Abstract
Barcoded semisynthetic nucleosomes combined with massively parallel sequencing provide an innovative new platform for analyzing the histone-recognition and histone-modifying activities of chromatin-associated proteins.
Once thought to simply function as scaffolds for DNA packaging in the nucleus, histone proteins and their post-translational modifications (PTMs) are now appreciated for their dynamic roles as regulators of DNA accessibility and genome function. Histone PTMs act in part through the recruitment of effector proteins that interpret the combinatorial PTM landscape decorating the chromatin template. Although our understanding of how histone PTMs are written, erased and read has rapidly evolved over the past two decades, it has been challenging to biochemically determine how these activities are regulated in the context of the nucleosome, the fundamental building block of chromatin. Combining the power of two established approaches in chromatin biochemistry—native chemical ligation coupled with recombinant nucleosome technology and high-throughput sequencing—Nguyen et al.1 now provide a robust and versatile strategy for defining the complex behavior of chromatin regulators using pooled libraries of barcoded designer nucleosomes.
On the heels of breakthrough discoveries that connected histone acetyltransferases and deacetylases to transcriptional regulation2,3, seminal structural and biochemical studies using modified histone peptides have identified the bromodomain as an acetyl-lysine reader motif and, as such, a key intermediate linking histone PTMs to gene activity4. Subsequent technological advances in high-throughput, microarray-based screening approaches using immobilized histone peptides or tagged putative reader domains further facilitated the identification of histone PTM readers and their targets, deorphaning the domains of several chromatin effectors, including malignant brain tumor domains5, Tudor5 and plant homeodomains6.
Despite these advances, much of our understanding regarding the interactions of effector proteins with the chromatin landscape comes from the characterization of single domains with modified histone peptides. However, many effectors contain multiple reader domains, and each nucleosome contains two pairs each of the core histone proteins (histones H2A, H2B, H3 and H4), with some nucleosomes containing variant histones (e.g., histones H2A.Z, H2A.X and H3.3) that have specialized functions in chromatin7. This level of organization, both within nucleosomes and effector proteins, creates the potential for a high degree of complexity in chromatin recognition. For example, a chromatin effector containing multiple binding domains could specifically recognize multiple PTMs in the same histone tail (i.e., cis interactions) or on different histone tails in the same or adjacent nucleosome (i.e., trans interactions). Indeed, a recent seminal study using semisynthetic nucleosomes demonstrated that the BPTF subunit of the NURF chromatin remodeling complex recognizes H4K16ac and H3K4me3 through trans interactions mediated by its bromodomain and plant homeodomain finger, respectively8. Nucleosome-based approaches therefore provide a more physiologically relevant context for studying the preferred PTM landscape of chromatin effectors, but these studies have been limited to a low-throughput, one nucleosome–one factor pace. As such, our understanding of chromatin interactions in the context of nucleosomal templates has lagged.
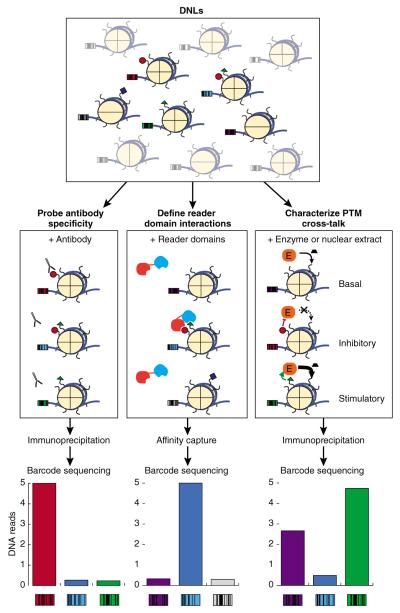
The approach described by Nguyen et al.1 streamlines previously developed expressed-protein ligation technology to rapidly generate an impressive library of semisynthetic nucleosomes harboring defined single and combinatorial PTMs on one or more histone tails. The notable advance in this new approach is the use of DNA with unique sequence tags (i.e., barcodes) to assemble the modified nucleosomes. These nucleosomes are pooled as DNA-barcoded nucleosome libraries (DNLs) that ultimately combine a competitive in-solution biochemical assay format with the sensitivity of in vitro chromatin immunoprecipitation, which is then coupled with high-throughput sequencing (ChIP-seq; Fig. 1). In addition to high-throughput characterization of the PTM specificity of histone reader domains and modifiers, DNL libraries offer a powerful and robust method to validate histone PTM-specific antibodies. Indeed, antibody specificity (or lack thereof) is a substantial concern in the chromatin field, particularly for accurate interpretation of genome-wide histone PTM analyses9.
Figure 1.
DNLs with modified histones can been used to scrutinize histone PTM antibody specificity, define nucleosomal interactions for individual and paired reader domains, and examine the histone-modifying activity of enzymes by identifying enriched nucleosomes through massively parallel sequencing of DNA barcodes.
Nguyen et al.1 use this platform to interrogate two well-studied histone readers, BPTF and BRD4, as well as the histone acetyltransferase p300. In addition to confirming known interactions of these factors8,10, the authors demonstrate an important role for histone H4 polyacetylation in recruiting these proteins. Notably, binding of p300 to polyacetylated H4 promotes further histone acetylation by this enzyme. They also observed this positive feedback loop on H3K4me3 nucleosomes, thereby affirming that trans-histone crossalk influences chromatin regulators such as p300.
The creation of DNL libraries is a dramatic step forward for chromatin investigation and will likely yield breakthrough discoveries in the near future. Although the power of this technology is well-illustrated in the Nguyen et al.1 report, semisynthetic nucleosome construction remains technically challenging, demanding a high proficiency in organic chemistry and biochemical approaches. It therefore remains to be seen whether this technology will be broadly applied in chromatin research. In addition, this semiquantitative technology may not be able to clarify all of the complex interactions that could theoretically occur, such as asymmetric PTM recognition (i.e., differentially modified histone H3 dimers in the same nucleosome) or trans internucleosomal interactions (i.e., adjacent nucleosomes harboring different PTMs). Future iterations of this DNL technology may one day overcome these limitations. Nonetheless, this new approach shows great promise for clarifying the complexities behind the recruitment and activity of chromatin regulators.
Footnotes
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
References
- 1.Nguyen UTT, et al. Nat. Methods. 2014;11:834–840. doi: 10.1038/nmeth.3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brownell JE, et al. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 3.Taunton J, Hassig CA, Schreiber SL. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 4.Dhalluin C, et al. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, et al. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, et al. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maze I, Noh KM, Soshnev AA, Allis CD. Nat. Rev. Genet. 2014;15:259–271. doi: 10.1038/nrg3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, et al. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs SM, Strahl BD. Epigenomics. 2011;3:247–249. doi: 10.2217/epi.11.23. [DOI] [PubMed] [Google Scholar]
- 10.Filippakopoulos P, et al. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]