Abstract
MicroRNAs (miRNAs) are short, non-coding RNAs that employ classic Watson-Crick base-pairing to identify their target genes, ultimately resulting in destabilizing their target mRNAs and/or inhibiting their translation. The role of miRNAs in a wide-range of human diseases, including those afflicting the kidney, has been intensely investigated. However there is still a vast dearth of knowledge regarding their specific mode of action and therapeutic effects in various kidney diseases. This review discusses the latest efforts to further our understanding of the basic biology of miRNAs, their impact on various kidney diseases and their potential as novel biomarkers and therapeutic agents. We initially provide an overview of miRNA biology and the canonical pathway implicated in their biogenesis. We will then discuss commonly employed experimental strategies for miRNA research and highlight some of the newly described state-of-the-art technologies to identify miRNAs and their target genes. Finally, we will carefully examine the emerging role of miRNAs in the pathogenesis of various kidney diseases.
Keywords: MicroRNA, Non-coding RNA, Kidney Disease, Kidney development, Diabetic nephropathy
Introduction
Non-coding RNAs (ncRNAs) comprise multiple classes of RNAs that by definition are not transcribed into proteins. To date, the most intensely studied ncRNAs are microRNAs (miRNAs). miRNAs are small (usually 19–24 nucleotides in length), non-coding, single-stranded RNAs that provide a new layer of gene regulation [1, 2] (Figure-1). They inhibit the expression of messenger RNAs (mRNAs) primarily by pairing with the 3'-UTR (three prime untranslated region) of their complementary mRNAs, causing translational repression and/or mRNA cleavage. miRNAs are largely evolutionarily conserved among species and commonly participate in the regulation of diverse cellular processes, including apoptosis, proliferation, hematopoiesis, and angiogenesis, [3].
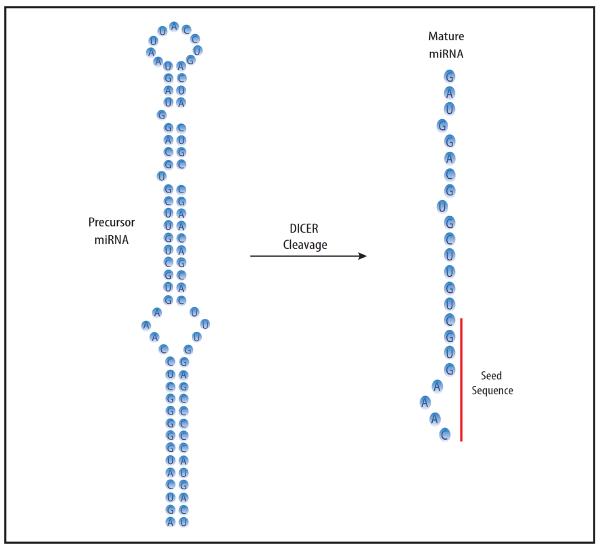
Figure 1. Schematic of miRNA maturation mediated by DICER.
The 70-80 nucleotide (nt) long double stranded, precursor-miRNA is cleaved by Dicer at the stem-loop sequence into a single stranded, 20-22 nt long mature miRNA. This mature miRNA is subsequently loaded into the RNA Induced Silencing Complex (RISC), where it exerts its gene regulatory activity. Highlighted in red is the 5–8 nt miRNA seed sequence, which is complementary to target mRNAs and is the responsible element for miRNA:mRNA interactions.
During the past decade, it has become increasingly apparent that miRNAs are causally linked to a variety of human diseases, including cancer, heart disease and diabetes [4]. The crucial role of miRNAs in the development of kidney diseases has been widely recognized. This is of no surprise considering that an individual microRNA can often regulate the expression of multiple target genes. Thus, dysregulation of a single microRNA can, in principle, influence an entire gene network and thereby contribute to complex disease phenotypes commonly observed in a variety of kidney diseases.
Discovery of miRNAs
One of the major advances in biology within the last decade has undoubtedly been the discovery of both small and large regulatory RNAs, including miRNAs which are among the best characterized group of these newly discovered regulatory class of RNAs [5].
Lee et al. initially described the first reported miRNA, lin4, as a novel, non-protein coding gene, which served as a regulator of developmental timing in Caenorhabditis elegans (C. elegans) [6]. Simultaneous reports by Lee et al., and Wightman et al., discovered that the protein coding gene, lin14, was negatively regulated by lin4, through interactions at the 3'-UTR of lin14 [6, 7]. Thereafter, Reinhart et al. reported a similar discovery of a small RNA in C. elegans, let7, whose target gene lin28 was also essential in maintaining proper developmental timing in C. elegans [8]. Both lin4 and let7 were considered to be unique and did not enjoy a great deal of attention for an extended period of time. However, the significance of these two supposedly isolated genes became relevant several years after their initial discovery when several groups almost simultaneously reported that this obscure family of non-protein coding genes, were indeed highly evolutionarily conserved among several species, including humans [9–12]. Shortly after, several groups convincingly demonstrated the basic steps of miRNA biogenesis, its modes of actions and ultimately its importance to human health and disease [2, 13].
The discovery of miRNAs in diverse species raised the question of what these tiny noncoding RNAs may be doing in the cell and whether they play any roles in disease processes. In this regard, Calin et al., made the first association between microRNA dysregulation and cancer. In their landmark study, miR-15 and -16, were found to be significantly upregulated in patients with B-cell leukemia [14]. These early association studies were followed up by other groundbreaking studies, where the effect of global microRNA deletion was investigated [15–18]. From these and other investigations, it was concluded that miRNAs are dysregulated in almost all types of human cancer and specific signatures of aberrantly expressed miRNAs harbor diagnostic, prognostic and therapeutic implications.
Biogenesis and mode of action
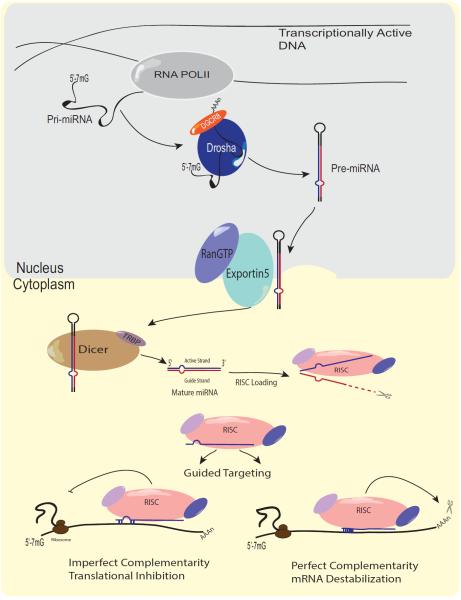
Mature miRNAs are evolutionarily conserved single-stranded RNAs. The generation of mature miRNAs is a multi-step process that starts with the initial transcription of their genes by RNA polymerase II into a long primary microRNA (pri-miRNA) (Figure-2). The nuclear pri-miRNA, which can be several hundred to several kilobases long, is then processed by two enzymes, Drosha and its binding partner, DGCR8 (DiGeorge syndrome critical region gene 8), into a 70-80 nucleotide (nt) long precursor miRNA (pre-miRNA) [19, 20]. The pre-miRNA is exported from the nucleus utilizing the Ran-Gap/Exportin-5 transport system into the cytoplasm [21] where it undergoes a final processing step by the RNAse III enzyme, Dicer. Dicer cleaves the pre-miRNA into a 20-22 nt long, double-stranded mature miRNA and subsequently facilitates loading of a single active strand of the mature miRNA into the RNA Induced Silencing Complex (RISC) [22]. Here the mature miRNA is bound to the Argonaute family of proteins, which expose the 5' seed sequence of the mature miRNA and direct the miRNAs targeting to the target mRNA [23]. At each step of miRNA biogenesis, several proteins contribute to the ability of Drosha and Dicer to accurately recognize sequence-specific features of pri-, pre and mature miRNAs to facilitate proper cleavage and maturation of biologically active miRNA. These different maturation steps are critical to maintain proper miRNA levels in order to prevent cellular abnormalities [24]. Once bound to its target mRNA, the RISC complex can facilitate several forms of transcriptional repression depending on the strength of the miRNA-mRNA interaction and seed-sequence/target site complementarity. Protein expression of the target mRNA can be blocked by miRNA/RISC complexes through several mechanisms, including ribosome stalling along on the mRNA in one instance, ribosomal dropoff, translation initiation repression, target mRNA degradation and mRNA deadenylation [25, 26]. Interestingly, miRNA-RISC targeting is not exclusive to the cytoplasm where the majority of mRNA is exported for protein translation. Indeed, miRNA-RISC shuttling between the cytoplasm and nucleus has been shown for several miRNAs, which elicit their activity within the nucleus to regulate the expression of pri-miRNAs and other target genes [23, 27, 28]. Additional nuclear roles of miRNAs have been demonstrated to include direct DNA binding of miRNAs to promoter regions of genes to functionally regulate gene transcription of certain genes, in either a gene activating or gene repressive manner [25].
Figure 2. miRNA biogenesis and mode of action.
Pri-miRNA are transcribed by RNA Polymerase II (Pol II), processed to include a 5'-7meG cap and a 3'-poly-A tail, and cleaved by the first member of the microprocessor complex, the RNAse III enzyme Drosha/DGCR8, into a 70-80 nt long pre-miRNA. This pre-miRNA is exported out of the nucleus via Exportin-5/RanGTP, cleaved by Dicer into a mature 20-22nt long miRNA and loaded into the RNA Induced Silencing Complex (RISC). RISC guides the single stranded mature, active miRNA to its target mRNAs and based on sequence complementarity to the mRNA target site and miRNA seed sequence may perform several gene regulatory functions, including ribosomal mediated translation inhibition or mRNA degradation/destabilization.
In 2008, it was discovered that miRNAs are also present in blood, where they can act much like hormones in an endocrine system [29, 30]. Indeed, circulating miRNAs can be transported in microparticles (exosomes, microvesicles) or alternatively can be directly released into blood in response to injury, chronic inflammation, or apoptosis [31]. Circulating miRNAs enter the bloodstream and are taken up by the cells via endocytosis where they can identify their targets and exert their effects on the recipient cells.
miRNA levels are meticulously regulated both at transcriptional and post-transcriptional levels. With regards to their transcriptional regulation, miRNAs have been shown to utilize either host gene promoters, or possess their own unique promoter elements upstream of the pri-miRNA putative transcription start sites [32]. Approximately 1/3 of intronic miRNAs are transcribed independently from their host genes [32, 33]. miRNA promoters harbor elements that resemble traditional Pol II promoters and typically contain enhancer-like elements upstream of transcriptional start sites [34, 35]. Furthermore, alterations to the histone code at miRNA promoters add an additional level of miRNA transcriptional regulation [36, 37].
Another layer of miRNA regulation is the existence of miRNA-transcription factor feedback circuitries. For example, O'Donnel et al., identified the oncogene c-Myc to be able to induce expression of a cluster of miRNAs on human chromosome 13, by binding to a putative promoter region upstream of the miR-17-92-20a cluster. Two of these miRNAs, miR-17-5p and miR-20a were shown to negatively regulate the transcription factor E2F1, a target of c-Myc [38]. Furthermore, transcription factor binding to miRNA promoters has been extensively described as critical in the regulation of several tissue specific cell fates. Specifically, a negative regulator of transcription and myogenesis, Yin Yang 1 (YY1) was shown to bind directly to several sites within the 20-kilobase region upstream of the miR-29b/c transcription start site. YY1 is a potent inhibitor of myogenesis. In this specific case, YY1 and miR-29b/c create a negative feedback loop where YY1 is negatively regulated by miR-29b/c, which serves to alleviate miR-29b/c repression during myogenesis and enhance the effect of miR-29b/c on differentiation [39].
Additionally, miRNAs and proteins comprising the miRNA processor complex (Drosha, Dicer, Argonaute, among others) are heavily regulated at the post-transcriptional level, and this serves as an additional layer influencing the final readout of miRNAs. Specifically, regulation of miRNA activity and stability can be influenced by post-translational modifications of Dicer and Argonaute proteins. Moreover, changes to accessory proteins that interact with Dicer and Argonaute may also affect the stability and final readout of miRNA activity under certain conditions [40].
Pre-miRNA and mature miRNAs may be further cleaved or modified with sequence specific alterations that could affect their stability or incorporation into RISC and thereby affect their final targeting potential [41]. For instance, adenylation of the 3' end of the mature miRNA increases stability [42, 43], whereas uridylation at the 3' end of miRNAs commonly decreases their stability [41, 44]. Interestingly, miRNA methylation seems to protect from 3'-end uridylation, [45] which would suggest a role for increased stability of mature miRNAs.
Differential profiling of miRNAs
Using high-throughput techniques (e.g., miRNA microarrays, Next-Gen Sequencing) differentially expressed miRNAs are routinely identified in a variety of human diseases. Although the use of miRNA microarrays for identifying differentially expressed microRNA profiling has been the most widespread method for many years, the emergence of next generation sequencing as a readily available research tool has recently challenged this enduring technique. We will next briefly review commonly used tools to identify miRNAs, and then describe standard methods to validate and assess the functional significance of miRNAs in physiological and pathological conditions.
miRNA microarray
A key aspect of discovery-based designs in miRNA research has been expression profiling for the global expression profile of miRNAs to gain insight into important miRNAs enriched or severely dysregulated. Microarray technology has proven to be a critical tool in this regard, and while not perfect, it has served as a robust means of identifying dysregulated miRNAs.
In order to increase sensitivity of miRNA microarrays, total RNA is initially enriched for small RNAs, since this class of RNA comprises a small proportion of total RNA. miRNAs are then 3'-end labeled with specific fluorophores and prepared for hybridization to miRNA arrays. These arrays are labeled with miRNA specific probes that exhibit specificity for individual miRNAs and can be designed for species-specific analysis. Several challenges, however, exist when profiling miRNAs with this method. For instance, a major shortcoming of the use of microarray is that since miRNA-binding affinity is different for each miRNA and its cognate probe, the probe hybridization for each miRNA may not be entirely specific or necessarily quantitative when assessed via miRNA microarray. Another important limitation with miRNA microarray technology is that sometimes-minor changes in miRNA levels, which could be biologically relevant, may not be detectable. This is of great interest since although less abundant miRNAs may not be detected based on microarrays; they may still serve as critical regulators of key genes in the context of a disease.
Next Generation Sequencing
The development of next-generation sequencing technologies has recently offered an unprecedented scale and depth of miRNA profiling. The high throughput miRNA RNA-Sequencing (miR-Seq) has been developed to robustly annotate and quantify miRNA expression levels with a high degree of precision and sensitivity. Next generation sequencing employs massively parallel sequencing, generating millions of reads per sequencing run. The increased sensitivity bypasses limitations of microarray technology and allows for quantification and discovery of miRNAs present in very small quantities.
Briefly, total enriched miRNAs are size selected by gel electrophoresis, subjected to end repair, amplification and finally platform specific adapters are ligated to the 3' and 5' ends of purified miRNA species and subjected to high throughput sequencing. These reads are mapped back to the genome of interest and abundance is quantified using several bioinformatics algorithms. The advent of readily accessible, high throughput sequencing platforms (Illumina, Solexa, Ion-Torrent) has greatly aided the research community with regards to the depth and sensitivity of global miRNA profiling and discovery. Currently, several companies (e.g., LC Sciences, Exiqon Inc., and ArrayStar Inc.) with expertise in noncoding RNA interrogation provide extensive miR-Seq services. A limiting step and a critical component of any successful miR-Seq projects is a strong bioinformatics infrastructure to accurately identify differentially regulated or novel miRNAs.
Validation of miRNAs
To validate the dysregulated miRNAs identified from miRNA profiling, northern blotting, real-time PCR and in situ hybridization are traditionally employed in pinpointing the size, tissue distribution and localization of specific miRNAs. These are especially useful in the kidney where there are several important cell compartments where miRNAs may be localized and differentially expressed. Following total RNA isolation, small RNAs including miRNAs are separated by gel electrophoresis and transferred to a nitrocellulose membrane. RNA is fixed to the membrane via UV crosslinking or baking. miRNA specific probes are used to detect the miRNA of interest and exist in two primary types: Poly-A tailed, hexamer labeled miRNA specific probes or LNA (miRCury, Exiqon) miRNA specific probes. Traditionally, both are 3' end labeled with 32P-ATP radioactive probe and detected using X-ray films or phosphorimager systems.
In situ hybridization is another powerful tool to determine the localization and relative expression levels of miRNAs in a myriad of cell compartments and tissues. This serves as an extremely useful tool in utilizing preserved patient samples to either quantify or determine the human pattern of expression of a specific miRNA of interest. To perform in situ hybridization assays, formalin fixed paraffin embedded or frozen tissue sections are treated with a series of steps to reduce miRNA diffusion and loss during the in situ procedure. Sequence specific miRNA probes are then hybridized based on calculated Tm for each specific probe. Locked Nucleic Acid (LNA) probes increase specificity and degree of detection and are a robust tool utilized in many in situ protocols. Probes can be labeled at the 5'-, 3'- ends with fluorophores to allow direct visualization following several wash steps, post-fixation and mounting. Alternatively, the probes may be labeled with small haptens like Digoxigenin (DIG), either unconjugated or conjugated with alkaline phosphatase. These can be coupled to signal amplification systems; most commonly in the case of miRNAs these include tyramide signal amplification systems [46, 47].
The most commonly used method to detect specific alterations in miRNA levels, however, remains the utilization of quantitative real-time PCR. Because miRNAs are short, designing highly specific probes for their detection poses a challenge. Two general approaches are employed when performing the reverse transcription, microRNA specific reverse transcription (RT) or universal reverse transcription. miRNA-specific RT utilizes stem-loop primers using a single stranded region complementary to the 3'-end of the mature miRNA and a double stranded universal primer region (stem) attached to this region that permits amplification of the specific miRNA under query. This miRNA specific RT product can be used to quantitative PCR by taking advantage of miRNA specific fluorescently labeled probes (Taq-Man, Life Tech) and a universal primer.
The second approach, universal reverse transcription, employs an approach to add poly A tails to the 3' end of all miRNAs present within a given RNA population, and subsequently these are amplified based on the Oligo(dT) universal primer which recognizes the 5' end of the primer attached to the miRNA. This cDNA template is then used in quantitative PCR with miRNA specific primers designed for quantitative PCR detection with SYBR Green Fluorescent Dyes. Development of these off-the shelf assays has greatly aided the entire scientific community in creating extremely accessible reagents to ask important questions related to miRNA research.
Identification of miRNAs' targets
miRNAs are one of three classes of small, noncoding RNA. The others include Piwi-Interacting-RNAs (piwiRNA), and small interfering RNAs (siRNA). All three classes recognize and bind to target mRNAs to inhibit gene expression.
Several bioinformatics and experimental approaches have traditionally been employed to identify novel and biologically relevant targets of miRNAs. Among these well established, open-source bioinformatics tools include TargetScan (www.targetscan.org), miRANDA (www.microrna.org) and Pictar (www.pictar.mdc-berlin.de) among many others. These sites use miRNA seed sequence to predict their targets. The so called seed sequence, a short continuous region of Watson-Crick base pairing between nucleotides 2–7 of the miRNA and complementary sequences in 3′ UTR regions of target mRNAs, determines the genes that could potentially be regulated by a specific miRNA.
The use of open-source bioinformatics tools, although indispensible in miRNA research, will only identify potential targets of a specific miRNA, and these potential targets must be each individually validated in the context of a stimulus and cell/tissue-specific conditions. More importantly, these computational resources will not validate the in vivo interaction between a specific miRNA and its potential target nor it will reveal novel targets for a miRNA. In this regard, experimental de novo identification of miRNA targets has been greatly aided by the ability to capture and quantify specific miRNA-mRNA interactions by a number of newly developed techniques. The most widely adapted technique, CLIP (UV Crosslinking and Immunoprecipitation) has been coupled with high throughput sequencing to aid in identification of novel miRNA binding sites throughout the genome. Several variations have been utilized but all rely on the same principle of purifying Argonaute-miRNA and Argonaute-mRNA interactions to generate genome wide interaction profiles of miRNA/mRNA interactions in vivo. The first widely employed method; HITS (High Throughput Sequencing)-CLIP established the feasibility of this technique to generate meaningful miRNA/mRNA interaction profiles [48]. A second technique commonly employed, iCLIP (individual-nucleotide resolution)-CLIP generates extremely precise data to help map miRNA-mRNA interactions at a single base pair level by circularization of the purified sequences followed by restriction digest linearization and library preparation. This technique identifies, with great precision, the site of the miRNA and mRNA interactions. [49]. Another method to precisely identify crosslinking sites and increase crosslinking efficiency, PAR (Photoactivatable-Ribonucleoside)-CLIP utilizes photoactivatable 4-thiouridine (4SU), which when excited by UV spectra at 365nm contributes to enhanced crosslinking efficiency. Sequenced cDNA prepared from these reactions is scored for the conversion of thymidine to cytidine, indicating a unique crosslinking site [50]. These interactions can be further verified by RNA-IP experiments, followed by RNA purification, cDNA synthesis and gene specific quantitative RT-PCR.
A powerful proteomic approach has recently been employed to identify novel targets of miRNAs. Stable Isotope Labeled Amino Acid Cell Culture (SILAC), employs the global incorporation of isotope labeled amino acids (Lysine, Arginine most typically), with a 13C6 or 15N4 into all proteins synthesized under those specific culture conditions. Following treatment with the miRNA of interest, these “heavy” protein populations are intermixed with cells cultured in “light” isotope conditions and treated with control mimics. Protein lysates are subjected to gel electrophoresis and Mass Spec analysis. Thereafter, relative peak intensities of each distinctly labeled peptide are used to quantify abundance for proteins in each experimental group. Heavy labeled proteins which exhibit significantly reduced peak intensity compared to light labeled controls, are subject to further bioinformatics analysis to identify the presence or absence of the desired miRNA target site. SILAC permits relative quantification of the proteome between “heavy” and “light” samples and is a powerful tool to identify unique miRNA targets acting at the level of protein translation, the ultimate final output of miRNA activity.
A commonly used method to validate the effect of miRNAs on a specific target is the use of gene reporter assays where the wild-type 3' UTR of the proposed mRNA target gene is cloned within the 3'-UTR of a gene reporter, most commonly luciferase. This construct along with the miRNA of interest is subsequently overexpressed within cells and reporter activity is then measured. Typically, inhibition of luciferase activity is observed in cells overexpressing the wild-type 3'-UTR and miRNA of interest, when compared to a non-targeting control miRNA. This would suggest that the mRNA could be a strong target of the miRNA [51].
An additional biochemical approach to verify the nature of a miRNA and potential target can be accomplished through the use of miRNA target site protectors. These are short oligonucleotides designed against a specific miRNA binding site within the 3'-UTR of the putative target mRNA. Once bound, the target site protector prevents interaction of the miRNA and its target mRNA, and hence it will determine whether a specific miRNA-dependent phenotype is due to a potential target of that miRNA. An advantage to this approach is to reduce ambiguity regarding the off-target effects of miRNA and to limit analysis to a single miRNA target [52, 53].
Manipulating microRNA levels in vitro and in vivo
A single miRNA may exert regulatory effects on a number of target genes and related biological pathways [54, 55]. The pleiotropic nature of miRNAs allows for robust and extremely penetrant gene regulation within the context of normal and pathological processes. This is indeed one of the main advantages of targeting miRNAs since each miRNA targets several coding and non-coding genes that are involved in many pathways critical in human pathologies. Furthermore, understanding the function of miRNAs lies in the ability to identify the direct and indirect targets of a specific miRNA and characterize numerous genes that a single miRNA can target by using several in vitro and in vivo tools described below.
Although we have just begun to understand the full range of biological functions of miRNAs, the ability of miRNAs to target multiple genes and pathways has created significant interest for their therapeutic development. Indeed, restoring or repressing miRNAs expression has proven extremely useful as potential therapeutic applications within various pathological states.
The traditional and dominant in vitro form of miRNA manipulation has been through the use of miRNA mimics to restore miRNA levels or antisense miRNAs to block miRNA function.
miRNA mimics are double stranded synthetic oligonucleotides manufactured as either the pre-miRNA or the mature miRNA, which are processed into a single stranded form in the cell to mimic the effect of miRNAs. Several strategies are under intense investigation in order to enhance the stability of miRNA mimics and protect them from degradation. These include, several lipid-based delivery reagents and formulations of inorganic and/or organic nanoparticles, namely modified polymers like polyethylenimine (PEI), PEG (polyethylglycol) and poly (lactide-co-glycolide) (PLGA). These, in theory, should allow for less frequent dosing and may avoid the need for continuous infusions [56].
Similarly, antisense oligonucleotides (anti-miRs) are commonly used to block the effect of miRNAs. Anti-miRs include locked nucleic acids (LNA anti-miRs), antagomirs and miRNA sponges. The anti-miRs are designed against the mature miRNA sequence and sequester miRNA from incorporation in to the RISC complexes. Several modifications mainly directed to the 2'-OH, 4'-hydrogens, and α-oxygen of the phosphate group and the amide linkage between ribose and base have been designed to overcome poor stability of miRNA for cell and tissue delivery. Examples of commonly used modifications include: Locked Nucleic Acid (LNA) modification which links the 4'-C of ribose with 2'-OH; modifications at the 2'-OH group, including 2'-O- Methoxyethyl and 2'-O-Methyl and 2'-Fluoro; and finally, the α-oxygen of the phosphate group which can be replaced with methylphosphonate, phosphothiorate and boranophosphate groups [56]. These modifications in vivo will protect against exonuclease activity, reduce the inflammatory response, increased tissue/cell uptake and enhanced targeting efficacy. Currently, there is a single LNA-Anti-miR that has progressed through clinical trials. Developed and tested by Santaris Bio, the liver specific LNA-Anti-miR-122, miravirsen, currently is being tested for the treatment of Hepatitis C.
Antagomirs are cholesterol-conjugated synthetic RNAs with a 2'-O-methoxyethyl linkage and phosphorothioate modification, whereas miRNA sponges are multiple short tandem repeats of the miRNA target site cloned as the 3'UTR of a reporter gene within a plasmid, retrovirus or lentivirus. Once expressed within the target cell type or tissue, miRNAs preferentially bind the sponge and are prevented by interacting with RISC, thereby sequestering their ability to inhibit true endogenous mRNA targets [57].
Extensive studies have convincingly shown the feasibility and therapeutic efficacy of miRNA inhibition within the kidney from several groups [58–60]. However, miRNA replacement within the kidney in vivo is less well investigated. There are several studies, on the other hand, that have shown the feasibility of miRNA replacement in other model systems. For instance, Liu et al., investigated the therapeutic efficacy of miR-34 replacement within prostate tumors with increased expression of CD44, a key marker of malignant prostate cancer stem cells. They found that coupling miR-34 with a lipid based delivery agent administered systemically via tail vein injection to NOD-SCID mice was sufficient to reduce tumor burden, metastatic potential and increase overall survival probability compared to mice injected with non-specific controls. Similarly, in a separate study, systemic miR-34a and miR-143/145 restoration was used to effectively reduce tumor burden in mice with pancreatic cancer xenografts [61]. These putative miRNA tumor suppressors were coupled with lipid based nanoparticles (nanovectors) and systemically administered to mice harboring either subcutaneous or orthotopic xenografts. Tumor growth was blocked and increased apoptosis, and decreased proliferations were achieved in these tumor models. Presently, MRX34 (Mirna Therapeutics), a liposome formulated miR-34 mimic, is the first miRNA mimic to enter clinical trials for patients with unresectable primary liver cancer or advanced metastatic cancer with liver involvement.
A potential downside effect of miRNA manipulation in vitro and in vivo is the off target effects induced by modulation of multiple miRNA targets. Specifically, antagomirs may target related miRNAs of a specific cluster or family, since many of these miRNAs exhibit strong sequence homology. Similarly, miRNA replacement therapy may result in modest downregulation of several genes, in addition to the intended target that may have unwanted consequences. Lastly, the tissue distribution of pharmacological miRNA manipulation suggests that other tissues, besides the desired tissue, may incorporate and affect miRNA levels and activity in a negative manner.
miRNAs & kidney homeostasis
Thus far, we have described the framework for understanding what miRNAs are, how they elicit their potent gene regulatory activity and how we detect and target specific miRNAs to unravel precise mechanisms related to interesting miRNAs and observed phenotypes. We will now shift our focus to understand the impact of miRNAs on kidney-related research in physiological and pathological conditions. Several groups highlighted the importance of miRNAs to normal kidney homeostasis when they set out to genetically delete Drosha and Dicer in podocytes [62–65]. Taken together, these observations demonstrated that a global knockdown of miRNA levels leads to significant proteinuria and podocyte effacement as early as 3 weeks, followed by glomerulopathy, reduced slit diaphragm protein (nephrin and podocin) expression, proteinuria, and renal failure [62–65].
Several additional studies investigated the role of Dicer in specific kidney compartments. For instance, Patel et al., demonstrated that Dicer deletion in maturing renal tubules and the ureteric bud stalk up until embryonic day 17.5 in mice leads to severe tubular and glomerular cysts, mediated in part by the downregulation of miR-200 family members, of which miR-200b/c;-429 were shown to target Pkd1 [66]. Consistent with the severe phenotype observed upon Dicer deletion in the developing kidney, conditional deletion of Dicer in Juxtaglomerular cells reduced the number of Juxtaglomerular cells present in the adult kidney accompanied by a significant decrease in renin expression (Ren1 and Ren2), with the subsequent reduced plasma renin levels, and reduced blood pressure. Finally, Dicer deletion in Juxtaglomerular cells gave rise to vascular abnormalities and striped fibrosis [67]. Conversely, conditional deletion of Dicer in adult renal proximal tubules does not result in a severe phenotype; in fact, these mice exhibit normal kidney development, morphology, histology and function. Interestingly, when stressed by ischemia (kidney ischemia/reperfusion model), these mice exhibited renoprotective features, namely improved kidney function, histology and increased survival [68]. These studies serve to highlight the diverse effect of miRNAs in different kidney cell types.
We now explore the role of several well-established miRNAs involved in the pathophysiology of a variety of kidney diseases. These selected miRNAs have been summarized in Table 1, and while not an exhaustive list provide an introductory framework to understand the importance of miRNAs within the pathophysiology of several kidney diseases.
Table 1.
Select miRNAs and their role in different kidney diseases
| miRNA: | Disease: | Expression Changes: | Key Targets: |
|---|---|---|---|
| miR-193a | Focal Segmental Glomerulosclerosis | ↑ | WT-1 |
| miR-31 | ↓ | NOTCH1, P53 | |
| miR-192 | Diabetic Nephropathy | ↑ | SIP1 |
| miR-377 | ↑ | PAK1, SOD | |
| miR-29a/b | ↓ | Collagens | |
| miR-29c | ↑ | SPRY1 | |
| miR-93 | ↓ | VEGFA | |
| miR-21 | Fibrosis | ↑ | PPARα, MPV171 |
| miR-22 | ↓ | BMP7, BMP6, BMPR1B | |
| miR-17 | Polycystic Kidney Disease | ↑ | PKD 2 |
| miR-17/92 Family | ↑ | PKD 1 | |
| PKD 2 | |||
| Oncomir-1 | Wilm's Tumor | ↑ | - |
| miR-370 | ↑ | STAT3 | |
| miR-562 | ↓ | EYA1 | |
| miR-185 | ↓ | SIX1 |
Focal segmental glomerulosclerosis (FSGS)
Several studies have investigated the potential role of miRNAs as biomarkers to predict FSGS. However, until only very recently, there have been only a few functional studies investigating the impact of miRNA dysregulation on the pathogenesis and progression of FSGS.
While searching for miRNAs that could affect breast tumorigenesis, Gebeshuber et al., unexpectedly identified that conditional overexpression of miR-193a is able to phenocopy features of FSGS [69]. To determine whether the cause of miR-193a-induced FSGS is an aberrantly secreted factor, the authors transplanted a single transgenic kidney into a wild-type animal and saw selective induction of FSGS in the transgenic kidney alone. The reciprocal transplantation experiments confirmed that miR-193a-induced-FSGS does not result from secretion of a pathogenic factor, but rather from exclusive overexpression of miR-193a. To delineate the mechanism which caused the induction of FSGS, the authors compared significantly downregulated RNA-Seq gene expression signatures from glomeruli of transgenic mice against a list of previously described genes downregulated in human cases of FSGS. From this analysis, Wilm's tumor gene (WT-1) was shown to be a direct target of miR-193a. Importantly, podocyte-specific deletion of WT-1 resulted in a similar phenotype observed upon miR-193a overexpression. Finally, the authors discovered that miR-193a was significantly overexpressed in patients diagnosed with several histological subtypes of FSGS when compared to either healthy individuals or individuals with other primary glomerular diseases (IgA Nephropathy, Minimal Change Disease, Membranous Nephropathy) [69]. This detailed study merits further work to determine if miR-193a is a prime therapeutic target for patients with FSGS.
In a separate study, Wu et al., recently discovered that members of the miR-30 family underwent significant downregulation in human podocytes treated with several factors known to induce podocyte injury, including TGF-β (Transforming growth factor beta), LPS (Lipopolysaccharide) or PAN (Puromycin Aminonucleoside). Interestingly, the miR-30 is among the most highly expressed miRNAs within the podocyte. This significant downregulation seems to contribute to TGF-β or PAN-induced podocyte cytoskeletal damage and apoptosis. These features associated with FSGS were attenuated upon overexpression of miR-30 family members, accomplished via glucocorticoid-mediated induction of miR-30 [70].
Diabetic nephropathy
Diabetic nephropathy (DN) is the most common cause of end stage renal disease (ESRD) in the United States. There is increasing experimental data to implicate miRNAs as essential in the progression of DN and possibly they may also serve as key targets of therapy.
The Natarajan group were the first to identify a group of differentially dysregulated miRNAs in glomeruli from type 1 (STZ) and type 2 diabetic mice (db/db), as well as mesangial cells cultured under high glucose conditions [71]. From this screening, they focused on miR-192, which has become one of the more prominently studied miRNAs within the DN field. miR-192 was shown to regulate proliferation of kidney mesangial cells via downregulation of SIP1 (Survival of Motor Neuron Interacting Protein-1). Repression of SIP1 by miR-192 was shown to increase collagen in response to TGF-β1/Smad signaling. They continued on to comprehensively dissect the role of miR-192 at a molecular level in several subsequent studies [72]. Namely, they have shown that inhibition of miR-192 with exogenously administered LNA-miR-192 antagomirs in STZ diabetic mice is renoprotective against features of DN [58]. These studies were later corroborated, when global genetic deletion of miR-192 proved to be similarly renoprotective in diabetic mice [73].
Recently, several reports, including data from our own laboratory, have further characterized the role of miRNAs in the homeostasis of podocytes and glomerular endothelial cells in different models of diabetic nephropathy. We have shown that two miRNAs, miR-93 and miR-29c, are significantly dysregulated within the diabetic environment in podocytes and glomerular endothelial cells, in vitro and in vivo. Using comparative miRNA microarrays, both miR-93 and miR-29c were identified as signature miRNAs in hyperglycemic conditions [59, 74]. miR-93 levels were significantly lower in glomeruli from Type 2 diabetic db/db mice compared with controls and in high glucose-treated podocytes and renal microvascular endothelial cells. In contrast, miR-29c levels were significantly increased in glomeruli obtained from diabetic db/db mice. VEGF-A was identified as a relevant target of miR-93 with a known role within the kidney. Further, a well-conserved miR-93 binding site was identified within the 3'-UTR of vegf-a. Using transgenic mice containing VEGF-LacZ bicistronic transcripts, we demonstrated that inhibition of miR-93 by peptide-conjugated morpholino oligomers elicited increased expression of VEGF [74]. Meanwhile, miR-29c was found to induce podocyte apoptosis. Overexpression of miR-29c decreased the levels of Spry1 protein, a direct target of miR-29c, and promoted activation of Rho kinase. Importantly, knockdown of miR-29c by a specific antisense oligonucleotide significantly reduced albuminuria and kidney mesangial matrix accumulation in the db/db mouse model in vivo [59]. Alternatively, several studies have found the miR-29 family to be repressed under hyperglycemic conditions, most notably miR-29a and –b in tubular epithelial cells and mesangial cells [75]. These contrasting studies could be, in part, understood by cell type and stimulus specific differences among studies, since many of the studies conducted demonstrating reduced miR-29 family expression were carried out in mesangial and tubular epithelial cells, and within the context of the TGF-β1 signaling cascade a well known pathogenic mediator of diabetic nephropathy and tubulointerstitial fibrosis.
Finally, Wang et al., found miR-377 to be a key miRNA upregulated by high glucose and TGF-β1 of mesangial cells and in Type 1 diabetes. They demonstrated that miR-377 overexpression contributed to increased Fibronectin 1 expression and oxidative stress, via repression of target Pak1 and Superoxide Dismutase (SOD) [76].
Kidney Fibrosis
Many studies investigating the role of miRNA in kidney fibrosis have focused on the activated myofibroblasts in primarily two models of murine kidney fibrosis, the unilateral ureteral obstruction (UUO) model and the ischemia-reperfusion (IR) model.
Among several miRNAs that have emerged as central regulators of renal fibrosis, miR-21, a TGF-β1-induced miRNA, has been one that has attracted a great deal of attention. miR-21 was initially shown to play a role in heart fibrosis [77]. Elevated miR-21 levels were similarly observed in idiopathic lung fibrosis and inhibition of miR-21 in mouse models of lung fibrosis ameliorated this phenotype [78]. The role of miR-21 in kidney fibrosis has been demonstrated in several ways: 1) miR-21 expression levels were dramatically induced in patients in patients with CKD, [79, 80] 2) miR-21 expression was dramatically elevated in several experimental models of kidney fibrosis, including the UUO, I/R and DN models of fibrosis [60, 81, 82], and 3) a more recent study investigating the role of miR-21 deletion in mouse models of fibrosis (UUO and IRI) revealed that miR-21 deletion significantly improves features of interstitial fibrosis. Indeed, miR-21 contributes to kidney fibrosis by targeting PPARα, a key regulator of lipid metabolic pathways, and Mpv171, an inhibitor of mitochondrial specific reactive oxygen species [83].
Our own group recently identified miR-22 as a master regulator of kidney fibrosis by targeting several BMP family members. Based on several screens of putative BMP-7/6 miRNA regulators, we demonstrated that miR-22 is a potent regulator of BMP-7/6 genes at both the protein and mRNA levels. To investigate the functional significance of this targeting cascade, we used miR-22 knockout mice where we found that key features of fibrosis were significantly attenuated using UUO model of kidney fibrosis. At the molecular level, we identified several BMP responsive elements (BREs) within the proximal promoter region of the miR-22 host gene, and demonstrated that BMP-7/6 are directly able to induce miR-22 expression [84].
Polycystic Kidney Disease
Several miRNAs have been described to target both PKD1 (Polycystin 1) and PKD2 (Polycystin 2). Among these miRNA, the miR-17-92 family has been highlighted as key mediators of the pathogenesis and progression of PKD. miR-17 was the first miRNA implicated to regulate the expression of PKD2 [85]. Shortly thereafter, two independent groups revealed that Bicaudal 1 (Bicc1) mutant mice developed fluid filled cysts within the kidney, reminiscent of PKD. It was demonstrated that the gene Bicc1 regulates mRNA stability and translation efficiency of PKD2. Furthermore, Bicc1 antagonizes the repression of PKD2 by miR-17, resulting in increased PKD2 expression [86, 87].
Recently, Patel et al., followed up on their earlier Dicer knockout studies within the kidney to demonstrate significant up regulation of the entire miR-17-92 family (miR-17, -18a, -19a, -19b, -20a, -92a) in several mouse models of PKD. They showed that kidney tubular-specific overexpression of the family leads to cyst development within the kidney, and kidney-specific genetic ablation of the family leads to reduced cyst development in a mouse model of PKD. These actions were mediated in part, through PKD1, PKD2 and hepatocyte nuclear factor-1β (HFN1 β) [88]. These results suggest that this family of miRNAs may serve as a potential therapeutic target for patients with PKD.
The specific role for miRNAs within autosomal recessive polycystic kidney disease (ARPKD) has also been described. Lee et al., reported that miR-15a has been associated with hepatic cystogenesis in the PCK cholangiocyte cell line derived from the rat model of ARPKD and in liver tissue from patients with polycystic liver disease [89]. Further studies are required to relate these findings to the role of miR-15a in ARPKD. Finally, miR-365-1 has been recently shown to inhibit the expression of Polycystic Kidney and Hepatic Disease 1 (PKHD1) [90], however, further studies are required to fully elucidate the role miR-365-1 within ARPKD.
Wilm's Tumor
Recently, several mutations have been identified in Dicer1, the key miRNA-processing enzyme, in patients with Wilms Tumor, heavily implicating the broad role of miRNA dysregulation in Wilms Tumor. Most notably, the Oncomir-1 is significantly increased in patients with Wilms Tumor, and this correlates with elevated expression levels of the transcriptional regulator of Oncomir-1, E2F3 [91]. Similarly, the oncogenic transcription factor STAT3, was capable of inducing miR-370 expression, which in turn inhibits expression of WTX (Wilm's Tumor Gene on the X Chromosome), a commonly inactivated tumor suppressor in Wilms Tumor [92].
Interestingly, a separate study identified the kidney specific miR-562 within the 2q37.1 region to be reduced in Wilms Tumor patients. The target of miR-562, EYA1 (Eyes Absent Homolog 1) is critical in kidney development, and its misregulation during development in these patients may contribute to tumor pathobiology [93].
Finally, Six1 is a critical transcription factor that coordinates several developmental programs, including programs within the kidney. Six1 is frequently upregulated in adult and pediatric malignancies, including Wilms tumor. Imam et al., found that expression of a putative miRNA regulator of Six1, miR-185, is frequently reduced in these malignancies, including Wilms tumor. Several experiments revealed that restoration of miR-185 expression is capable of reducing tumorigenicity of cancer cells in vitro and in vivo [94]. While good progress has been made, thus far, there is still a need to better understand the role of miRNAs in this rare patient population.
Renal Cell Carcinoma (RCC)
Extensive studies have been dedicated to unraveling the role of miRNAs in RCC, including comprehensive analysis from the Cancer Genome Atlas consortium (TCGA) [95]. Most notably, miR-21, miR-34a, miR-200c, miR-215, and miR-17-5p have been characterized as promising biomarkers and possibly therapeutic targets for RCC. However, additional in vivo work with a robust mouse model or xenograft model of RCC is needed to further validate many of these initial findings primarily made in in vitro conditions.
Future challenges
Within a relatively short time frame, kidney researchers have made impressive strides in delineating the potential impact of a number of miRNAs as key regulators of physiological and pathological processes in the kidneys. The relatively stable nature of miRNAs and the many recent advancements in their isolation, purification and detection methods has made the feasibility of utilizing miRNA as biomarkers in human disease samples, an impending reality. Indeed, several groups have investigated the release and quantification of miRNAs in the blood and urine of patients with different kidney diseases as new potential biomarkers [96, 97]. These studies are promising and continued exploration into the possibility of circulating miRNAs as predictive factors for kidney disease is important. However, despite significant progress, many challenges still remain before we can successfully employ miRNAs as potential therapeutic targets. For example, the effect of a miRNA can be cell- and stimulus-specific, where the same miRNA can have different targets in the same organism with potentially opposite effects in different cells. Therefore, modulating miRNAs may result in beneficial effects in one cell type but harmful effects in another. Furthermore, another substantial challenge associated with miRNA-targeted approach is the delivery of miRNAs. Delivery systems and the ability to manipulate miRNAs in humans will continue to be the critical elements to bringing miRNAs from bench to bedside.
Acknowledgements
This work was supported by grants from NIDDK RO1DK091310 and RO1DK078900 (F.R.D).
Footnotes
Disclosures: The authors have nothing to disclose.
References
- 1.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy D. Breakthrough of the year. Science. 2002;298:2283. doi: 10.1126/science.298.5602.2283. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 8.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 9.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 17.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Asli NS, Pitulescu ME, Kessel M. MicroRNAs in organogenesis and disease. Curr Mol Med. 2008;8:698–710. doi: 10.2174/156652408786733739. [DOI] [PubMed] [Google Scholar]
- 20.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature reviews Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 21.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Gene Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 23.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 25.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Am Acac Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Ann Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 27.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 28.Liang H, Zhang J, Zen K, Zhang CY, Chen X. Nuclear microRNAs and their unconventional role in regulating non-coding RNAs. Protein Cell. 2013;4:325–330. doi: 10.1007/s13238-013-3001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Am Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 31.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 32.Long YS, Deng GF, Sun XS, Yi YH, Su T, Zhao QH, Liao WP. Identification of the transcriptional promoters in the proximal regions of human microRNA genes. Mol Biol Rep. 2011;38:4153–4157. doi: 10.1007/s11033-010-0535-y. [DOI] [PubMed] [Google Scholar]
- 33.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- 35.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Nervi C, Fazi F, Grignani F. Oncoproteins, heterochromatin silencing and microRNAs: a new link for leukemogenesis. Epigenetics : official journal of the DNA Methylation Society. 2008;3:1–4. doi: 10.4161/epi.3.1.5651. [DOI] [PubMed] [Google Scholar]
- 38.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 41.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 42.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Gene Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu S, Sun YH, Chiang VL. Adenylation of plant miRNAs. Nucleic Acids Res. 2009;37:1878–1885. doi: 10.1093/nar/gkp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3'-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renwick N, Cekan P, Masry PA, McGeary SE, Miller JB, Hafner M, Li Z, Mihailovic A, Morozov P, Brown M, Gogakos T, Mobin MB, Snorrason EL, Feilotter HE, Zhang X, Perlis CS, Wu H, Suarez-Farinas M, Feng H, Shuda M, Moore PS, Tron VA, Chang Y, Tuschl T. Multicolor microRNA FISH effectively differentiates tumor types. J Clin Invest. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rooij E. The art of microRNA research. Circ Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 52.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 53.Staton AA, Giraldez AJ. Use of target protector morpholinos to analyze the physiological roles of specific miRNA-mRNA pairs in vivo. Nat Protoc. 2011;6:2035–2049. doi: 10.1038/nprot.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 55.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Meth. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Putta S, Lanting L, Sun G, Lawson G, Kato M, Natarajan R. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol. 2012;23:458–469. doi: 10.1681/ASN.2011050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long J, Wang Y, Wang W, Chang BH, Danesh FR. MicroRNA-29c is a signature microRNA under high glucose conditions that targets Sprouty homolog 1, and its in vivo knockdown prevents progression of diabetic nephropathy. The J Biol Chem. 2011;286:11837–11848. doi: 10.1074/jbc.M110.194969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong X, Chung AC, Chen HY, Dong Y, Meng XM, Li R, Yang W, Hou FF, Lan HY. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia. 2013;56:663–674. doi: 10.1007/s00125-012-2804-x. [DOI] [PubMed] [Google Scholar]
- 61.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Neprol. 2008;19:2150–2158. doi: 10.1681/ASN.2008020233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi S, Yu L, Chiu C, Sun Y, Chen J, Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P, Bottinger EP. Podocyte-selective deletion of dicer induces proteinuria and glomerulosclerosis. J Am Soc Nephrol. 2008;19:2159–2169. doi: 10.1681/ASN.2008030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho J, Ng KH, Rosen S, Dostal A, Gregory RI, Kreidberg JA. Podocyte-specific loss of functional microRNAs leads to rapid glomerular and tubular injury. J Am Soc Nephrol. 2008;19:2069–2075. doi: 10.1681/ASN.2008020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, Johnstone DB, Zavadil J, Chong MM, Littman DR, Holzman LB, Barisoni L, Skolnik EY. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int. 2011;80:719–730. doi: 10.1038/ki.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel V, Hajarnis S, Williams D, Hunter R, Huynh D, Igarashi P. MicroRNAs regulate renal tubule maturation through modulation of Pkd1. J Am Soc Nephrol. 2012;23:1941–1948. doi: 10.1681/ASN.2012030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, Carretero O, Sigmund CD, Gomez RA. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21:460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol. 2010;21:756–761. doi: 10.1681/ASN.2009070718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gebeshuber CA, Kornauth C, Dong L, Sierig R, Seibler J, Reiss M, Tauber S, Bilban M, Wang S, Kain R, Bohmig GA, Moeller MJ, Grone HJ, Englert C, Martinez J, Kerjaschki D. Focal segmental glomerulosclerosis is induced by microRNA-193a and its downregulation of WT1. Nat Med. 2013;19:481–487. doi: 10.1038/nm.3142. [DOI] [PubMed] [Google Scholar]
- 70.Wu J, Zheng C, Fan Y, Zeng C, Chen Z, Qin W, Zhang C, Zhang W, Wang X, Zhu X, Zhang M, Zen K, Liu Z. Downregulation of microRNA-30 facilitates podocyte injury and is prevented by glucocorticoids. J Am Soc Nephrol. 2014;25:92–104. doi: 10.1681/ASN.2012111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acac Sci USA. 2007;104:3432–3437. doi: 10.1073/pnas.0611192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato M, Dang V, Wang M, Park JT, Deshpande S, Kadam S, Mardiros A, Zhan Y, Oettgen P, Putta S, Yuan H, Lanting L, Natarajan R. TGF-beta induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. SciSignal. 2013;6:ra43. doi: 10.1126/scisignal.2003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deshpande SD, Putta S, Wang M, Lai JY, Bitzer M, Nelson RG, Lanting LL, Kato M, Natarajan R. Transforming growth factor-beta-induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathy. Diabetes. 2013;62:3151–3162. doi: 10.2337/db13-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Long J, Wang Y, Wang W, Chang BH, Danesh FR. Identification of microRNA-93 as a novel regulator of vascular endothelial growth factor in hyperglycemic conditions. J Biol Chem. 2010;285:23457–23465. doi: 10.1074/jbc.M110.136168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, Koh P, Thomas M, Jandeleit-Dahm K, Gregorevic P, Cooper ME, Kantharidis P. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Q, Wang Y, Minto AW, Wang J, Shi Q, Li X, Quigg RJ. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB journal : official publication of the Federation of American Societies for Exp Biol. 2008;22:4126–4135. doi: 10.1096/fj.08-112326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 78.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szeto CC, Ching-Ha KB, Ka-Bik L, Mac-Moune LF, Cheung-Lung CP, Gang W, Kai-Ming C, Kam-Tao LP. Micro-RNA expression in the urinary sediment of patients with chronic kidney diseases. Dis Markers. 2012;33:137–144. doi: 10.3233/DMA-2012-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zawada AM, Rogacev KS, Muller S, Rotter B, Winter P, Fliser D, Heine GH. Massive Analysis of cDNA Ends (MACE) and miRNA expression profiling identifies proatherogenic pathways in chronic kidney disease MACE and miRNA profiling in CKD. Epigenetics. 2013;9:161–172. doi: 10.4161/epi.26931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong X, Chung AC, Chen HY, Meng XM, Lan HY. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol. 2011;22:1668–1681. doi: 10.1681/ASN.2010111168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dey N, Das F, Mariappan MM, Mandal CC, Ghosh-Choudhury N, Kasinath BS, Choudhury GG. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem. 2011;286:25586–25603. doi: 10.1074/jbc.M110.208066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chau BN, Xin C, Hartner J, Ren S, Castano AP, Linn G, Li J, Tran PT, Kaimal V, Huang X, Chang AN, Li S, Kalra A, Grafals M, Portilla D, MacKenna DA, Orkin SH, Duffield JS. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med. 2012;4:121ra118. doi: 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Long J, Badal SS, Wang Y, Chang BH, Rodriguez A, Danesh FR. MicroRNA-22 is a master regulator of bone morphogenetic protein-7/6 homeostasis in the kidney. J Biol Chem. 2013;288:36202–36214. doi: 10.1074/jbc.M113.498634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun H, Li QW, Lv XY, Ai JZ, Yang QT, Duan JJ, Bian GH, Xiao Y, Wang YD, Zhang Z, Liu YH, Tan RZ, Yang Y, Wei YQ, Zhou Q. MicroRNA-17 post-transcriptionally regulates polycystic kidney disease-2 gene and promotes cell proliferation. Molr Biol Rep. 2010;37:2951–2958. doi: 10.1007/s11033-009-9861-3. [DOI] [PubMed] [Google Scholar]
- 86.Piazzon N, Maisonneuve C, Guilleret I, Rotman S, Constam DB. Bicc1 links the regulation of cAMP signaling in polycystic kidneys to microRNA-induced gene silencing. J Mol Cell Biol. 2012;4:398–408. doi: 10.1093/jmcb/mjs027. [DOI] [PubMed] [Google Scholar]
- 87.Tran U, Zakin L, Schweickert A, Agrawal R, Doger R, Blum M, De Robertis EM, Wessely O. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development. 2010;137:1107–1116. doi: 10.1242/dev.046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Patel V, Williams D, Hajarnis S, Hunter R, Pontoglio M, Somlo S, Igarashi P. miR-17~92 miRNA cluster promotes kidney cyst growth in polycystic kidney disease. Proc Natl Acad Sci USA. 2013;110:10765–10770. doi: 10.1073/pnas.1301693110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee SO, Masyuk T, Splinter P, Banales JM, Masyuk A, Stroope A, Larusso N. MicroRNA15a modulates expression of the cell-cycle regulator Cdc25A and affects hepatic cystogenesis in a rat model of polycystic kidney disease. J Clin Invest. 2008;118:3714–3724. doi: 10.1172/JCI34922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duan J, Huang H, Lv X, Wang H, Tang Z, Sun H, Li Q, Ai J, Tan R, Liu Y, Chen M, Duan W, Wei Y, Zhou Q. PKHD1 post-transcriptionally modulated by miR-365-1 inhibits cell-cell adhesion. Cell Biochem Funct. 2012;30:382–389. doi: 10.1002/cbf.2795. [DOI] [PubMed] [Google Scholar]
- 91.Kort EJ, Farber L, Tretiakova M, Petillo D, Furge KA, Yang XJ, Cornelius A, Teh BT. The E2F3-Oncomir-1 axis is activated in Wilms tumor. Cancer Res. 2008;68:4034–4038. doi: 10.1158/0008-5472.CAN-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao X, Liu D, Yan X, Zhang Y, Yuan L, Zhang T, Fu M, Zhou Y, Wang J. Stat3 inhibits WTX expression through up-regulation of microRNA-370 in Wilms tumor. FEBS Lett. 2013;587:639–644. doi: 10.1016/j.febslet.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 93.Drake KM, Ruteshouser EC, Natrajan R, Harbor P, Wegert J, Gessler M, Pritchard-Jones K, Grundy P, Dome J, Huff V, Jones C, Aldred MA. Loss of heterozygosity at 2q37 in sporadic Wilms tumor: putative role for miR-562. Clin Cancer Res. 2009;15:5985–5992. doi: 10.1158/1078-0432.CCR-09-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Imam JS, Buddavarapu K, Lee-Chang JS, Ganapathy S, Camosy C, Chen Y, Rao MK. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29:4971–4979. doi: 10.1038/onc.2010.233. [DOI] [PubMed] [Google Scholar]
- 95.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woo YM, Park JH. microRNA biomarkers in cystic diseases. BMB Rep. 2013;46:338–345. doi: 10.5483/BMBRep.2013.46.7.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kato M, Castro NE, Natarajan R. MicroRNAs: potential mediators and biomarkers of diabetic complications. Free Radic Biol Med. 2013;64:85–94. doi: 10.1016/j.freeradbiomed.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]