SUMMARY
Efforts to construct synthetic networks in living cells have been hindered by the limited number of regulatory components that provide wide dynamic range and low crosstalk. Here, we report a new class of de-novo-designed prokaryotic riboregulators called toehold switches that activate gene expression in response to cognate RNAs with arbitrary sequences. Toehold switches provide a high level of orthogonality and can be forward-engineered to provide average dynamic range above 400. We show that switches can be integrated into the genome to regulate endogenous genes and use them as sensors that respond to endogenous RNAs. We exploit the orthogonality of toehold switches to regulate 12 genes independently and to construct a genetic circuit that evaluates 4-input AND logic. Toehold switches, with their wide dynamic range, orthogonality, and programmability, represent a versatile and powerful platform for regulation of translation, offering diverse applications in molecular biology, synthetic biology, and biotechnology.
INTRODUCTION
Synthetic biology seeks to apply engineering design principles to predict and control the behavior of living systems. Synthetic gene networks have been used to construct a wide range of biological devices, including molecular counters (Friedland et al., 2009), oscillators (Elowitz and Leibler, 2000), toggle switches (Gardner et al., 2000), logic gates (Auslaender et al., 2012; Moon et al., 2012; Siuti et al., 2013; Win and Smolke, 2008), cell classifiers (Xie et al., 2011), and analog signal processors (Daniel et al., 2013). Despite these developments, an underlying problem in synthetic biology remains the limited number of composable, high-performance parts for constructing genetic circuits and difficulties that arise when integrating multiple components into a large, complex synthetic network (Purnick and Weiss, 2009). Unlike electronic circuit elements, which can be electrically and spatially insulated from each other, biological components can interact with one another in the complex cellular environment and suffer from unwanted crosstalk between components. Limitations imposed by the number and orthogonality of available biological circuit components hinder the construction of more complex circuits that can operate robustly in living cells. Consequently, new classes of regulatory components that offer wide dynamic range, low system crosstalk, and design flexibility represent a much-needed, enabling step towards fully realizing the potential of synthetic biology in areas such as biotechnology and medicine (Khalil and Collins, 2010).
RNA-based regulatory elements offer a potential solution to this component bottleneck. Biological parts constructed from RNA take advantage of predictable Watson-Crick base pairing to control cellular behavior, and can harness sophisticated software tools for predicting RNA-RNA interactions. Nature has already developed a wide assortment of RNA-based parts operating at the transcriptional and post-transcriptional level (Brantl and Wagner, 2000; Gultyaev et al., 1997; Winkler et al., 2002). Starting from these natural systems, researchers have developed a number of engineered RNA regulatory elements, including riboregulators that control translation and transcription in response to cognate RNAs (Callura et al., 2012; Callura et al., 2010; Isaacs et al., 2004; Lucks et al., 2011; Mutalik et al., 2012; Rodrigo et al., 2012), and mRNA transducers that convert signals from small molecule or protein ligands to protein outputs (Bayer and Smolke, 2005; Culler et al., 2010; Qi et al., 2012; Win and Smolke, 2008).
Engineered riboregulators consist of cognate pairs of RNAs: a transducer strand that regulates translation or transcription, and a trans-acting RNA that binds to the transducer to modulate its biological activity. Riboregulator designs can be classified according to the initial RNA-RNA interaction that drives hybridization between the transducer and trans-acting RNAs. Reactions initiated between loop sequences in both RNAs are termed loop-loop interactions, while those that occur between a loop sequence and an unstructured RNA are termed loop-linear (Takahashi and Lucks, 2013).
A common limitation for riboregulators has been their dynamic range (Liu et al., 2012). Previous prokaryotic translational riboregulators have typically modulated biological signals by up to a maximum of ~55-fold for activators (Callura et al., 2012) and up to ~10-fold for repressors (Mutalik et al., 2012). In contrast, protein-based transcriptional regulators have demonstrated dynamic ranges over an order of magnitude higher, with widely-used inducible promoters regulating protein expression over 350-fold (Lutz and Bujard, 1997) and sigma factor-promoter pairs providing up to 480-fold modulation (Rhodius et al., 2013). Despite the inherent programmability of RNA-based systems, efforts at constructing large sets of orthogonal riboregulators have been limited to libraries of at most seven parts with crosstalk levels of ~20; (Takahashi and Lucks, 2013). Typical RNA-based regulators employ interaction domains consisting of ~30-nts, which corresponds to a sequence space of over 1018 potential regulatory elements. Thus, the sheer diversity of possible RNA-based parts suggests that previous devices have not come close to realizing the potential of highly orthogonal regulation.
Much of this discrepancy arises from the significant sequence constraints imposed on riboregulators engineered thus far (Figure 1A). Like natural riboregulators, engineered riboregulators of translation have invariably used base pairing to the RBS to prevent ribosome binding, thereby preventing translation (Callura et al., 2012; Isaacs et al., 2004; Mutalik et al., 2012; Rodrigo et al., 2012). Since repression is caused by RBS binding, trigger RNAs that activate translation are engineered to contain an RBS sequence to displace the repressing sequence, which in turn reduces the potential sequence space for the riboregulator.
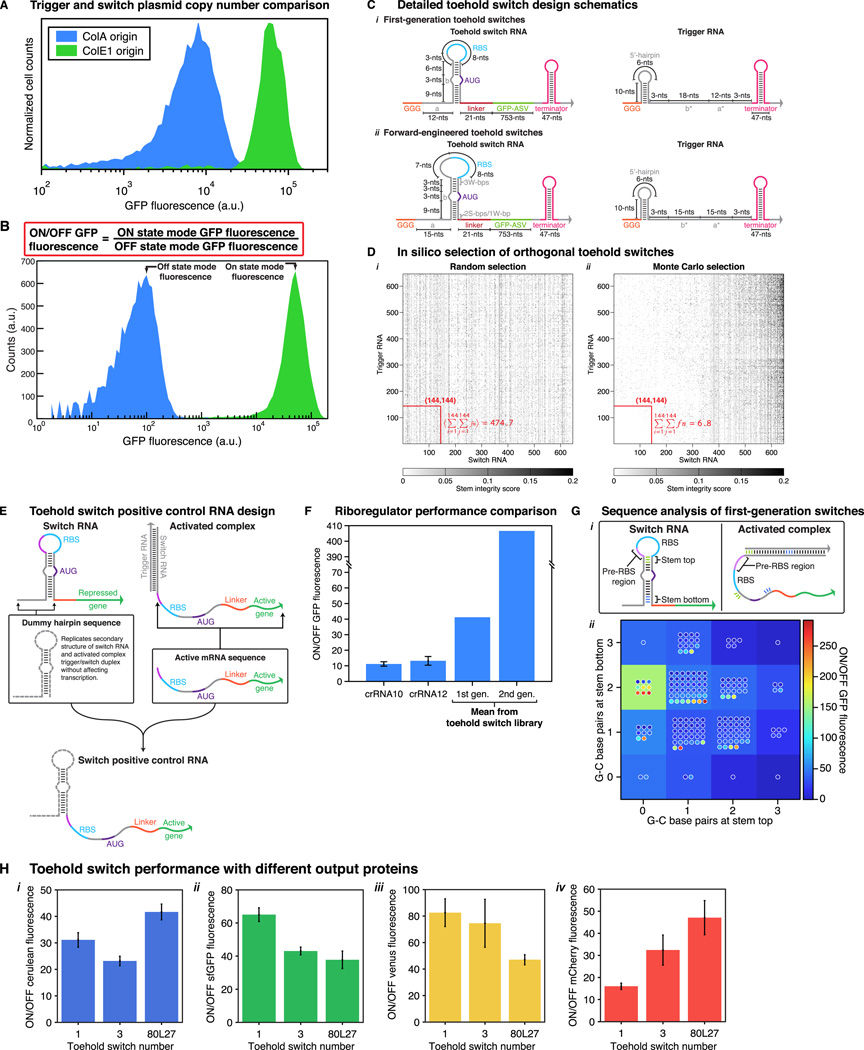
Figure 1. Toehold switch design and in vivo characterization.
(A,B) Design schematics of conventional riboregulators (A) and toehold switches (B). Variable sequences are shown in gray, while conserved or constrained sequences are represented by different colors. (A) Conventional riboregulator systems repress translation by base pairing directly to the RBS region. RNA-RNA interactions are initiated via a loop-linear or loop-loop interaction at the YUNR-loop in an RNA hairpin. (B) Toehold switches repress translation through base pairs programmed before and after the start codon (AUG), leaving the RBS and start codon regions completely unpaired. RNA-RNA interactions are initiated via linear-linear interaction domains called toeholds. The toehold domain a binds to a complementary a* domain on the trigger RNA. Domains a and b are 12- and 18-nts, respectively. (C) Flow cytometry GFP fluorescence histograms for toehold switch number 2 compared to E. coli autofluorescence and a positive control. Autofluorescence level measured from induced cells not bearing a GFP-expressing plasmid. (D) GFP mode fluorescence levels measured for switches in their ON and OFF states in comparison to positive control constructs and autofluorescence. (E) ON/OFF GFP fluorescence levels obtained three hours after induction for 168 first-generation toehold switches. Inset: ON/OFF GFP fluorescence measured for toehold switches of varying performance levels at different time points following induction. See also Figure S1, Table S1.
Previous riboregulators have also relied on U-turn loop structures to drive loop-loop and looplinear interactions between RNAs (Figure 1A) (Callura et al., 2012; Isaacs et al., 2004; Lucks et al., 2011; Takahashi and Lucks, 2013). U-turn loops are common RNA structural motifs formed by tertiary interactions that have been identified in ribozymes, ribosomal RNAs, and transfer RNA anti-codon loops (Gutell et al., 2000). While recent work has begun to show that loops with canonical U-turn sequences are not essential for riboregulators (Mutalik et al., 2012; Rodrigo et al., 2012), the engineered systems reported to date have continued their reliance on the loop-mediated RNA interactions from natural systems. Although these loop interactions have been selected by evolution in nature, alternative approaches employing linear-linear RNA interactions are amenable to rational engineering and exhibit more favorable reaction kinetics and thermodynamics, factors that could be exploited to increase riboregulator dynamic range.
To address these fundamental limitations, we designed a new class of de-novo-designed riboregulators that enable post-transcriptional activation of protein translation through mechanisms employed in artificial systems rather than natural ones. Based on their interaction mechanism and near-digital signal processing behavior, we term these new riboregulator systems toehold switches. Unlike conventional riboregulators, our synthetic riboregulators take advantage of toehold-mediated linear-linear interactions developed in vitro (Dirks and Pierce, 2004; Yin et al., 2008; Yurke et al., 2000) to initiate RNA-RNA strand displacement interactions. Furthermore, they rely on sequestration of the region around the start codon to repress protein translation, eschewing any base pairing to the RBS or start codon itself to regulate translation. As a result, toehold switches can be designed to activate protein translation in response to a trigger RNA with an arbitrary sequence, enabling substantial improvements in component orthogonality. The absence of binding to the RBS and use of thermodynamically favorable linear-linear interactions also enables facile tuning of translational efficiency via RBS engineering. Consequently, these systems routinely enable modulation of protein expression over two orders of magnitude.
Here we demonstrate the utility of toehold switches by validating over 100 functional systems in E. coli. We exploit the expanded RNA sequence space afforded by the new riboregulator class to construct libraries of components with unprecedented orthogonality, including a set of 26 systems that exhibit less than 12; crosstalk. Heuristic design principles are used to generate forward-engineered switches that exhibit average ON/OFF ratios exceeding 400, a dynamic range typically reserved for protein-based transcriptional regulators. We also use toehold switches to sense endogenous RNAs in vivo and to regulate endogenous gene expression by integrating them into the genome. For potential applications in synthetic biology, we demonstrate that toehold switches can be used to regulate a dozen components in the cell at the same time, and to construct a genetic circuit that computes a 4-input AND expression. The versatility, dynamic range, orthogonality, and programmability of toehold switches provide them with the potential to become a powerful new platform for sensing and programming the internal states of living cells. We anticipate these devices will be enabling tools for constructing increasingly complex genetic circuits in synthetic biology.
RESULTS
Design of a new biological device from first principles
A riboregulator that activates gene expression must switch from a secondary structure that prevents translation to a configuration that promotes translation upon binding of a cognate trans-acting RNA. We developed toehold switches based on previous reports describing the influence of secondary structure on mRNA translation and knowledge of RNA thermodynamics and kinetics.
Although the Shine-Dalgarno sequence is an important factor in determining the efficiency of translation from a given mRNA, studies have found that secondary structure in regions nearby the start codon also play a critical role (Kudla et al., 2009). Furthermore, genome-wide analyses have revealed strong biases towards low secondary structures around the start codon of mRNAs from a panel of hundreds of bacterial genomes (Bentele et al., 2013). Thus, we ensured that toehold switches sequestered the region around the start codon to repress translation, rather than binding to either the RBS or the start codon.
Instead of using loop regions to initiate interactions, we recognized the design advantages afforded by linear-linear nucleic acid interaction strategies developed in vitro and focused in particular on toehold-based strand displacement reactions (Yurke et al., 2000). In these systems, interactions between reaction species are kinetically controlled through hairpins or multi-stranded complexes that feature exposed single-stranded domains called toeholds. These domains serve as reaction initiation sites for input nucleic acid species and do not require U-turn structural motifs to increase their accessibility. Such toehold mechanisms have been extensively used to engineer complex dynamic systems utilizing reconfigurable DNA hairpins in test tubes (Dirks and Pierce, 2004; Yin et al., 2008) and RNA hairpins in fixed tissues (Choi et al., 2010), and for complex in vitro information processing systems (Qian and Winfree, 2011; Qian et al., 2011; Yin et al., 2008).
Toehold switch systems are composed of two RNA strands referred to as the switch and trigger (Figure 1B). The switch RNA contains the coding sequence of the gene being regulated. Upstream of this coding sequence is a hairpin-based processing module containing both a strong ribosome binding site and a start codon that is followed by a common 21-nt linker sequence coding for low molecular weight amino acids added to the N-terminus of the gene of interest. A single-stranded toehold sequence at the 5’ end of the hairpin module provides the initial binding site for the trigger RNA strand. This trigger molecule contains an extended single-stranded region that completes a branch migration process with the hairpin to expose the RBS and start codon, thereby initiating translation of the gene of interest.
The hairpin processing unit functions as a repressor of translation in the absence of the trigger strand. Unlike previous riboregulators, the RBS sequence is left completely unpaired within the 11-nt loop of the hairpin. Instead, the bases immediately before and after the start codon are sequestered within RNA duplexes that are 6-bp and 9-bp long, respectively. The start codon itself is left unpaired in the switches we tested, leaving a 3-nt bulge near the midpoint of the 18-nt hairpin stem. Since the repressing domain b (Figure 1B) does not possess complementary bases to the start codon, the cognate trigger strand in turn does not need to contain corresponding start codon bases, thereby increasing the number of potential trigger sequences. The sequence in the hairpin added after the start codon was also screened for the presence of stop codons, as they would prematurely terminate translation of the gene of interest when the riboregulator was activated. We employed a 12-nt toehold domain at the 5’ end of the hairpin to initiate its interaction with the cognate trigger strand. The trigger RNA contains a 30-nt single-stranded RNA sequence that is complementary to the toehold and stem of the switch RNA.
In silico design of toehold switches
We used the NUPACK nucleic acid sequence design package (Zadeh et al., 2011) to generate libraries of de-novo-designed translational activators satisfying the desired device parameters (see Supplemental Information Section S6 and Figure S1C). We initially designed a set of 24 toehold switches (see Table S1 for RNA sequences) to gauge in vivo performance.
We then designed an extended library of toehold switches containing orthogonal elements selected for low crosstalk with the rest of the library. The orthogonal library was generated by assembling a set of 646 unique toehold switch designs and simulating all 417,316 pairwise interactions in the set to evaluate crosstalk interactions (see Section S6.4 and Figure S1D). A Monte Carlo algorithm was used to identify a subset of 144 switches that exhibited the lowest level of overall component crosstalk (see Table S1 for RNA sequences). The resulting library of orthogonal regulators provided a large set of components to independently regulate translation in vivo, and comprised devices well separated from one another in the sequence space to provide valuable information regarding sequence-dependent effects.
In vivo component validation
The toehold switches were tested in E. coli BL21 Star DE3 with the switch and trigger RNAs expressed from separate medium and high copy plasmids, respectively. Although BL21 Star DE3 is an RNase-deficient strain, we found that toehold switches also performed well in strains with wild-type RNase levels (see Section S10, Figure S2C, Table S3). Expression of both strands was induced using IPTG, which triggered production of both RNA species through T7 RNA polymerase. GFP was used to characterize switch output performance via flow cytometry.
Representative flow cytometry histograms of GFP output from toehold switch number 2 are shown in Figure 1C. ON state fluorescence measured from cells expressing both the switch and its cognate trigger was near the fluorescence intensity of control constructs with unrepressed GFP expression. OFF state fluorescence from cells expressing the switch and a non-cognate trigger revealed a slight increase in GFP levels over E. coli autofluorescence. The mode fluorescence value from these histograms was used to calculate the ON/OFF ratios of each toehold switch design (see Experimental Methods, Section S3 and Figure S1B). Cell autofluorescence was not subtracted from either the ON or OFF state fluorescence for determination of ON/OFF ratios.
Figure 1D presents the mode GFP fluorescence obtained from three high-performance toehold switches in their ON and OFF states. The OFF state fluorescence of the three switches was near the background autofluorescence levels measured for induced cells not expressing GFP. The ON state fluorescence for the toehold switches was comparable to the fluorescence obtained from positive controls that recapitulated the secondary structure of the toehold switch in its activated state (see Section S7, Figure S1E, and Table S1). The activated toehold switches all showed higher GFP output than a standard GFP expression construct featuring a completely single-stranded region upstream of the RBS and gene, likely as a result of the increased mRNA stability afforded by the secondary structures of the switch RNA and the trigger/switch RNA complex.
Of the 168 first-generation systems tested, 20 exhibit ON/OFF ratios exceeding 100 and nearly two thirds display at least an ON/OFF level greater than 10 (Figure 1E). In comparison, we also characterized the widely used engineered riboregulators (Isaacs et al., 2004) crRNA 10 and 12 in identical conditions. These earlier riboregulators exhibited lower dynamic range with ON/OFF values of 11 ± 2 and 13 ± 4 for crRNA systems 10 and 12, respectively (see Section S8 and Figure S1F). Time-course measurements revealed system activation within one hour of induction and ON/OFF ratios increased over time with continued production of GFP (Figure 1E, inset). The switches were also successful in regulating four different output proteins (see Section S10.1 and Figure S1H).
Evaluation of toehold switch orthogonality
To measure the orthogonality of the devices, we performed additional in silico screening to isolate a subset of 26 that displayed extremely low levels of predicted crosstalk (see Section S6.4) and assayed in E. coli all 676 pairwise combinations of switch and trigger plasmids. Figure 2A displays images of GFP fluorescence from colonies of E. coli induced on LB plates (see Section S5). Clearly visible is the strong emission from cognate switch and trigger pairs along the diagonal of the grid with the final switch at index 26 displaying lower fluorescence as a result of its low ON/OFF ratio. Low fluorescence levels are observed for the off-diagonal elements featuring non-cognate trigger/switch RNA pairs.
Figure 2. Assessment of toehold switch orthogonality.
(A) GFP fluorescence from colonies of E. coli expressing 676 pairwise combinations of switch and trigger RNAs. GFP-expressing colonies are visible as green points along the diagonal in cells containing cognate switch and trigger strands. Off-diagonal, non-cognate components have low fluorescence. (B) Crosstalk measured by flow cytometry for all trigger-switch combinations. Crosstalk was determined by taking GFP output measured for a given trigger-switch combination and dividing it by the GFP output measured for the switch with its cognate trigger. (C) Comparison of overall library dynamic range and orthogonal library size for the toehold switches and previous riboegulators. The overall library dynamic range corresponds to the minimum ON/OFF ratio to expect in a network employing this library of switches. See also Table S2.
We used flow cytometry to quantify GFP output from all trigger-switch interactions (See Section S4). Crosstalk was calculated by dividing the GFP fluorescence obtained from a noncognate trigger and a given switch RNA by the fluorescence of the switch in its triggered state (Figure 2B). The full set of 26 switches tested displayed less than 12; crosstalk. Since the number of switches in an orthogonal set is defined by its threshold crosstalk level, we identified orthogonal subsets for a range of different crosstalk thresholds (see Section S11 and Table S2). For instance, a subset of 18 of the toehold switches exhibited less than 2; subset-wide crosstalk.
A relevant metric for assessing orthogonal library performance is the reciprocal of the threshold crosstalk level. For example, the set of 18 switches with less than 2; crosstalk has a reciprocal crosstalk threshold of 50. This metric corresponds to the minimum output dynamic range to expect in a network employing this library of 18 switches. Figure 2C plots this library dynamic range metric against the maximum orthogonal subset size for the toehold switches as well as a number of previous riboregulator systems.
Forward engineering of new high-performance devices
In-silico-designed riboregulators forward engineered to exhibit high performance in vivo have the potential to significantly reduce the time required for generating new genetic circuits. The first-generation toehold switches fell short of this goal since they displayed large variations in output characteristics (Figure 1E). To determine the design features that yielded high-performance switches, we performed detailed analyses of the first-generation library to uncover sequence-dependent factors that affected switch performance (see Section S9, S12; Figure S1G, S2A; Table S1). We integrated these findings into the design of second-generation toehold switches forward engineered for wide dynamic range.
The forward-engineered switches incorporate four design changes as shown in Figure 3A (see Figure S1A for direct comparisons). Modifications were made to the sequences at the top of the switch RNA stem, the loop of the switch RNA was increased, and the binding site of the trigger RNA was shifted away from the RBS. These three design changes increased the strength of the switch RNA RBS to increase its ON state protein production. The length of the toehold domain was also increased to promote binding between switch and trigger RNAs.
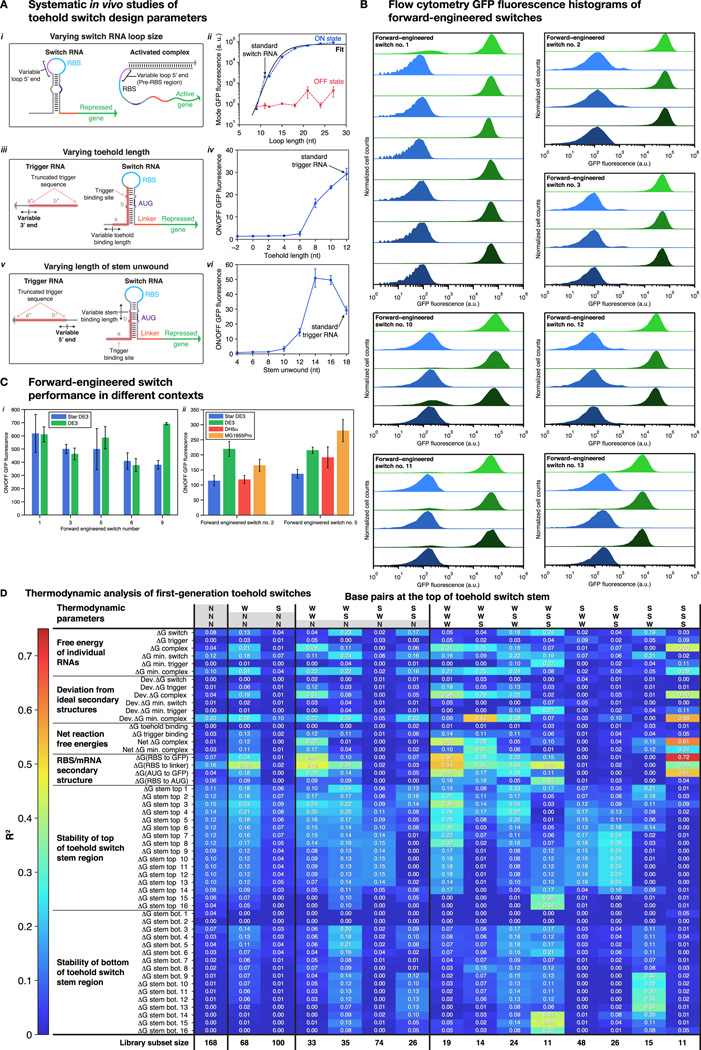
Figure 3. Forward engineering and thermodynamic analysis of toehold switches.
(A) Schematic of the design modifications made for the forward-engineered switches compared to the firstgeneration switches. (B) ON/OFF GFP fluorescence ratios obtained for the set of 13 forwardengineered toehold switches after three hours of induction. Dashed black line marks the mean ON/OFF fluorescence measured for the 168 first-generation toehold switches. Inset: Timecourse measurements for forward-engineered switches number 6 and number 9. (C) Percentage of first-generation and forward-engineered library components that had ON/OFF ratios that exceeded the value defined on the x-axis. (D) Correlation between ;GRBS-linker and ON/OFF ratio measured for the 68 first-generation toehold switches that had an A-U base pair at the top of the hairpin stem. Inset: Schematic showing the RNA sequence range in the trigger-switch complex used to define ;GRBS-linker. (E) Strong correlation (R2 = 0.79) between ;GRBS-linker and ON/OFF ratio measured for the complete set of forward-engineered switches. See also Figure S2, Table S3.
We designed and evaluated a set of 13 of these second-generation toehold switches (see Table S3 for RNA sequences). Figure 3B presents the ON/OFF fluorescence ratios for the forward-engineered systems regulating GFP after 3 hours of induction (see Figure S2B for flow cytometry data). There is a dramatic increase in ON/OFF fluorescence for almost all the systems tested, with 12 out of 13 switches exhibiting a dynamic range comparable to or higher than the highest performance toehold switch from the initial library (ON/OFF ratio 290 ± 20). These systems exhibit an average ON/OFF ratio of 406 compared to 43 for the first-generation design. This mean ON/OFF ratio rivals the dynamic range of protein-based regulators, and it does so using a highly programmable system design without requiring any evolution or largescale screening experiments. Furthermore, even the lowest performance second-generation switch displayed an ON/OFF ratio of 33 ± 4, which is still sufficient for many logic operations. Hourly time course measurements reveal activation of forward-engineered switches after 1 or 2 hours of induction (Figure 3B, inset). Furthermore, ON state fluorescence increased steadily over 4 hours, yielding ON/OFF levels well over 600 for the switches. Additional testing confirmed successful forward-engineered switch operation in different strains of E. coli and using the endogenous E. coli RNA polymerase (see Section S10, Figure S2C, Table S3).
We quantified the effectiveness of our rational engineering strategy by calculating the percentage of forward-engineered designs with ON/OFF ratios exceeding a given minimal level and comparing them to the same evaluation performed on the library of 168 first-generation toehold switches (Figure 3C). The yield of high-performance switches is higher for the forward-engineered devices for all ON/OFF ratios tested.
Thermodynamic parameter correlated with hermodynamic switch performance
We conducted extensive analyses to identify thermodynamic parameters that were linked to the ON/OFF performance of the toehold switches. Candidate thermodynamic factors were rapidly screened by computing linear regressions against subsets of the first-generation toehold switches satisfying particular sequence constraints (see Section S13, Figure S2D, and Table S3). The screening process identified a single parameter ;GRBS-linker that was correlated with the ON/OFF ratios for the subset of 68 first-generation switches with an A-U base pair at the top of their stem. A linear fit of ;GRBS-linker to the logarithm of their ON/OFF ratios yielded a coefficient of determination R2 = 0.40 (Figure 3D). This ;GRBS-linker term is defined as the free energy of the sequence running from the RBS region to the end of the linker (Figure 3D, inset). It reflects the amount of energy required by the ribosome to unwind the RBS and early-mRNA region as it binds and begins translation of the output gene. A fit of ;GRBS-linker to the ON/OFF ratio for the forward-engineered switches yielded a much stronger correlation with R2 = 0.79 (Figure 3E). Importantly, we found that this single thermodynamic factor could explain the lone low-performance, forward-engineered toehold switch.
Toehold switches triggered by mRNAs and endogenous RNAs
The capacity of toehold switch to accept trigger RNAs with arbitrary sequences enables them, in principle, to be activated by functional mRNAs (Figure 4A). However, the fixed sequences of potential mRNA triggers present significant challenges for effective system activation. Unlike synthetic trigger RNAs designed de novo to be single-stranded, strong secondary structures abound within mRNAs, complicating toehold binding and decreasing the thermodynamics driving the branch migration process. The cognate toehold sequences defined by the trigger mRNA can also exhibit base pairing both internally and with sequences downstream of the hairpin module, and thus pose similar challenges to activation.
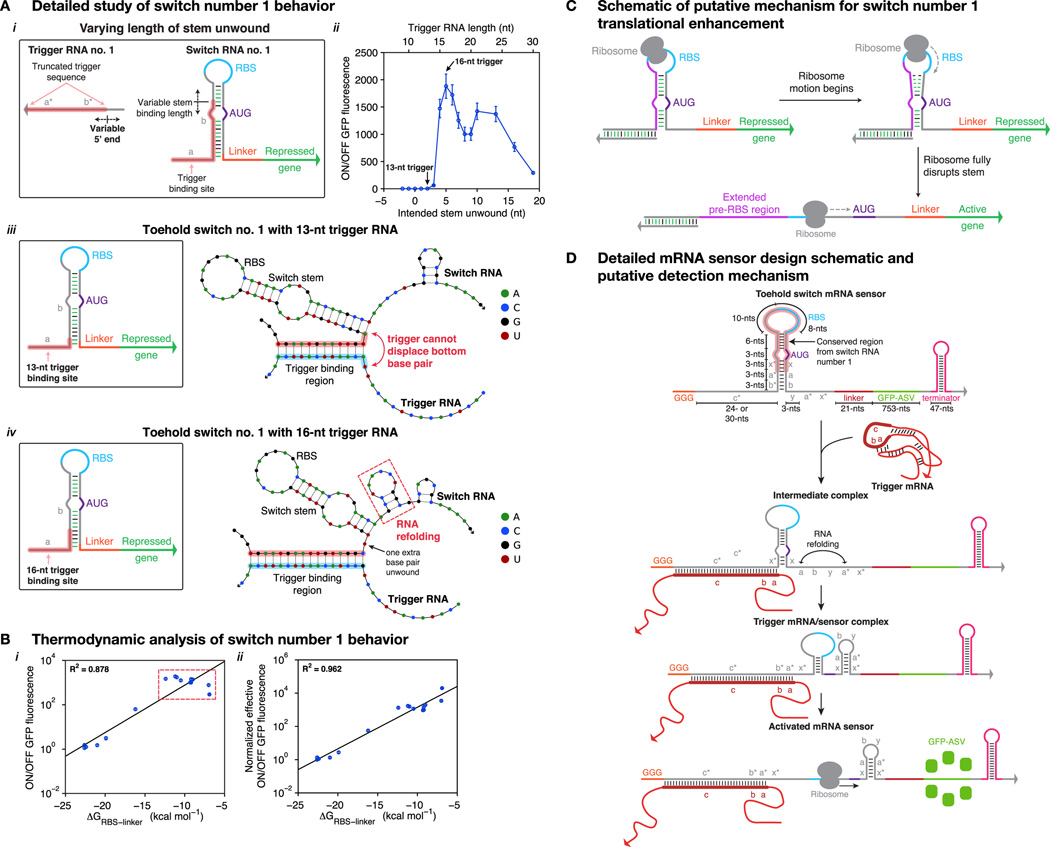
Figure 4. Toehold switch activated by mRNA and endogenous small RNA triggers.
(A) Design schematic and putative activation pathway of the toehold switch mRNA sensors. (B) Mode GFP and mCherry fluorescence obtained from flow cytometry of three mCherry sensors in their repressed and activated states, as well as positive and negative controls. (C) ON/OFF GFP fluorescence ratios for a series of toehold switches activated by the mCherry mRNA, and cat and aadA mRNAs, which confer chloramphenicol and spectinomycin resistance, respectively. (D) Endogenous and synthetic gene networks used for sensing the ryhB sRNA. (E) Transfer function for the ryhB sensor (purple curve) as a function of ryhB inducer concentration. Output of a constitutive GFP expression cassette is shown for comparison (green curve). See also Figure S3, Table S4.
To counter these effects, we increased the toehold domain length of the mRNA-sensing switches from 12- or 15-nts to ; 24-nts. This modification shifted the importance of single-stranded regions for binding initiation from the trigger mRNA to the toehold switch itself. We also incorporated a common sequence element from a high performance first-generation switch into the sensor designs (see Section S14.1, Figures S3A–C, and Table S4) and adopted a programmed RNA refolding mechanism to decrease the energetic barrier to switch activation (Figure 4A; see Section S14.2 and Figure S3D). Candidate sensors for a given mRNA or endogenous RNA were designed and screened in silico to identify those with the greatest probability of success (see Section S14.3, and Table S4 for RNA sequences).
We first validated sensors for detecting the mCherry mRNA. These sensors express GFP only upon binding to mCherry transcripts enabling simultaneous monitoring of sensor output via GFP and transcription of the trigger species through mCherry fluorescence. Figure 4B displays the GFP and mCherry fluorescence levels measured for three mCherry-responsive switches in the absence or presence of their cognate trigger mRNA. Fluorescence levels from control measurements are also shown for comparison. Negative controls reflect E. coli autofluorescence measured in either fluorescence channel, while positive controls were obtained from cells expressing unrepressed GFP and mCherry from medium and high copy vectors, respectively. These results show that the switches strongly activate expression of GFP only upon transcription of the mCherry mRNA, and provide low GFP expression in the absence of the mCherry trigger. ON/OFF GFP fluorescence values obtained from the three mCherry mRNA sensors are shown in Figure 4C. In addition, we validated two sensors for detecting antibiotic resistance conferring mRNAs: chloramphenicol acetyltransferase (cat) and aadA (spectinomycin resistance) (Figure 4C).
We also designed a toehold switch sensor to detect the endogenous E. coli small RNA (sRNA) ryhB. RyhB is a 90-nt transcript that down-regulates iron-associated genes in conditions where iron levels are low (Masse and Gottesman, 2002). To characterize the sensors, cells were transformed with plasmids constitutively expressing the ryhB-responsive toehold switch regulating GFP (Figure 4D; see Section S15 and Table S4). The ryhB sRNA was induced by adding the iron-chelating compound 2,2’-bipyridyl to the growth media (Figure 4D), which is known to rapidly stimulate expression of ryhB (Masse and Gottesman, 2002). We measured sensor output one hour after induction by the chelator using flow cytometry. The sensor transfer function shows a steady increase in GFP expression as 2,2’-bipyridyl levels increase to 0.3 mM, beyond which levels plateau (Figure 4E). In contrast, a GFP positive control construct demonstrated decreasing output as 2,2’-bipyridyl levels increased.
Synthetic regulation of endogenous genes
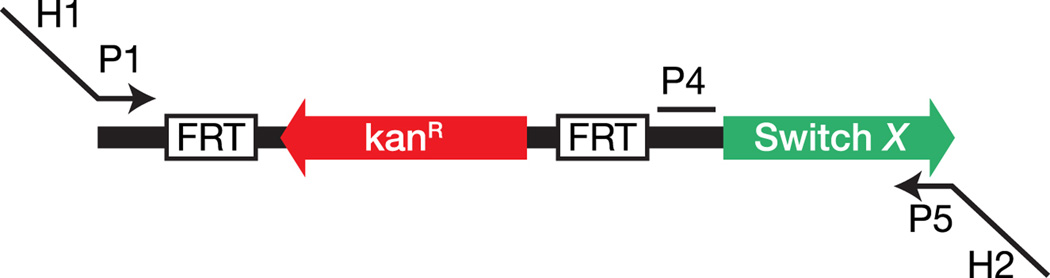
Toehold switches can be integrated into the genome to regulate translation of endogenous genes. Template genome-editing plasmids were constructed that contained a high-performance second-generation switch adjacent to a kanamycin resistance marker flanked by a pair of FRT sites (Figure 5A; see Section S16, Table S5, and Figure S4). Linear DNA fragments were amplified from these plasmids and inserted upstream of targeted chromosomal genes using λ Red recombination (Datsenko and Wanner, 2000). The resulting E. coli strain retains a functional copy of the targeted gene in its chromosome; however, it is deactivated as a result of the co-transcribed switch RNA module. This repressed gene can be activated post-transcriptionally by expression of a cognate trigger.
Figure 5. Synthetic regulation of endogenous genes.
(A) Integration of switch modules into the genome upstream of the targeted gene (gene B) at sites H1 and H2 using λ Red recombination. Switch-edited gene B is translationally repressed, but can be activated via the cognate trigger RNA. (B) Images of uidA::Switch A and uidA::Switch B spread onto X-Gluc plates with different trigger RNAs. uidA expression like the wild-type (top left) is only observed with cognate trigger RNAs as seen by blue/green color change. (C) Images of lacZ::Switch C with different combinations of IPTG and aTc chemical inducers. lacZ::Switch C only activates with aTc-induced expression of trigger C in conditions where lacZ transcription is induced by IPTG. Wild-type lacZ (top left) is activated whenever IPTG is present. (D) Motility assays for cheY::Switch D on soft agar plates. cheY::Switch D is only able to move away from the point of inoculation at the plate center when trigger D is induced with IPTG. See also Figure S4, Table S5.
We inserted switches upstream of the genes uidA, lacZ, and cheY. The genes uidA and lacZ produce the enzymes β-glucoronidase and β-galactosidase, respectively. Cells expressing these enzymes can be readily identified by their blue/green color on plates containing the corresponding substrates X-Gluc and X-Gal. We constructed two strains with synthetic uidA regulation by integrating switches A and B into the chromosome (uidA::Switch A and uidA::Switch B, respectively). Figure 5B displays these strains upon expression of different trigger RNAs as well as a control strain with the wild-type uidA genotype. As expected, uidA::Switch A only exhibits the blue/green wild-type phenotype upon expression of trigger A. Similarly, uidA::Switch B activates β-glucoronidase only with cognate trigger B.
The edited strain lacZ::Switch C provides more complicated behavior since the lac operon is regulated at the transcriptional level by lactose or chemical analogs such as IPTG. Thus, lacZ::Switch C requires both lactose/IPTG and trigger RNA C to turn on expression of β-galactosidase. This behavior results in a genetic AND circuit combining transcriptional and post-transcriptional regulation. We tested this AND circuit by expressing different trigger RNAs using inducible promoters responsive to anhydrotetracycline (aTc). Figure 5C provides images of lacZ::Switch C transformed with different trigger plasmids for all four combinations of the two chemical inputs (i.e. IPTG and aTc) of the AND circuit. In the absence of IPTG, none of the strains show any change in color since expression of the lac operon is strongly repressed. For a plate containing IPTG and no aTc, the wild-type lacZ strain (upper left quadrant) becomes blue/green in color, while the lacZ::Switch C strains with different triggers do not change in color since the trigger RNAs are not being expressed. When the AND condition is satisfied by adding both IPTG and aTc to the plate, lacZ::Switch C exhibits the blue/green color change with trigger RNA C as expected, while those expressing triggers A and B are unchanged.
Lastly, we conditionally regulated the E. coli chemotaxis gene cheY. CheY::Switch D was transformed with plasmids that expressed triggers inducibly via IPTG. Only cells expressing trigger RNA D with IPTG demonstrated significant motility, while those expressing non-cognate trigger RNA A or lacking the IPTG inducer were unable to move from the point of inoculation (Figure 5D).
Multiplexed regulation
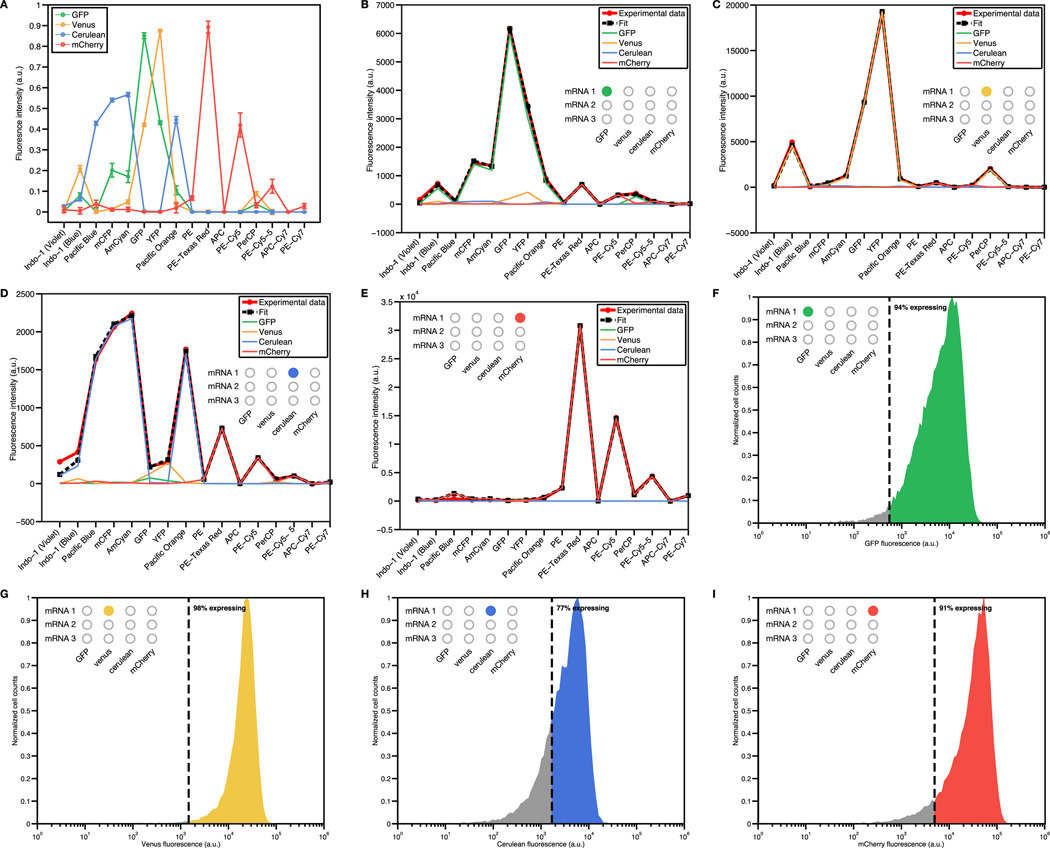
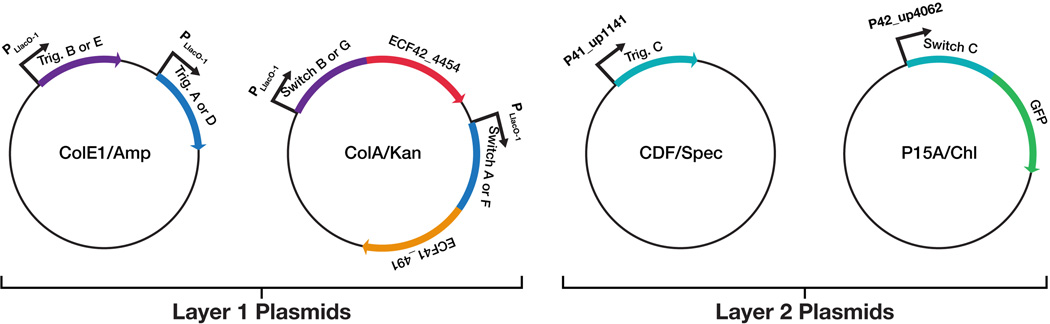
To demonstrate the full multiplexing capabilities of toehold switches, we expressed twelve toehold switches in the same cell and independently confirmed their activity via flow cytometry. We used four different fluorescent proteins (GFP, venus, cerulean, and mCherry) as reporters and constructed three plasmids to express each of the reporter proteins (Figure 6A; see Section S17 and Table S6). Each of these proteins was regulated using a different switch from the second-generation library. The resulting constructs were expressed from a single T7 promoter as ~3.4-kb polycistronic mRNAs resulting in a total synthetic network size of over 10-kb.
Figure 6. Simultaneous regulation of gene expression by twelve toehold switches.
(A) Schematics of plasmids and ~3.4-kb polycistronic mRNAs used for multiplexing studies. Each reporter has its own switch RNA that can be independently activated by its cognate trigger RNA. (B) Percentage of cells expressing each of the four reporters for a set of 24 different trigger RNA combinations. Gray and colored circles are used to identify the particular trigger RNA being expressed and the corresponding switch RNA. See also Figure S5, Table S6.
Figure 6B presents the outcome of the multiplexing experiments with the output of each reporter represented in terms of the percentage of cells expressing the reporter. A cell was determined to be expressing a given reporter if its fluorescence exceeded a threshold level held constant for all the plots in Figure 6B (see Section S17.3 and Figure S5). The first two rows of Figure 6B display the output from all twelve of the switches activated separately from a single expressed trigger RNA. In all twelve cases, significant expression is only observed from the intended reporter with limited crosstalk in the three other channels.
We also tested activation of all two- and three-color combinations of reporter proteins and observed all the expected output color combinations (Figure 6B). We detected some unintended leakage of cerulean and GFP for trigger combinations activating GFP/venus and venus/cerulean, respectively. Since single trigger measurements of each of these triggers displayed low leakage, we attribute much of the observed leakage in these two-trigger experiments to imperfect compensation of the flow cytometry data caused by the spectral overlap between GFP, venus, and cerulean (see Figure S5A). We also successfully activated all four output proteins by simultaneously expressing four trigger RNAs and observed low system leakage from two non-cognate trigger RNAs (Figure 6B, two bottom-right panels).
Implementation of a 4-input AND circuit
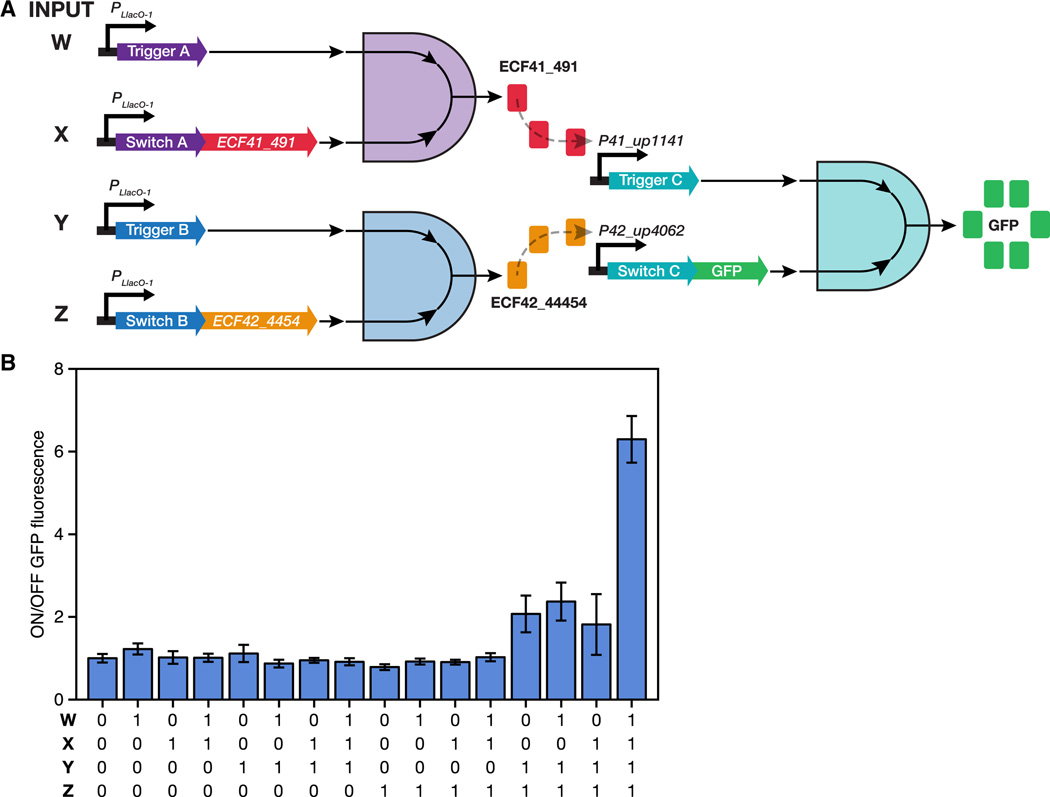
Toehold switches are readily integrated with existing biological components to build sophisticated genetic programs. We constructed a layered 4-input AND gate consisting of three toehold switches coupled to two orthogonal transcription factors and a GFP reporter (Figure 7A). In this circuit, toehold switch RNA pairs act as two independent input species that must both be present before a 2-input logical AND expression evaluates as TRUE. The first computational layer in the circuit consists of two 2-input AND toehold switch gates, which each produce a transcription factor. A pair of extracytoplasmic function (ECF) sigma factors were used as the transcription factors in the circuit since they had previously been reported to be highly orthogonal (Rhodius et al., 2013). The sigma factors produced from the first layer then activate transcription of the toehold switch RNAs in the second computational layer, which in turn lead to expression of a GFP reporter. Similar layered circuits have previously been constructed using transcription factors that required a second chaperone protein for full activity (Moon et al., 2012).
Figure 7. A layered 4-input AND circuit.
(A) Design schematic for the 4-input AND circuit consisting of three 2-input AND gates formed by three toehold switches, two orthogonal transcription factors (ECF41_491 and ECF42_4454), and a GFP reporter. (B) Complete 16- element truth table for the 4-input AND system. GFP expression from the sole logical TRUE output case with all input RNAs are expressed (far right) is significantly higher than the logical FALSE output cases where one or more input RNAs is absent. See also Figure S6, Table S7.
To validate the circuit, we constructed plasmids to express all 16 combinations of the four input trigger and switch RNAs (see Section S18, Table S7, and Figure S6). The full truth table for the 4-input AND computation is shown in Figure 7B with ON/OFF levels calculated by normalizing GFP fluorescence to that measured for the case in which none of the input bits are expressed. GFP output when all four input bits are present is significantly higher than all other input permutations, as expected for a functional 4-input AND computation.
DISCUSSION
Origins of increased toehold switch performance compared to previous riboregulators
Toehold switches represent a versatile and powerful new platform for regulating translation at the post-transcriptional level. They combine a high degree of component orthogonality with system dynamic range comparable to protein-based transcriptional regulators (Lutz and Bujard, 1997; Rhodius et al., 2013). Of note, the dynamic range of the toehold switches is significantly greater than that observed for previous engineered riboregulators. Earlier efforts to design riboregulators with higher performance have been hindered by three general misconceptions. (1) Since natural riboregulators have served as the starting point for new riboregulators, it has been implicitly assumed that engineered riboregulators cannot be substantially modified from their parent natural systems without abolishing their functionality. This approach employing small sequence perturbations parallels efforts in engineering new protein-based parts. However, it neglects the considerable advantage in in silico simulation and design available for engineering RNAs compared to proteins. This underlying assumption has in turn led to: (2) an emphasis on loop-loop and loop-linear RNA interaction mechanisms and (3) the use of RNA binding to the RBS region to repress translation.
Although application of any of these criteria for engineered riboregulators does not necessarily preclude them from functioning well as biological parts, toehold switches demonstrate that each of these design rules is not required for high-performance devices. First, toehold switches do not have any direct natural riboregulator counterparts, and thus they have no natural analogues to narrow the sequence space of potential devices. Relieving this constraint enabled us to generate large libraries of potential devices and explore a more diverse array of sequences compared to previous riboregulators. Furthermore, this new approach allowed RNA sequences to be designed de novo with ideal secondary structures for optimal device performance. Thus, trigger RNA transcripts feature single-stranded domains for binding to the switch RNA, and the ensuing trigger-switch RNA complex is engineered to have low secondary structure nearby the start codon to encourage efficient translation by the ribosome to maximize the device ON state.
Second, toehold switch RNA interactions are mediated through linear-linear interactions rather than loop-mediated ones. These reactions can provide faster kinetics than loop-based reactions, and the long ; 12-nt toehold of the switch RNA provides stronger thermodynamics than the shorter loop domains of earlier riboregulators. These features cause a higher percentage of the total switch RNAs present in the cell to be triggered, and thus producing the output protein. We found that the fraction of activated switch RNAs was approximately 100; based on comparison measurements with unrepressed versions of the switch RNA (Figure 2AB). In contrast to earlier reports, thermodynamic analyses of toehold switch performance did not reveal significant correlations between riboregulator ON/OFF levels and the free energy of the switch-trigger interaction nor the free energy of toehold-trigger binding (Mutalik et al., 2012). These observations suggest that RNA-RNA interactions for the toehold switches are so strongly thermodynamically favored that other factors are presently limiting device performance in cells.
Third, toehold switches do not rely on direct binding to the RBS to repress translation. Instead, the RBS is enclosed within a loop and repression is achieved by secondary structures immediately before and after the start codon. This design feature increased the space of potential trigger RNA sequences and it provided a straightforward means of enhancing ON state translation levels simply by adding A-rich sequences to the loop upstream of the RBS. Importantly, these modifications did not require corresponding changes to the trigger RNA, which enabled facile interpretation of the effects of modifications to this pre-RBS region. Taken together, all these design features enabled the toehold switch to detect arbitrary trigger RNAs and translate any output protein.
Our results suggest that existing riboregulators can be improved substantially by adopting some of the features of our toehold switches. The length of loop domains in loop-mediated interactions can be increased to improve reaction thermodynamics. Furthermore, the unpaired region upstream of the RBS in translational riboregulators can be lengthened to enhance protein output and in turn device dynamic range.
Constraints on toehold switch trigger RNAs and output proteins
The designs we have chosen for the toehold switches place some restrictions on the sequences of trigger RNAs. Inclusion of the 3-nt bulge at the start codon AUG position of the switch RNA precludes the trigger RNA from having an AUG sequence at positions programmed to hybridize with this bulge. The switch RNA also possesses a 9-bp stem after the start codon, which must not code for an in frame stop codon. This restriction in turn imposes sequence conditions on the trigger, which must unwind this region of the switch RNA stem. The toehold switches also add 11 residues from the switch stem to the linker to the N-terminus of the regulated protein, which could be problematic for an output protein sensitive to changes in its N-terminus.
In practice, the conditions imposed on the trigger RNA and output protein can be avoided with a few modifications to the toehold switch design. Constraints on the trigger RNA are a byproduct of the base-pairing conditions specified for the switch RNA stem and the trigger-switch complex. These particular secondary structures are not strictly required for switch operation. For instance, forward-engineered switch number 5 has only a 1-nt bulge in its stem, but provides ON/OFF > 400. We expect that design modifications that add and subtract base pairs from the switch RNA will allow the toehold switches to modulate gene expression while providing sufficient design flexibility to eliminate stop and start codon constraints on the trigger sequence. The toehold switches can also be modified to incorporate the coding sequence of the output protein directly into the switch RNA stem, thus making them compatible with any protein sensitive to N-terminal modifications.
Engineered regulators with very low system crosstalk
The capacity of toehold switches to respond to diverse trigger RNAs enabled us to use in silico techniques to design large libraries of orthogonal components. Comprehensive evaluations of in vivo trigger-switch RNA pairwise interactions revealed a set of 26 toehold switches with 12; crosstalk levels. The largest previously reported orthogonal riboregulator set consisted of seven transcriptional attenuators displaying 20; crosstalk (Takahashi and Lucks, 2013). In this case, crosstalk level of 20; meant that the set of 42 off-target RNA sense-antisense interactions attenuated transcription by at most 20;. Such crosstalk results in an upper bound in overall library dynamic range of 5 (Figure 3C). Earlier orthogonal translational activators and repressors have been limited to sets of four (Callura et al., 2012) and six (Mutalik et al., 2012), respectively, at 20; crosstalk. For proteins, an engineered library of five orthogonal eukaryotic transcription factors crosstalk of ~30; was reported (Khalil et al., 2012). Recently, part mining was used to engineer a set of four orthogonal chimeric sigma factors with ~10; crosstalk and a subset of three with ~2; crosstalk (Rhodius et al., 2013). Thus to our knowledge, these toehold switches constitute the largest set of engineered orthogonal regulatory elements, RNAor protein-based, reported to date.
Crosstalk experiments also revealed smaller sets of orthogonal switches exhibiting substantial improvements in overall dynamic range, including a set of 18 with 50-fold predicted minimum dynamic range (<2; crosstalk) and a set of 5 with >200-fold dynamic range (<0.5; crosstalk). Subsets of orthogonal toehold switches of comparable size to previously reported riboregulator libraries therefore exhibit minimum dynamic range over an order of magnitude larger than the earlier RNA-based systems. At this point, the ultimate size of the orthogonal libraries of toehold switches is limited by throughput of our crosstalk assay, not design features intrinsic to the devices themselves.
RNA-based, forward-engineered components for synthetic biology
We anticipate that our large library of orthogonal toehold switches will enable new applications and capabilities for synthetic biology. Their combination of wide dynamic range, low leakage levels, orthogonality, and rapid response are ideal for implementing time-sensitive logic circuitry in living systems. Furthermore, the protein-like dynamic range of the toehold switches is achieved at a lower metabolic cost than protein-based systems, since all regulatory components are composed of RNA and translation only occurs if the switches are activated. As synthetic genetic networks grow in complexity and impose greater burdens on the host, the lower metabolic footprint of the toehold switches will become an increasingly important advantage in their favor. Moreover, their low leakage levels suggest they can be used as constitutively expressed passive monitoring or sensor systems that trigger a downstream response only in desired situations.
Our success in using in silico design tools to reliably produce new high-performance, orthogonal toehold switches represents an important step forward for synthetic biology. The process of constructing new synthetic gene networks generally requires multiple design-build-test cycles before the desired network behavior is achieved. Consequently, new devices like the toehold switch that routinely provide functional in-silico-designed components will reduce the number of design cycles required for optimization by improving the components tested with each passing cycle. Improvements in device design can be used for decreasing development time, but will also facilitate construction of more complex genetic networks. In particular, in silico design tools currently enable simulation of multiple interacting RNAs, which could be used for carrying out complex logic operations (Qian and Winfree, 2011) from programmed RNA networks. In these higher-order computations, the programmability of toehold switches provides them with a distinct advantage over protein-based regulators that are currently less amenable to forward engineering. We also expect that additional improvements to toehold switch design performance can be obtained by incorporating the effects of the ;GRBS-linker term into future in silico design algorithms.
Toehold switches as RNA-to-protein signal transducers
The ability of toehold switches to activate translation in response to arbitrary RNA sequences, including full-length mRNAs, enables them to act as universal RNA-to-protein signal transducers. This new capability provides a generalizable approach to interfacing endogenous RNA networks with artificial genetic networks for synthetic biology and a non-destructive means of monitoring RNAs in living cells for answering fundamental questions in biology. Many future applications in synthetic biology will require genetic circuits that can sense the current status of the cell and operate on this information to modulate their activity. For instance, this capability can be used in metabolic engineering to construct self-optimizing networks that detect cellular stress levels or pathway-associated RNAs to tune network activity up or down for maximizing yields of useful chemical products. For biological studies, toehold switch signal transducers can be used to simultaneously monitor the levels of multiple untagged, endogenous RNA species by producing different output fluorescent proteins.
Potential role of similar systems in nature
The protein-like dynamic range of toehold switches challenges the widely held view that RNA-based systems offer weaker regulation than their protein counterparts (Liu et al., 2012). The performance of these riboregulators also suggests that some of their operating mechanisms may already play a role in natural systems. Although to our knowledge analogous systems have yet to be found in nature, our results indicate that a re-examination of bacterial genomes in a directed search for these specific regulatory mechanisms may be in order. Conversely, the absence of well-known natural systems that exploit the design principles of toehold switches could also imply that these synthetic devices are not easy to evolve. In particular, we hypothesize that the loop regions of loop-mediated riboregulators are more insulated from interactions with nearby bases than the unprotected single-stranded regions of toehold switch trigger RNAs. Thus, single-stranded trigger RNAs must sample a greater number of potential sequences to evolve to their intended function than natural loop-mediated trigger RNAs.
EXPERIMENTAL PROCEDURES
Extended experimental methods are available online
Strains and growth conditions
The following E. coli strains were used in this study: BL21 Star DE3 (F− ompT hsdSB (rB−mB−) gal dcm rne131 (DE3); Invitrogen), BL21 DE3 (F− ompT hsdSB (rB−mB−) gal dcm (DE3); Invitrogen), MG1655Pro (F− λ− ilvG- rfb-50 rph-1 SpR lacR tetR), and DH5α (endA1 recA1 gyrA96 thi-1 glnV44 relA1 hsdR17(rK−mK+) λ−). All strains were grown in LB medium with appropriate antibiotics at 37°C (see also Section S1).
Plasmid construction
All DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. Plasmids were constructed and purified using standard molecular biology techniques. Table S1 contains the primers and sequences used for experiments (see also Section S2).
Flow cytometry measurements and analysis
Flow cytometry was performed using a BD LSRFortessa cell analyzer equipped with a high-throughput sampler. Cells were typically diluted by a factor of ~65 into phosphate buffered saline (PBS) and sampled from 96-well plates. Forward scatter (FSC) was used for trigger and ~20,000 individual cells were analyzed. Error levels for the fluorescence measurements of ON state and OFF state cells were calculated from the standard deviation of measurements from at least three biological replicates. The relative error levels for the ON/OFF fluorescence ratios were then determined by adding the relative errors of ON and OFF state fluorescence in quadrature (see also Section S3 and S4).
Supplementary Material
HIGHLIGHTS.
Toehold switches detect RNAs with arbitrary sequences, including endogenous RNAs
A library of 26 orthogonal toehold switches provides < 12; component crosstalk
Rational RNA sequence design yields switches with > 400-fold mean dynamic range
Toehold switches can be integrated into the genome and complex genetic circuits
ACKNOWLEDGEMENTS
The authors acknowledge J. Kim, C. Myhrvold, K. Pardee, and X. Chen for critical reading of the manuscript and helpful discussions; J.M.L. Ho for preliminary experiments, plasmid preparation, and helpful discussions. This work was supported by a DARPA Living Foundries grant (HR001112C0061) to P.A.S., P.Y., and J.J.C.; an NIH Director's New Innovator Award (1DP2OD007292), an NIH Transformative Research Award (1R01EB018659), an Office of Naval Research (ONR) Young Investigator Program Award (N000141110914), ONR grants (N000141010827, N000141310593, N000141410610), a National Science Foundation (NSF) Faculty Early Career Development Award (CCF1054898), an NSF Expedition in Computing Award (CCF1317291), NSF grant (CCF1162459) and Wyss Institute funds to P.Y.; Defense Threat Reduction Agency grant HDTRA1-14-1-0006 to J.J.C.; ONR MURI grant N000141110725 to J.J.C. and P.A.S.; and the Howard Hughes Medical Institute (J.J.C.). A provisional patent based on this work has been filed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes extended experimental methods, 6 figures, and 7 tables.
AUTHOR CONTRIBUTIONS
A.A.G. conceived the project, designed the system, designed and performed the experiments, analyzed the data, and wrote the paper. P.A.S. and J.J.C. assisted in experiment design, data interpretation, and manuscript preparation. P.Y. conceived, designed, and supervised the study, interpreted the data, and wrote the paper. All authors commented on and approved the paper.
REFERENCES
- Auslaender S, Auslaender D, Mueller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- Bentele K, Saffert P, Rauscher R, Ignatova Z, Bluethgen N. Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 2013;9:675. doi: 10.1038/msb.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S, Wagner EGH. Antisense RNA-mediated transcriptional attenuation: an in vitro study of plasmid pT181. Mol. Microbiol. 2000;35:1469–1482. doi: 10.1046/j.1365-2958.2000.01813.x. [DOI] [PubMed] [Google Scholar]
- Callura JM, Cantor CR, Collins JJ. Genetic switchboard for synthetic biology applications. Proc. Natl. Acad. Sci. USA. 2012;109:5850–5855. doi: 10.1073/pnas.1203808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl. Acad. Sci. USA. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, Pierce NA. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010;28:1208–1212. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler SJ, Hoff KG, Smolke CD. Reprogramming Cellular Behavior with RNA Controllers Responsive to Endogenous Proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Rubens JR, Sarpeshkar R, Lu TK. Synthetic analog computation in living cells. Nature. 2013;497:619–623. doi: 10.1038/nature12148. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RM, Pierce NA. Triggered amplification by hybridization chain reaction. Proc. Natl. Acad. Sci. USA. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic Gene Networks That Count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- Gultyaev AP, Franch T, Gerdes K. Programmed cell death by hok/sok of plasmid R1: Coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hok family of mRNAs. J. Mol. Biol. 1997;273:26–37. doi: 10.1006/jmbi.1997.1295. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Cannone JJ, Konings D, Gautheret D. Predicting U-turns in Ribosomal RNA with Comparative Sequence Analysis. J. Mol. Biol. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- Isaacs FJ, Dwyer DJ, Ding CM, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat. Rev. Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AS, Lu TK, Bashor CJ, Ramirez CL, Pyenson NC, Joung JK, Collins JJ. A Synthetic Biology Framework for Programming Eukaryotic Transcription Functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-Sequence Determinants of Gene Expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Qi L, Lucks JB, Segall-Shapiro TH, Wang D, Mutalik VK, Arkin AP. An adaptor from translational to transcriptional control enables predictable assembly of complex regulation. Nat. Methods. 2012;9:1088–1094. doi: 10.1038/nmeth.2184. [DOI] [PubMed] [Google Scholar]
- Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl. Acad. Sci. USA. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491:249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalik VK, Qi L, Guimaraes JC, Lucks JB, Arkin AP. Rationally designed families of orthogonal RNA regulators of translation. Nat. Chem. Biol. 2012;8:447–454. doi: 10.1038/nchembio.919. [DOI] [PubMed] [Google Scholar]
- Purnick PEM, Weiss R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- Qi L, Lucks JB, Liu CC, Mutalik VK, Arkin AP. Engineering naturally occurring transacting non-coding RNAs to sense molecular signals. Nucleic Acids Res. 2012;40:5775–5786. doi: 10.1093/nar/gks168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Winfree E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- Qian L, Winfree E, Bruck J. Neural network computation with DNA strand displacement cascades. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- Rhodius VA, Segall-Shapiro TH, Sharon BD, Ghodasara A, Orlova E, Tabakh H, Burkhardt DH, Clancy K, Peterson TC, Gross CA, et al. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σ s, and promoters. Mol. Syst. Biol. 2013;9 doi: 10.1038/msb.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo G, Landrain TE, Jaramillo A. De novo automated design of small RNA circuits for engineering synthetic riboregulation in living cells. Proc. Natl. Acad. Sci. USA. 2012;109:15271–15276. doi: 10.1073/pnas.1203831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 2013;31:448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- Takahashi MK, Lucks JB. A modular strategy for engineering orthogonal chimeric RNA transcription regulators. Nucleic Acids Res. 2013;41:7577–7588. doi: 10.1093/nar/gkt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- Yin P, Choi HMT, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-fuelled molecular machine made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.