously developed informatics tool (Hsu et al., 2013). To select an effective sgRNA for each gene, we individually expressed each sgRNA as well as Cas9 in a cancer cell line (Neuro-2a) and quantified the editing efficiency using the SURVEYOR nuclease assay. Most of these sgRNAs were capable of inducing indels in specific targeted loci (Figure S3A). One sgRNA was chosen for each of the three genes and was then used to generate a single vector containing U6 expression cassettes for all three sgRNAs. Transient transfection of this single vector resulted in significant indels in all three genes (35% KRAS, 44% p53, and 30% LKB1) (Figure S3B).
Whereas p53 and LKB1 mutations in patient tumors are often loss-of-function in nature, mutations in KRAS are often missense mutations that result in a gain of function (TCGA-Network, 2014). To model a missense gain-of-function KRAS mutation, we designed an HDR donor template, which consists of an 800 base-pair (bp) genomic sequence homologous to a region encompassing the first exon of the mouse KRAS gene. This HDR donor encoded: (1) a glycine (G) to aspartate (D) mutation in the 12th amino acid position (G12D), resulting in the oncogenic KRASG12D mutation; (2) 11 synonymous single-nucleotide changes to aid the distinction between the donor and wild-type sequences; and (3) protospacer-adjacent motif (PAM) mutations to prevent donor DNA cleavage by Cas9. To test the efficiency of HDR-mediated missense KRAS mutation, we used deep sequencing to assess the rate of G12D incorporation in Neuro-2a cells in vitro. Four percent of the sequencing reads identically matched the donor sequence, which included the G12D mutation and synonymous mutations (Figure S3C). Collectively, these data indicate that a single vector can stimulate HDR events to induce KRASG12D mutations as well as facilitate efficient editing of the p53 and LKB1 genes in vitro.
Cas9-Mediated Genome Editing of p53 LKB1, and KRAS in the Lung
After confirming that Cas9 was expressed in the lung after induction by Cre recombinase (Figures S4A and S4B), we evaluated the efficiency of AAV-mediated gene delivery (Figure 4A). A Firefly luciferase (Fluc) expression vector was packaged using two different serotypes of AAV (AAV6-Fluc and AAV9-Fluc), both of which are capable of transducing the lung efficiently (Bell et al., 2011; Halbert et al., 2002; Limberis and Wilson, 2006). The viruses were delivered using both intranasal (i.n.) and intratracheal (i.t.) methods (Figures 4A, 4B, and S4C). Using in vivo luminescence imaging, we detected luciferase activity in the lung 1 week after i.t. delivery of AAV9-Fluc, but not AAV6-Fluc. Both AAV6-Fluc and AAV9-Fluc showed positive signal in the lung by 3 weeks, but AAV9 generated a higher intensity (Figure S4C). Based on these data, we chose i.t. delivery of AAV9 vectors for all subsequent in vivo studies.
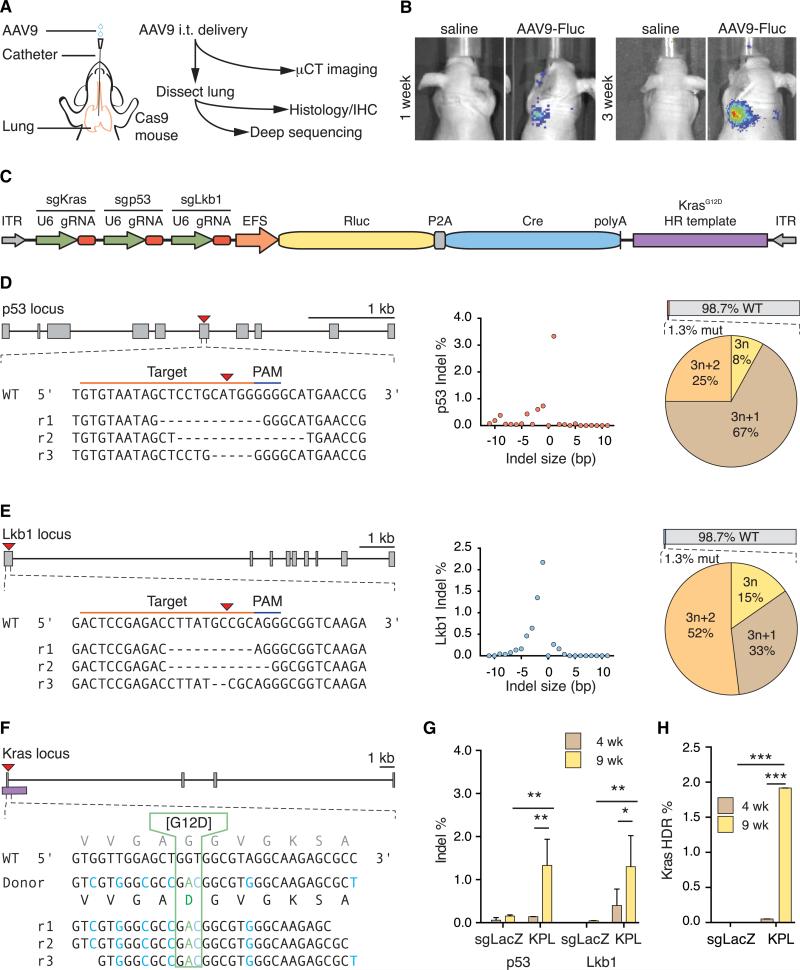
Figure 4. In Vivo AAV9-KPL Delivery and Mutation Analysis.
(A) Schematic of intratracheal (i.t.) delivery of AAV9 into the lung of a Cre-dependent Cas9 mouse and experimental flow.
(B) Luciferase imaging of nude mice injected with either AAV9-Fluc or saline, showing efficient AAV9-mediated expression in vivo in the lung.
(C) Schematic of the AAV-KPL vector.
(D and E) (Left) sgRNA designs for targeting the mouse p53 (D) and LKB1 (E) loci and representative Illumina sequencing reads (rn) from Cre-dependent Cas9 mice injected with AAV9-KPL, showing indel formation at the target site. (Middle) Size distribution of indels found at the target site. (Right) Indel analysis from whole lung (top) and the phase characteristics of edited alleles (bottom). p53 and LKB1 loci scale bars, 1 kb.
(F) sgRNA and HDR donor design for targeting the mouse KRAS locus for G12D incorporation and representative Illumina sequencing reads (rn) from Cre-dependent Cas9 mice injected with AAV9-KPL. Green text indicates the G12D mutation, whereas blue text indicates the intended synonymous mutations, showing successful generation of the KRASG12D mutation. KRAS locus scale bar, 1 kb.
(G) p53 and LKB1 indel analysis of whole lung from Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ, showing significant indel formation only in AAV9-KPL-injected mice. Data are plotted as mean ± SEM. *p < 0.05; **p < 0.005.
(H) KRASG12D mutation analysis of whole lung from Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ, showing significant G12D incorporation only in AAV9-KPL-injected mice. The data are plotted as the mean ± SEM. ***p < 0.0005.
See also Figures S3, S4, and S5.
We generated a single AAV vector integrating the KRASG12D HDR donor, the U6-sgRNA cassettes for KRAS, p53, and LKB1, as well as an expression cassette containing Cre recombinase and Renilla luciferase (AAV-KPL, Figure 4C). The AAV-KPL vector was packaged using AAV9 (AAV9-KPL) and delivered i.t. into Cre-dependent Cas9 mice, with a relatively low dose of virus for sparse targeting. Four weeks later, lungs were harvested from AAV9-KPL-transduced mice and AAV9-sgLacZ-transduced controls and were characterized by Illumina sequencing. We identified indels in p53 and LKB1 at the predicted cutting site, with 0.1% p53 indels and 0.4% LKB1 indels in the whole lung (Figures 4D, 4E, and 4G). A large fraction of these indels potentially disrupted the endogenous gene function because they were mostly out of frame (i.e., 3n+1bp or 3n+2bp in length) (Figures 4D and 4E). Furthermore, we detected 0.1% KRASG12D HDR events in the genomes of the lung cells, as indicated by the synonymous-SNP-barcoded G12D reads (Figures 4F and 4H). The frequency of indels in p53, and LKB1 and targeted KRASG12D mutations increased over time, as we detected 1.3% p53 indels, 1.3% LKB1 indels and 1.8% KRASG12D mutations in the whole lung at 9 weeks postdelivery (Figures 4G–4H). This was potentially due to the selective growth advantage of mutant cells. These data suggest that delivery of AAV9-KPL into the lungs of Cre-dependent Cas9 mice generates the KRASG12D mutation and putative loss-of-function mutations in p53 and LKB1.
Cas9-Mediated Mutations in p53, LKB1, and KRAS in the Lung Induced Tumor Formation
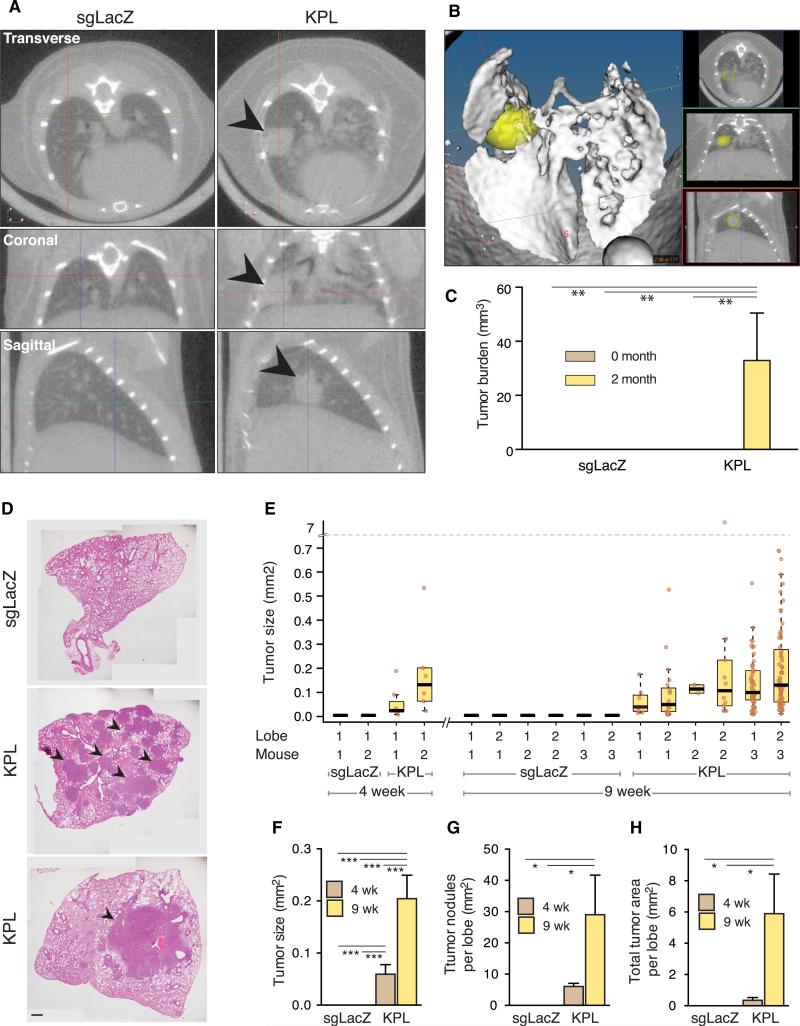
To investigate the phenotypic effect of delivery of AAV9-KPL, injected animals were imaged by micro-computed tomography (μCT). Two months postinjection, all (5 out of 5 = 100%) AAV9-KPL-treated animals developed nodules in the lung, whereas none (0/5 = 0%, Fisher’s exact test, one-tail, p = 0.008) of the AAV9-sgLacZ-treated animals showed detectable nodules (Figures 5A and 5B and Movie S1). Quantification showed that the average total tumor burden at 2 months was ~33 mm3 (Figure 5C), which was close to 10% of the total lung volume (Mitzner et al., 2001).
Figure 5. In Vivo Tumor Formation in AAV9-KPL-Injected Mice.
(A) Lung mCT images of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 2 months posttransduction, showing tumor formation (indicated by the arrowhead) only in AAV9-KPL injected mice.
(B) Lung μCT 3D rendering of Cre-dependent Cas9 mice injected with AAV9-KPL 2 months posttransduction, showing tumor formation (indicated by a yellow oval).
(C) Major tumor burden quantification of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ, showing significant tumor burden in AAV9-KPL-injected mice. Data are plotted as mean ± SEM. **p < 0.005.
(D) Representative lung H&E images of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks posttransduction, showing heterogeneous tumor formation in AAV9-KPL-injected mice. Arrowheads highlight a representative subset of tumors within the lungs of AAV9-KPL injected mice. Scale bar, 500 μm.
(E) Lung tumor size quantification of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks posttransduction. Data are plotted as a boxplot, with each box representing the group's median, upper, and lower quantiles, and 95% confidence interval. Data points of individual tumors were overlaid as brown dots.
(F–H) Average tumor size (F), average nodules per lobe (G), and total tumor area per lobe (H) quantification of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks posttransduction, showing the range of tumor heterogeneity in AAV9-KPL injected mice. Data are plotted as mean ± SEM. *p < 0.05, **p < 0.005, ***p < 0.0005.
See also Figures S3, S4, and S5.
In detail, we observed multiple tumor nodules in the lung of AAV9-KPL-treated, but not AAV9-sgLacZ-treated, mice (Figure 5D). Moreover, certain lobes were dominated by one large tumor, whereas others had many tumors of various sizes (Figures 5D–5G). The sizes of tumors significantly increased over time (Figure 5E). The total tumor area per lobe also significantly increased over time (Figure 5H). The variation in tumor size suggests dynamic tumor initiation and growth across lobes and animals, which is reminiscent of the complexity observed in human tumors (Herbst et al., 2008).
Pathological and Genetic Analysis of Tumors Generated via p53, LKB1, or KRAS Mutations
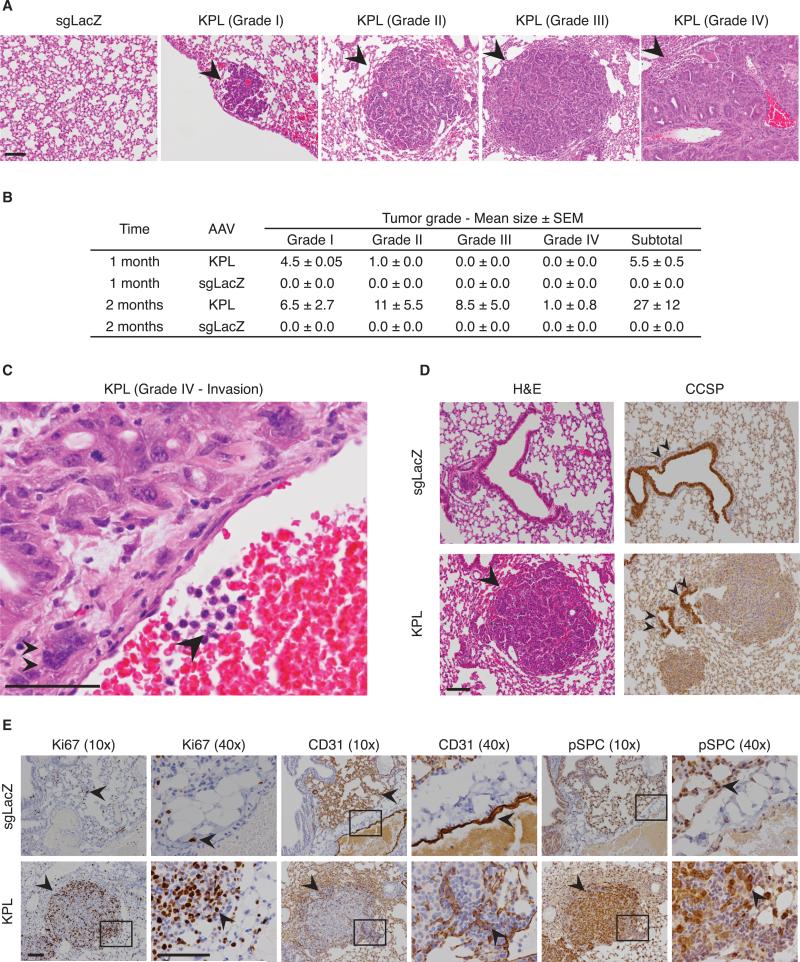
To understand the pathology of tumors formed by injection of the AAV9-KPL vector, we performed hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC). Pathology showed that lungs of AAV9-KPL-treated mice developed multiple grade I and grade II bronchial alveolar adenomas at 1 month postinjection (Figure S4D). In patients, this tumor type is thought to arise from type II pneumocytes (also called alveolar type II cells) at the junction between bronchial and terminal bronchial (Park et al., 2012). These tumors progressed to grade III lung adenocarcinomas in 2 months and occasionally became invasive grade IV adenocarcinomas (Figures 6A–6C and Table S2). None of the AAV9-sgLacZ mice showed any tumors detectable by histology (t test, one-tailed, p = 0.004) (Figures 5D, 6A, and 6B). Clara cell secretory protein (CCSP) staining showed that almost all tumors were adjacent to the clara cells of bronchials (Figure 6D). In addition, most tumors (178/182 = 95.7%) stained positive for pro-surfactant C (pSPC), a marker for type II pneumocytes (Figure 6E), implying that most of these tumors originated from this cell type. Many tumor cells also stained positive for Ki67, an indicator of active cell cycle (Figure 6E), signifying more rapid proliferation in these tumor cells as compared to adjacent normal tissue. All tumors grades II to IV contained CD31-positive endothelial cells (Figure 6E), suggesting that these tumors induced angiogenesis. These data suggest that AAV9-KPL delivery generates a spectrum of tumors from bronchial alveolar adenomas to invasive adenocarcinomas in the lungs of Cre-dependent Cas9 mice in less than 2 months.
Figure 6. Histopathology of Tumors Formed within AAV9-KPL-Injected Mice.
(A) Representative lung H&E images of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks posttransduction, showing a spectrum of grade I to grade IV tumors. Black arrowheads highlight tumors. Scale bar, 100 μm.
(B) Table of tumor grade statistics.
(C) Representative lung H&E and IHC images of Cre-dependent Cas9 mice injected with AAV9-KPL 9 weeks posttransduction, showing grade IV adenocarcinoma with signs of invasion (arrowhead) and aneuploidy (double arrowheads). Scale bar, 100 μm.
(D) Representative lung H&E and IHC images of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks posttransduction. H&E images show tumor formation only in AAV9-KPL-injected mice. Clara cell secretory protein (CCSP), a marker for Clara cells, staining shows a tumor adjacent to these cells (double arrowheads). Scale bar, 100 μm.
(E) Representative IHC images of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks posttransduction. Ki67, a marker for proliferating cells (arrow), staining showing extensive proliferation in tumors found within AAV9-KPL injected Cre-dependent Cas9 mice. CD31, a marker for endothelial cells (arrow), staining showing embedded CD31-positive endothelial cells. Positive for prosurfactant C (pSPC), a marker for type II pneumocytes, staining suggests tumors originate from this cell type. Scale bar, 200 μm.
See also Figures S3, S4, and S5 and Table S2.
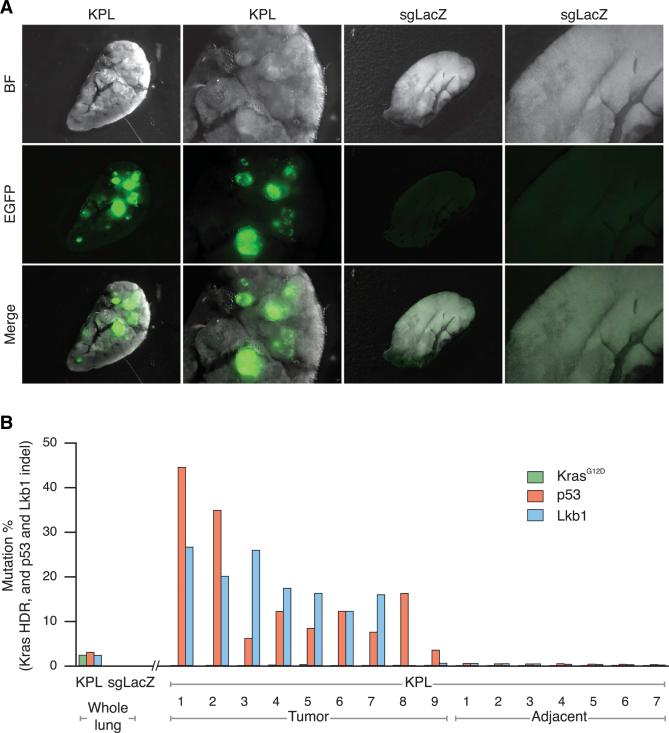
We dissected the largest EGFP-positive tumors and adjacent tissues without visible tumors from the lungs of AAV9-KPL-treated mice (Figure 7A) and performed captured Illumina sequencing of KRAS, p53, and LKB1 loci. We detected KRASG12D mutations and p53 and LKB1 indels in dissected tumors with low background in adjacent tissues (Figure 7B). The frequencies of p53 or LKB1 indels in dissected tumors from AAV9-KPL-treated animals were highly enriched over the whole lobe, adjacent tissues, and tissues from AAV9-sgLacZ controls, suggesting that p53 and/or LKB1 loss of function were driving the formation of these particular tumors. Interestingly, the frequency of KRASG12D mutations was lower in these dissected tumors compared to whole-lobe tissue (Figure 7B). Moreover, KRASG12D mutations were much lower than the frequencies of p53 and LKB1 indels. At the whole-lobe level, we observed 0.1% KRASG12D HDR at 4 weeks and 1.8% at 9 weeks (Figure 4H), suggesting that KRASG12D mutation frequency increases over time in whole tissue but is not enriched in the large tumors (Figure 7B). We did, however, observe a KRASG12D-dominated tumor in one out of three small tumors laser captured from lung sections, with 65% KRASG12D HDR reads (Figures S5A and S5B). These data suggest that the largest and fastest-growing dissected tumors do not depend on oncogenic KRAS for growth, either because p53;LKB1 double-mutant tumors outcompete KRASG12D-containing tumors or because activating KRASG12D by HDR in this model has longer latency. This heterogeneity of mutations is reminiscent of mosaicism of multiple mutations in the clinic, with different patients having different combinations (Figures S5C and S5D).
Figure 7. Mutational Analysis of Individual Tumors.
(A) Representative stereomicroscope lung images of Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks post-transduction, showing EGFP-positive tumors only within the lung of an AAV9-KPL injected mouse.
(B) KRAS, p53, and LKB1 mutational analysis of whole lung and individual tumors dissected from Cre-dependent Cas9 mice injected with either AAV9-KPL or AAV9-sgLacZ 9 weeks post-transduction, showing p53 and LKB1 mutations predominate in fast-growing tumors. The data are plotted as the percent of Illumina sequencing reads containing KRASG12D HDR, p53 indels, or LKB1 indels at the target site in whole lung, dissected tumors, or adjacent tissues.
DISCUSSION
The pair of Cre-dependent and constitutive Cas9-expressing mice extend the existing CRISPR-Cas9 toolbox to facilitate powerful in vivo genome editing applications. The ability to introduce covalent modifications in the genome of somatic cells enables the study of gene function for many areas of biology and provides significant time savings over conventional transgenic technologies. In this study, we demonstrated that Cas9 mice can be used to introduce indels in the brain using AAV-mediated sgRNA expression; restrict Cas9 expression to cellular subtypes using tissue-specific Cre drivers; mutate genes in primary immune cells using lentivirus; mutate genes in pulmonary and cardiovascular endothelium using nanoparticle-mediated sgRNA delivery (Extended Text and Figures S6 and S7); and model dynamics of multiple mutations in lung cancer. The viral and nanoparticle sgRNA expression reagents are modular and can be easily modified to target many tissue types and virtually any gene of interest with combinations of loss-of-function and/or gain-of-function mutations. Though we cannot completely rule out the possibility of off-target effects for the chosen sgRNAs, the Cas9 mice provide a unique early-stage screening platform for identifying candidate causal mutations, which can be further validated using independent same-gene-targeting sgRNAs and complementary conventional genetic models.
The ability to achieve multiplex genetic perturbations using the Cas9 mouse also enables the interrogation of multigenic effects. In this study, we applied the Cas9 mouse to study the competition between multiple genetic lesions in tumorigenesis. The use of Cas9 mice in conjunction with multiplex sgRNA delivery allowed us to easily introduce multiple genetic lesions in the same animal to more closely recapitulate the nature of mutation accumulation in evolving tumors. These types of tumor models have been particularly challenging to generate due to the exponential increase in technical difficulty associated with large numbers of genetic crosses. As multigene interactions play a critical role in virtually any biological process, multiplex genetic perturbation using the Cas9 mouse will likely find many applications beyond cancer biology.
In addition to using the Cas9 mouse to study individual or small sets of genes, a particularly exciting category of future application will be in vivo high-throughput genetic screens. A variety of RNA interference-based in vivo screens has already been applied to identify genes involved in tumor suppression (Schramek et al., 2014), stem-cell renewal (Chen et al., 2012), host determinants of virus replication (Varble et al., 2013), and regulators of oncogenic growth (Beronja et al., 2013). Given recent successes using Cas9 and large-scale sgRNA libraries for genetic screening (Findlay et al., 2014; Koike-Yusa et al., 2014; Sanjana et al., 2014; Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014), the Cas9 mouse may be directly combined with genome-wide or targeted sgRNA library to uncover novel biology.
EXPERIMENTAL PROCEDURES
Generation of the Cre-Dependent Cas9 mouse
Cre-dependent Cas9 knockin mice were generated by homologous recombination in R1 embryonic stem cells and implanted in C57BL6/J blastocysts using standard procedures, as described previously (Heyer et al., 2010; Nagy et al., 1993). Briefly, a codon-optimized 3 × FLAG-NLS-SpCas9-NLSP2A-EGFP expression cassette was cloned into the Ai9 Roas26 targeting vector (Madisen et al., 2010) and further verified by sequencing. Linearized targeting vector was electroporated into R1 embryonic stem cells followed by G418 and DTA selection for a week. Targeted single-ES cell colonies were screened by PCR with primers amplifying both recombinant arms. PCR products were sequenced to further validate correct insertion. Correctly targeted colonies were injected into C57BL6/J blastocysts for generating chimeric mice. High-percentage male chimeric mice were mated with C57 BL6/J female mice (Jackson Laboratory) to establish germline-transmitted founders. Genotypes of Cas9 mice were determined by amplifying a 4.5 kb product from purified mouse tail DNA (forward: GCAGCCTCTGTTCCACATA CAC; reverse: ACCATTCTCAGTGGCTCAACAA).
SgRNA Design
SgRNAs were designed using the CRISPRtool (http://crispr.mit.edu) to minimize potential off-target effects. SgRNA sequences and genomic primers are listed in Table S3.
Genomic DNA Extraction, Captured Illumina Sequencing, and Indel Analysis
Genomic DNA was extracted from both cells and tissue using Quick Extract DNA extraction solution (Epicenter) following the recommended protocol. Treated cells and tissues were used as a template for PCR for both captured sequencing and SURVEYOR nuclease assay using high-fidelity polymerases (Thermo Scientific) as previously described (Hsu et al., 2013). Genomic PCR products were subjected to library preparation using the Nextera XT DNA Sample Prep Kit (Illumina) or using customized barcoding methods. Briefly, low-cycle, first-round PCR was performed to amplify the target site. Second-round PCR was performed to add generic adapters, which were then used for a third round of PCR for sample barcoding. Samples were pooled in equal amounts and purified using QiaQuick PCR Cleanup (QIAGEN), quantified using Qubit (Life Technologies). Mixed barcoded library was sequenced on an Illumina MiSeq System.
Illumina Sequencing Analysis
Illumina sequencing reads were mapped to specific amplicons of KRAS, p53, or LKB1 as reference sequences using Burrows-Wheeler Aligner (Li and Durbin, 2010) with custom scripts. Insertions and deletions were called against reference using VarScan2 (Koboldt et al., 2012). Indel length distribution, indel phase, and donor allele frequencies were processed using custom scripts as described previously (Chen et al., 2014; Hsu et al., 2013). The rate of KRASG12D HDR was calculated as donor allele frequency based on the G12D mutations as well as the synonymous barcoded SNPs.
AAV1/2 DNA Vectors
The AAV vectors used for stereotaxic injection into the mouse brain were cloned between AAV serotype 2 ITRs and also included the human U6 promoter for noncoding sgRNA transcription, the ubiquitous Cbh promoter (a hybrid form of the CBA promoter) (Gray et al., 2011), HA-tagged Cre recombinase for recombination of loxP-SV40polyA-LoxP, WPRE, and human growth hormone polyA sequence. For nuclei sorting, a similar plasmid was cloned between AAV serotype 2 ITRs and also included the human U6 promoter for noncoding sgRNA transcription, the ubiquitous hSyn promoter, HA-tagged Cre recombinase for recombination of loxP-SV40polyA-LoxP, P2A, EGFP-KASH fusion for nuclei labeling, WPRE, and human growth hormone polyA sequence. Complete plasmid sequences and annotations are found in Data S1.
AAV1/2 Production
HEK293FT cells were transfected with the plasmid of interest, pAAV1 plasmid, pAAV2 plasmid, helper plasmid pDF6, and PEI Max (Polysciences, Inc. 24765-2) in Dulbecco's modified Eagle medium (DMEM from Life Technologies, 10569-010). At 72 hr posttransfection, the cell culture media was discarded. Then the cells were rinsed and pelleted via low-speed centrifugation. Afterward, the viruses were applied to HiTrap heparin columns (GE Biosciences 17-0406-01) and washed with a series of salt solutions with increasing molarities. During the final stages, the eluates from the heparin columns were concentrated using Amicon ultra-15 centrifugal filter units (Millipore UFC910024). Titering of viral particles was executed by quantitative PCR using custom Cre-targeted Taqman probes (Life Technologies).
Stereotactic Injection of AAV1/2
2–3 month old animals were anesthetized by intraperitoneally injection of ketamine (100 mg/kg) and xylazine (10 mg/kg). We also administered buprenorphine HCl (0.1 mg/kg) intraperitoneally as a pre-emptive analgesic. Once subject mice were in deep anesthesia, they were immobilized in a Kopf stereo-taxic apparatus using intra-aural positioning studs and a tooth bar to immobilize the skull. Heat is provided for warmth by a standard heating pad. We drilled a hole on the surface of the skull at 1.94 mm anterior to Bregma and 0.39 mm lateral for injection into the prefrontal cortex. Using a 33 G Nanofil needle and World Precision Instrument Nanofil syringe at a depth of [–2.95 mm, we injected 1 ul (1 × 1013 viral genome copies) AAV into the right hemisphere of the brain. Injection rates were monitored by the World Precision Instruments UltraMicroPump3. After injection, we closed the incision site with 6-0 Ethilon sutures (Ethicon by Johnson & Johnson). Animals were postoperatively hydrated with 1 ml lactated Ringer's solution (subcutaneous) and housed in a temperature controlled (37°C) environment until achieving ambulatory recovery. Meloxicam (1–2 mg/kg) was also administered subcutaneously directly after surgery. For downstream analysis, EGFP+ tissue was dissected under a stereotactic microscope.
AAV6 and AAV9 Vector Design
The single vector design was cloned utilizing ITRs from AAV2, human U6 promoter for noncoding sgRNA transcription, the short EFs promoter derived from EF1a, Renilla luciferase for in vivo luciferase imaging, P2A for peptide cleavage, Cre recombinase for recombination of LSL, a short polyA sequence, and an 800 bp KRASG12D homologous recombination donor template. Complete plasmid sequences and annotations are found in Data S1.
AAV6/9 Production and Purification
AAV6 and AAV9 were packaged and produced in HEK293FT cells and chemically purified by chloroform. In brief, HEK293FT cells were transiently transfected with the vector of interest, AAV serotype plasmid (AAV6 or AAV9), and pDF6 using polyethyleneimine (PEI). At 72 hr posttransfection, cells were dislodged and transferred to a conical tube in sterile DPBS. 1/10 volume of pure chloroform was added and the mixture was incubated at 37°C and vigorously shaken for 1 hr. NaCl was added to a final concentration of 1 M and the mixture was shaken until dissolved and then pelleted at 20,000 3 g at 4°C for 15 min. The aqueous layer was discarded while the chloroform layer was transferred to another tube. PEG8000 was added to 10% (w/v) and shaken until dissolved. The mixture was incubated at 4°C for 1 hr and then spun at 20,000 × g at 4°C for 15 min. The supernatant was discarded and the pellet was resuspended in DPBS plus MgCl2 and treated with Benzonase (Sigma) and incubated at 37°C for 30 min. Chloroform (1:1 volume) was then added, shaken, and spun down at 12,000 × g at 4C for 15 min. The aqueous layer was isolated and passed through a 100 kDa MWCO (Millipore). The concentrated solution was washed with DPBS and the filtration process was repeated. The virus was titered by qPCR using custom Taqman assays (Life Technologies) targeted to Cre.
Virus Delivery into the Lung
Intranasal and intratracheal delivery of adeno-associated virus were performed accordingly to previously established protocol (DuPage et al., 2009). In brief, 2- to 3-month-old animals were anesthetized using isoflurane and were set up in a biosafety cabinet. For intranasal delivery, previously titrated virus solution in 50 ml sterile saline was pipetted directly into one nostril of the mouse. For intratracheal delivery, a gauge-24 catheter was inserted to the trachea, and virus solution in 75 μl sterile saline was pipetted to the top of the catheter to allow animal to gradually breathe in the solution. A titer of 1 × 1011 viral genome copies was administered to each mouse. Animals after the procedure were kept warm using a heat lamp for recovery.
Immunostaining and Imaging
Mice were given a lethal dose of Ketamine/Xylazine and transcardially perfused with 0.9% saline followed by 4% paraformaldehyde using a peristaltic pump (Gilson) and fixed overnight. Tissue sectioning was performed on a vibratome (Leica VT1000S) at a thickness of 40 μm. Sections were rinsed three times in phosphate-buffered saline (PBS) with 0.1% Triton X-100 (PBSTx) and blocked with 5% normal goat serum (NGS) (Cell Signaling Technology) in PBS-Tx for 1 hr. Sections were incubated with primary antibodies diluted in PBS-Tx with 5% NGS overnight at 4°C on an orbital shaker. The following is a list of primary antibodies that were utilized: anti-cleaved caspase 3 (CC3) (1:1,000, Cell Signaling Technology, 9664), anti-yH2AX (1:1,000, Millipore, 05-636) anti-NeuN (1:800, Cell Signaling Technology, 12943), anti-GFP (1:1,600, Nacalai Tesque, GF090R), anti-parvalbumin (1:1,000, Sigma Aldrich, P3088), and anti-tyrosine hydroxylase (1:1,000, Immunostar, 22941). The following day, sections are washed three times in PBS-Tx and then incubated in PBS-Tx with the appropriate AlexaFluor 405, 488, 568 and/or 647 secondary antibody (1:400, Life Technologies) for 3 hours at room temperature on an orbital shaker. After the incubation, sections are rinsed with PBS-Tx three times and then mounted onto superfrost microscope slides (VWR). The sections are then coverslipped with VECTASHIELD HardSet Mounting Medium with DAPI (VECTOR Laboratories) and visualized under a confocal microscope (Zeiss LSM 710, Ax10 ImagerZ2, Zen 2012 Software).
Purification and FACS of Single Neuronal Nuclei
For labeling neuronal nuclei, we generated an AAV-U6-sgRNA-hSyn-Cre-P2A-EGFP-KASH vector. At 3 weeks postinjection, we dissected the EGFP+ injection site in the prefrontal cortex under a stereotactic microscope. As previously described (L.S. and M. Heidenreich, unpublished data), tissue was gently homogenized in 2 ml ice-cold homogenization buffer (HB) (320 mM Sucrose, 5 mM CaCl, 3 mM Mg(Ac)2, 10 mM Tris [pH 7.8], 0.1 mM EDTA, 0.1% NP40, 0.1 mM PMSF, 1 mM β-mercaptoethanol) using 2 ml Dounce homogenizer (Sigma Aldrich); 25 times with pestle A, followed by 25 times with pestle B. An additional 3 ml of HB was added and the mixture was put on ice for 5 min. Gradient centrifugation was performed using 5 ml 50% OptiPrep density gradient medium (Sigma Aldrich) containing 5 mM CaCl, 3 mM Mg(Ac)2, 10 mM Tris pH 7.8, 0.1 mM PMSF, 1 mM β-mercaptoethanol. The nuclei-containing mixture was gently added on the top of a 29% iso-osmolar OptiPrep solution in a conical 30 ml centrifuge tube (Beckman Coulter, SW28 rotor). Samples were centrifuged at 10,100 × g (7,500 rpm) for 30 min at 4°C. The supernatant was removed and the nuclei pellet was gently re-suspended in 65 mM β-glycerophosphate (pH 7), 2 mM MgCl2, 25 mM KCl, 340 mM sucrose, and 5% glycerol. Intact nuclei were labeled with Vybrant DyeCycle Ruby Stain (Life Technologies) and sorted using a BD FACSAria III. EGFP+ nuclei were sorted into individual wells of a 96 well-plate containing 5 μl of QuickExtract DNA Extraction Solution (Epicenter).
Mouse Dendritic Cells
Six- to eight-week old constitutive Cas9-expressing female mice were used for all DC experiments. Bone marrow cells were collected from femora and tibiae and plated at concentra tion of 2 × 105/ml on nontreated tissue culture dishes in RPMI medium (GIBCO, Carlsbad, CA, Invitrogen, Carlsbad, CA), supplemented with 10% FBS (Invitrogen), L-glutamine (Cellgro), penicillin/streptomycin (Cellgro), MEM nonessential amino acids (Cellgro), HEPES(Cellgro), sodium pyruvate (Cellgro), β- mercaptoethanol (GIBCO), and GM-CSF (20 ng/ml; Peprotech). SgRNAs targeting Myd88 and A20 were cloned into a guide-only lentiviral vector (Sanjana et al., 2014). At day 2, cells were infected with lentiviruses encoding guide RNAs. Cells were expanded in the presence of GM-CSF. At day 7, infected cells were selected by adding puromycin (Invitrogen) at 5 mg/ml. At day 9, 100 ng/ml LPS (Invivogen) was added 30 min prior to protein analysis or 3 hr prior to mRNA expression profiling. Flow cytometry for GFP detection was performed with BD Accuri C6. Western blot was done using anti-Myd88 (R&D Systems AF3109) and anti-actin (Abcam, ab6276) antibodies.
Nanostring nCounter Expression Measurement
DCs were processed and analyzed as previously described (Amit et al., 2009). In brief, 5 × 104 cells were lysed in TCL buffer (QIAGEN) supplemented with β-mercaptoethanol. 5% of the lysate was hybridized for 16 hr with a previously described Nanostring Gene Expression CodeSet (Geiss et al., 2008) and loaded into the nCounter Prep Station followed by quantification using the nCounter Digital Analyzer. Counts were normalized using control genes as previously described (Amit et al., 2009) and fold changes were calculated relative to cells transduced with sgRNA targeting GFP and nontargeted controls. Heat maps were created using GENE-E (http://www.broadinstitute.org/cancer/software/GENE-E/).
μCT Imaging and Processing
μCT imaging was performed using standard imaging protocol with a μCT machine (GE Healthcare). In brief, animals were anesthetized using isoflurane and were set up in the imaging bed with a nosecone providing constant isoflurane. A total of 720 views were acquired for each mouse using a soft-tissue-fast-scan setting. Raw image stacks were processed for lung reconstruction using the standard ROI tool (MicroView). Rendering and quantification were performed using render volume tool and measurement tool in MicroView. Tumor burden was calculated as the sum of the sizes in mm3 from all detectable tumors per mouse, with 3–4 mice per group.
Lung Histology
Mice were sacrificed by carbon dioxide asphyxiation. Lungs were dissected under a fluorescent stereoscope, fixed in 4% formaldehyde or 10% formalin overnight, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin (HE) for pathology. For tumor size quantification, images were taken tiling the whole lobe, merged into a single lobe, and tumors were manually outlined as region of interest (ROI) and were subsequently quantified using ImageJ (Schneider et al., 2012). Sections were de-waxed, rehydrated, and stained using standard immunohistochemistry (IHC) protocols as previously described (Chen et al., 2014). The following antibodies were used for IHC: anti-Ki67 (abcam ab16667, 1:100), anti-CCSP (Millipore, 1:500), anti-pSPC (Millipore AB3786, 1:500) and anti-CD31 (abcam, ab28364, 1:50). IHC was quantified in 10 randomly chosen low-magnification fields per lobe with 3 mice per group using ImageJ (Schneider et al., 2012).
TCGA Data Analysis
TCGA lung adenocarcinoma (LUAD) mutation data were downloaded from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). Mutation annotation format file of LUAD was reprocessed to show KRAS coding missense mutation as well as Tp53 and STK11 loss-of-function mutations in 227 patients.
Animal Work Statement
All animal work was performed under the guidelines of Division of Comparative Medicine (DCM), with protocols (0411-040-14, 0414-024-17 0911-098-11, 0911-098-14 and 0914-091-17) approved by Massachusetts Institute of Technology Committee for Animal Care (CAC), and were consistent with the Guide for Care and Use of Laboratory Animals, National Research Council, 1996 (institutional animal welfare assurance no. A-3125-01).
Supplementary Material
ACKNOWLEDGMENTS
We thank the entire Zhang laboratory and Sharp laboratory. We thank J.T. Ting, I. Tirosh, N. Sanjana, M. Muzumdar, S. Jones, Y. Li, and colleagues in the Broad and Koch Institutes for technical assistance and discussions. We thank the Swanson Biotechnology Center for their support (Applied Therapeutics and Whole Animal Imaging, Bioinformatics and Computing, ES Cell and Transgenics, Microscopy, Flow Cytometry, and Histology, in particular). R.J.P. is supported by a National Science Foundation Graduate Research Fellowship under grant number 1122374. S.C. is a Damon Runyon Cancer Research Fellow (DRG-2117-12). Y.Z. is supported by the Simons Center for the Social Brain at MIT, postdoctoral fellowship. L.S. is a European Molecular Biology Organization (EMBO) and Foundation for Polish Science (FNP) Fellow. J.E.D. is supported by the NDSEG, NSF, and MIT Presidential Graduate Fellowships. M.J. is supported by fellowships of the Swiss National Science Foundation for advanced researchers (SNF) and the Marie Sklodowska-Curie IOF. R.J.X. and D.G. are supported by the Helmsley CharitableTrust and DK43351. A.R. is supported by NHGRI CEGS P50 HG006193, HHMI and the Klarman Cell Observatory. N.H. is supported by NHGRI CEGS P50 HG006193. R.L. and D.A.G. are supported by the NIH Centers of Cancer Nanotechnology Excellence grant U54CA151884, the Controlled Release grant EB000244, and the National Heart, Lung, and Blood Institute, National Institutes of Health, as a Program of Excellence in Nanotechnology (PEN) Award, contract HHSN268201000045C. G.F. is supported by the McGovern Internal Funding Poitras Gift 1631119, the Stanley Center, the SFARI/Simons Foundation 6927482, and the Nancy Lurie Marks Family Foundation 6928117. P.A.S. is supported by United States Public Health Service R01-CA133404 from the National Institutes of Health; by an MIT-Harvard Center for Cancer Nanotechnology Excellence Grant U54 CA151884 from the National Cancer Institute; by a generous gift from the Marie D. and Pierre Casimir-Lambert Fund; by an SkTech/MIT Initiative Grant from the Skolkovo Foundation; and partially by the Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute. F.Z. is supported by the NIMH through a NIH Director's Pioneer Award (DP1-MH100706), the NINDS through a NIH Transformative R01 grant (R01-NS 07312401), NSF Waterman Award, the Keck, Damon Runyon, Searle Scholars, Klingenstein, Vallee, Merkin, and Simons Foundations, and Bob Metcalfe. CRISPR reagents are available to the academic community through Addgene, and associated protocols, support forum, and computational tools are available via the Zhang lab website (http://www.genome-engineering.org). The Cre-dependent Cas9 (JAX Stock Number: 024857) and constitutive Cas9-expressing (JAX Stock Number: 024858) mice are available via the Jackson Laboratory.
Footnotes
AUTHOR CONTRIBUTIONS
R.J.P. and F.Z. conceived of the study and oversaw experiments related to Figures 1 and 3–7. R.J.P., S.C., P.A.S., and F.Z. conceived of the cancer modeling application and oversaw experiments related to Figures 4–7. R.J.P., Y.Z., G.F., and F.Z. conceived of the generation of the Cas9 mouse and oversaw experiments related to Figure 1. O.P., T.M.E., M.J., N.H., and A.R. conceived of the ex vivo dendritic cell application and oversaw data related to Figure 2. R.J.P., J.E.D., S.J., R.L., D.G.A., and F.Z. conceived of the systemic nanoparticle-mediated sgRNA delivery application and oversaw data related to Figures S6 and S7. D.B.G. and R.X. conceived of the validation of Cas9 expression in various immune cells and oversaw data related to Figure S2. R.J.P. performed most of the experiments related to Figures 1 and 3. R.J.P. and S.C. performed most of the experiments related to Figures 4– 7. R.J.P. and Y.Z. generated the Rosa26 Cas9 knockin ESC clones, and Y.Z. conducted the electrophysiological characterizations (Figures 1C–1H). M.Y. managed the mouse colony and assisted with AAV production, craniotomy, and immunohistochemistry related to Figures 1 and 3. L.S. assisted with nuclei sorting (Figures 1H–1J) and experimental methodologies related to Figures 1 and 3. H.R.K. assisted with vector cloning and sample processing related to Figures 1 and 3–7. J.E.D. formulated 7C1 nanoparticles, injected mice, and processed tissues related to Figure S6 and S7. O.P., T.M.E., and M.J. performed dendritic cell experiments related to Figure 2. D.B.G. performed flow cytometry experiments validating Cas9 expression (Figure S2). S.J. performed flow cytometry experiments to isolate endothelial cells (Figures S6 and S7). R.J.P., S.C., P.A.S., and F.Z. wrote the manuscript with input from all authors.
ACCESSION NUMBERS
Sequencing data are deposited in NCBI Sequence Read Archive (SRA) under accession PRJNA260178.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Results, Extended Experimental Procedures, seven figures, three tables, one movie, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j. cell.2014.09.014.
REFERENCES
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K, Agbandje-McKenna M, Wilson JM. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J. Clin. Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beronja S, Janki P, Heller E, Lien WH, Keyes BE, Oshimori N, Fuchs E. RNAi screens in mice identify physiological regulators of oncogenic growth. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, Chandrasegaran S. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Mol. Cell. Biol. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Chen T, Heller E, Beronja S, Oshimori N, Stokes N, Fuchs E. An RNA interference screen uncovers a new molecule in stem cell self-renewal and long-term regeneration. Nature. 2012;485:104–108. doi: 10.1038/nature10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Xue Y, Wu X, Le C, Bhutkar A, Bell EL, Zhang F, Langer R, Sharp PA. Global microRNA depletion suppresses tumor angiogenesis. Genes Dev. 2014;28:1054–1067. doi: 10.1101/gad.239681.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier N, Mertins P, Artyomov MN, Shalek AK, Iannacone M, Ciaccio MF, Gat-Viks I, Tonti E, DeGrace MM, Clauser KR, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147:853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, Cowan CA, Rader DJ, Musunuru K. Permanent Alteration of PCSK9 With In Vivo CRISPR-Cas9 Genome Editing. Circ. Res. 2014;115:488–492. doi: 10.1161/CIRCRESAHA.115.304351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell. 2012;47:810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Gray SJ, Foti SB, Schwartz JW, Bachaboina L, Taylor-Blake B, Coleman J, Ehlers MD, Zylka MJ, McCown TJ, Samulski RJ. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 2011;22:1143–1153. doi: 10.1089/hum.2010.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AJF. Introduction to genetic analysis. Tenth Edition W.H. Freeman and Co.; New York: 2012. [Google Scholar]
- Halbert CL, Allen JM, Miller AD. Efficient mouse airway transduction following recombination between AAV vectors carrying parts of a larger gene. Nat. Biotechnol. 2002;20:697–701. doi: 10.1038/nbt0702-697. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N. Engl. J. Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer MP, Feliciano C, Peca J, Feng G. Elucidating Gene Function through use of Genetically Engineered Mice. In: Starkey M, Elaswarapu R, editors. Genomics. John Wiley & Sons, Ltd; Hoboken, NJ: 2010. pp. 211–248. [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, de Villiers J, Weber F, Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell. 1985;43:695–703. doi: 10.1016/0092-8674(85)90242-9. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel.C., Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M, Keller B, Makalou N, Sutton RE. Systematic determination of the packaging limit of lentiviral vectors. Hum. Gene Ther. 2001;12:1893–1905. doi: 10.1089/104303401753153947. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat. Rev. Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberis MP, Wilson JM. Adeno-associated virus serotype 9 vectors transduce murine alveolar and nasal epithelia and can be readministered. Proc. Natl. Acad. Sci. USA. 2006;103:12993–12998. doi: 10.1073/pnas.0601433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg J, Usoskin D, Bengtsson H, Gustafsson A, Kylberg A, Söderström S, Ebendal T. Transgenic expression of Cre recombinase from the tyrosine hydroxylase locus. Genesis. 2004;40:67–73. doi: 10.1002/gene.20065. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang LH, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- Mitzner W, Brown R, Lee W. In vivo measurement of lung volumes in mice. Physiol. Genomics. 2001;4:215–221. doi: 10.1152/physiolgenomics.2001.4.3.215. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo: a laboratory manual. Third Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2003. [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Saiwai H, Kumamaru H, Kubota K, Harada A, Yamaguchi M, Iwamoto Y, Ohkawa Y. Flow cytometric sorting of neuronal and glial nuclei from central nervous system tissue. J. Cell. Physiol. 2011;226:552–558. doi: 10.1002/jcp.22365. [DOI] [PubMed] [Google Scholar]
- Park WY, Kim MH, Shin DH, Lee JH, Choi KU, Kim JY, Park Y, Lee CH, Sol MY. Ciliated adenocarcinomas of the lung: a tumor of non-terminal respiratory unit origin. Mod. Pathol. 2012;25:1265–1274. doi: 10.1038/modpathol.2012.76. [DOI] [PubMed] [Google Scholar]
- Rudin N, Sugarman E, Haber JE. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmbeck R, Reimann J, Kochanek S, Kreppel F. The immunogenicity of adenovirus vectors limits the multispecificity of CD8 T-cell responses to vector-encoded transgenic antigens. Mol. Ther. 2008;16:1609–1616. doi: 10.1038/mt.2008.141. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Adiconis X, Gertner RS, Gaublomme JT, Raychowdhury R, Schwartz S, Yosef N, Malboeuf C, Lu D, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA-Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014 doi: 10.1038/nature13385. Published online July 9, 2014. http://dx.doi.org/10.1038/nature.13385. [DOI] [PMC free article] [PubMed]
- Varble A, Benitez AA, Schmid S, Sachs D, Shim JV, Rodriguez-Barrueco R, Panis M, Crumiller M, Silva JM, Sachidanandam R, tenOever BR. An in vivo RNAi screening approach to identify host determinants of virus replication. Cell Host Microbe. 2013;14:346–356. doi: 10.1016/j.chom.2013.08.007. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg RA. The biology of cancer. Garland Science; New York: 2007. [Google Scholar]
- Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol. Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014 doi: 10.1038/nature13589. Published online August 6, 2014. http://dx.doi.org/10.1038/nature.13589. [DOI] [PMC free article] [PubMed]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.