PARP inhibitors (PARPi) are being used in patients with BRCA1/2 mutations; however, doubly deficient BRCA1−/−53BP1−/− tumors become resistant to PARPis. 53BP1 and its known downstream effectors, PTIP and RIF1, lack enzymatic activities directly implicated in DNA repair. Wang et al. uncovered a nuclease, Artemis, as a PTIP-binding protein that trims DNA ends, promotes NHEJ, and directly competes with the HR repair pathway. Loss of Artemis restores PARPi resistance in BRCA1-deficient cells.
Keywords: PTIP, Artemis, 53BP1, BRCA1, DNA repair, PARP inhibition
Abstract
PARP inhibitors (PARPis) are being used in patients with BRCA1/2 mutations. However, doubly deficient BRCA1−/−53BP1−/− cells or tumors become resistant to PARPis. Since 53BP1 or its known downstream effectors, PTIP and RIF1 (RAP1-interacting factor 1 homolog), lack enzymatic activities directly implicated in DNA repair, we decided to further explore the 53BP1-dependent pathway. In this study, we uncovered a nuclease, Artemis, as a PTIP-binding protein. Loss of Artemis restores PARPi resistance in BRCA1-deficient cells. Collectively, our data demonstrate that Artemis is the major downstream effector of the 53BP1 pathway, which prevents end resection and promotes nonhomologous end-joining and therefore directly competes with the homologous recombination repair pathway.
Eukaryotic cells have two main repair pathways for DNA double-stranded breaks (DSBs); namely, the nonhomologous end-joining (NHEJ) and homologous recombination (HR) pathways. Which pathway is selected for DSB repair under certain circumstance is critically important for genome maintenance (for review, see Chapman et al. 2012). In particular, understanding the detailed regulation of DNA repair pathway choice will help us design better therapies for cancer patients. For example, a major breakthrough in the targeted treatment of BRCA1/2 mutant cancers is the use of PARP inhibitors (PARPis), which display “synthetic lethality” with BRCA1 or BRCA2 deficiency (Bryant et al. 2005; Audeh et al. 2010; Chan and Mok 2010; Tutt et al. 2010). Unexpectedly, loss of 53BP1 renders these BRCA1-deficient tumors resistant to PARPis, suggesting that 53BP1 and BRCA1 compete with each other to influence the pathway choice for DSB repair (Bouwman et al. 2010; Bunting et al. 2010; Jaspers et al. 2013). However, despite extensive studies in the past decade, it remains elusive precisely how 53BP1 acts in DNA repair, since it does not possess any enzymatic activities that would be required for DNA repair. The current hypothesis is that 53BP1 is an adaptor protein that does not act directly in DNA repair. Instead, it may recruit other proteins to the sites of DNA breaks to facilitate NHEJ repair, which competes with HR repair. In support of this hypothesis, several research groups demonstrated that RIF1 (RAP1-interacting factor 1 homolog), is at least one of the key factors that acts downstream from 53BP1 and counteracts BRCA1’s functions in DNA repair (Chapman et al. 2013; Di Virgilio et al. 2013; Escribano-Diaz et al. 2013; Feng et al. 2013; Zimmermann et al. 2013). However, RIF1 loss seems insufficient to rescue the HR defect in BRCA1-deficient cells. A more recent study showed that ablation of PTIP provides additional or sustained resection required for rescuing HR repair in BRCA1-deficient cells, indicating that PTIP and RIF1 may inhibit different resection steps during HR repair (Callen et al. 2013). Unfortunately, RIF1 and PTIP again are adaptor proteins, since they also lack any enzymatic activities that would be involved in DNA repair.
We thus decided to further explore the 53BP1-dependent pathway, focusing on the identification of downstream effector proteins that directly participate in DNA repair. PTIP is known to be involved in DNA repair but also has a separate role in transcriptional regulation through its association with MLL2/MLL3 histone methyltransferase complexes (Cho et al. 2007; Munoz et al. 2007; Patel et al. 2007; Gong et al. 2009). In this study, we uncovered a nuclease, Artemis, as a PTIP-binding protein. Artemis is an evolutionally conserved nuclease (for review, see Cattell et al. 2010). Defects in Artemis lead to a severe combined immunodeficiency disorder (SCID) known as radiosensitive SCID (RS-SCID) syndrome (Moshous et al. 2001). Artemis has endonuclease activity in vivo and is a known component of the NHEJ pathway (Ma et al. 2002). Here, we show that Artemis is the major downstream effector of the 53BP1 pathway, which trims DNA ends and promotes NHEJ and therefore directly competes with the HR repair pathway.
Results and Discussion
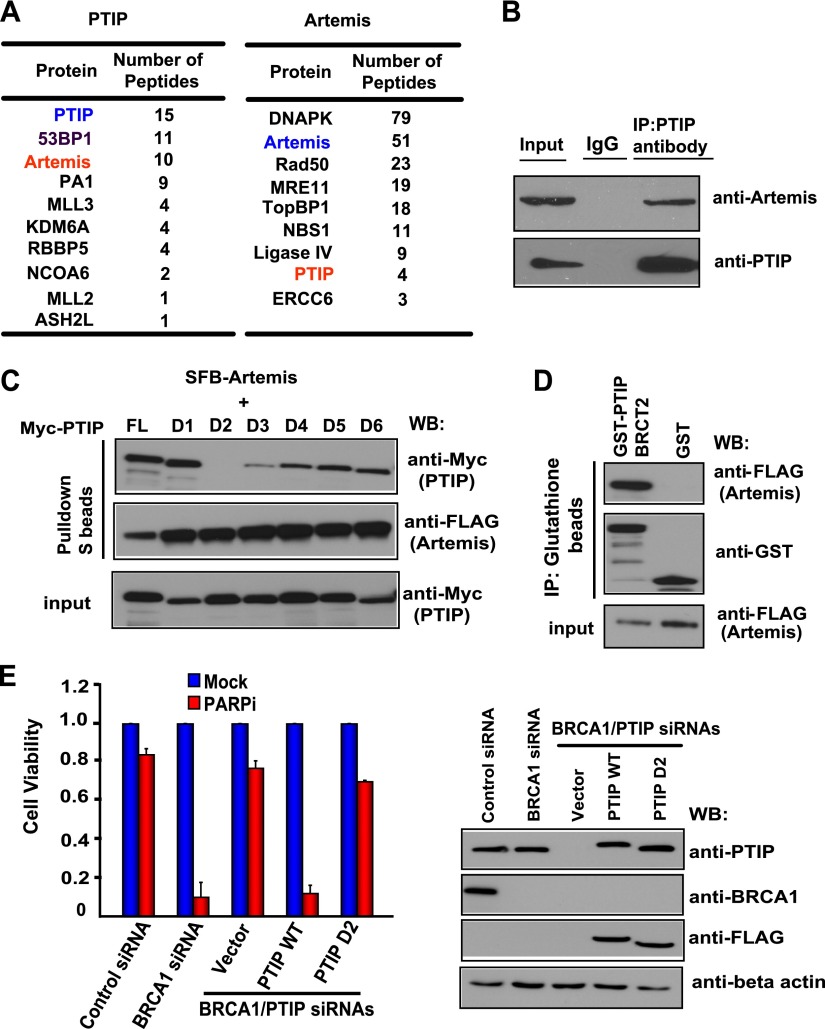
PTIP plays important roles in the cellular response to DNA damage (Munoz et al. 2007; Gong et al. 2009). However, PTIP lacks any enzymatic activity that would be directly involved in DNA repair. Thus, we used a modified tandem affinity purification (TAP) coupled with mass spectrometry (TAP-MS) approach that was established in our laboratory (Gong et al. 2010; Wang et al. 2011, 2013) to identify novel PTIP-associated proteins. Excitingly, MS analysis revealed not only several known PTIP-binding proteins—including 53BP1, PA1, and components of histone methyltransferase complexes (e.g., MLL3 and NCOA6)—but also a nuclease, Artemis (also called SNM1C), as major PTIP-associated proteins (Fig. 1A). Furthermore, we identified PTIP as an Artemis-associated protein in a reverse TAP-MS analysis (Fig. 1A). We further confirmed this interaction by coimmunoprecipitation experiment (Fig. 1B; Supplemental Fig. S1). Using Myc-tagged wild-type PTIP and a series of PTIP BRCT domain deletion mutants (D1–D6, each with a deletion of one of the six BRCT domains in PTIP) that we previously generated (Gong et al. 2009), we showed that deletion of the second BRCT domain (D2) abolished the PTIP/Artemis interaction, indicating that the BRCT2 domain is important for the binding of PTIP to Artemis (Fig. 1C). Furthermore, using a bacterially expressed and purified GST-fused BRCT2 of PTIP protein, we showed that the PTIP BRCT2 binds to Artemis in vitro (Fig. 1D), indicating that the BRCT2 domain of PTIP is necessary and sufficient for its binding to Artemis.
Figure 1.
PTIP interacts with Artemis through its BRCT2 domain. (A) Lists of PTIP or Artemis-associated proteins identified by mass spectrometric analysis. (B) Endogenous PTIP interacts with Artemis. HeLa cell lysates were prepared and immunoprecipitated with PTIP antibody. Immunoprecipitates were blotted using antibodies as indicated. (C) The BRCT2 domain of PTIP is required for PTIP/Artemis interaction. 293T cells were transfected with plasmids encoding SFB-tagged Artemis together with plasmids encoding deletion mutants of Myc-tagged PTIP. Precipitation reactions were conducted using S protein beads and then subjected to Western blotting using the indicated antibodies. D1–D6 are PTIP BRCT domain deletion mutants, each of which has a deletion of one of the PTIP BRCT domains. (D) PTIP BRCT2 specifically binds to Artemis. Beads coated with bacterially expressed GST-fused PTIP BRCT2 were incubated with cell lysates containing exogenously expressed SFB-tagged Artemis. Immunoblotting experiments were carried out using the indicated antibodies. (E) The BRCT2 domain of PTIP counteracts HR repair. HeLa cells were reconstituted with empty vector, siRNA-resistant PTIP wild type, or the PTIP D2 mutant. Next, HeLa cells as well as these reconstituted cells were transfected with the indicated siRNAs and treated with 1 μM PARPis. Colony formation was quantified relative to colonies formed in untreated cells from the same setting. (Left panel) Data are represented as the mean ± SEM (n = 3). (Right panel) Knockdown efficiency of PTIP or BRCA1 was confirmed by Western blotting with the indicated antibodies.
A recent study demonstrated that PTIP acts downstream from 53BP1, and PTIP deficiency restored PARPi resistance in a BRCA1-deficient background (Callen et al. 2013). Indeed, we showed that codepletion of PTIP and BRCA1 led to cellular resistance to PARPis (Fig. 1E). Moreover, reconstitution of wild-type PTIP resensitized these cells to PARPis, but reconstitution with the BRCT2 deletion mutant of PTIP failed to do so (Fig. 1E). These data indicated that the BRCT2 domain of PTIP, which is required for its interaction with Artemis, is important for the function of PTIP in counteracting HR repair.
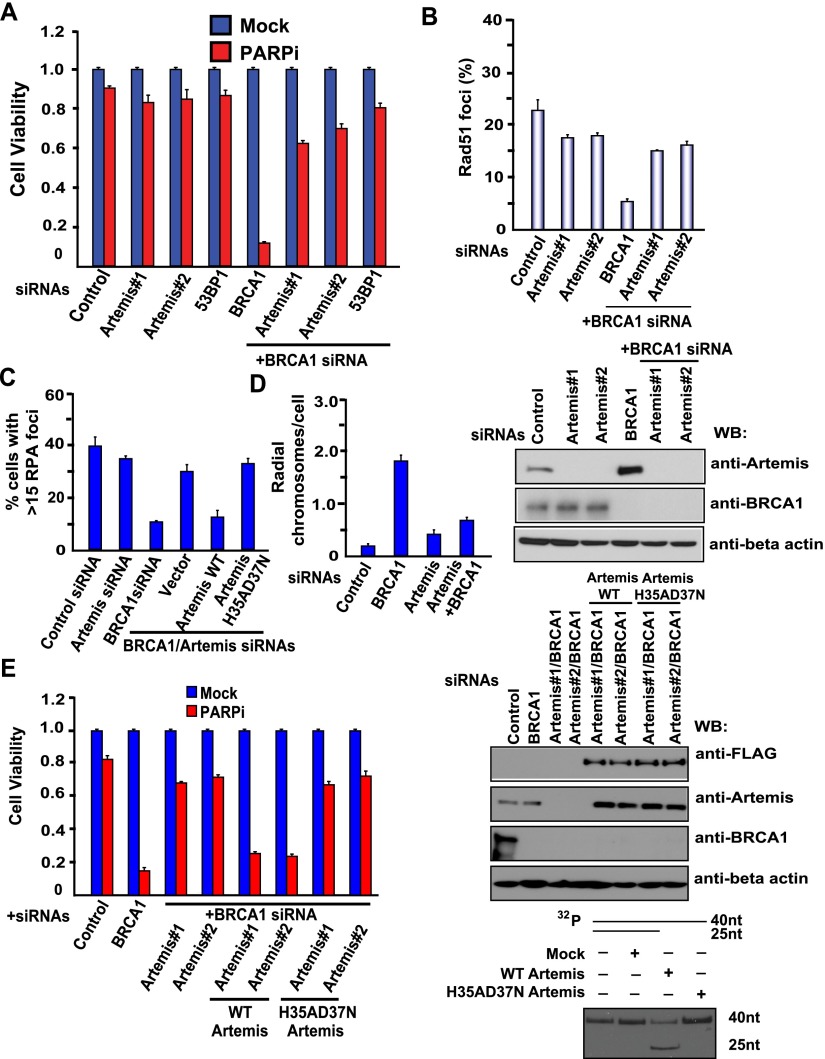
Since Artemis possesses endonuclease activities (Ma et al. 2002), we reasoned that Artemis may use its nuclease activity to trim DNA ends and therefore facilitate NHEJ and prevent DNA end resection and HR repair. Indeed, we showed that loss of Artemis renders BRCA1-deficient cells resistant to PARP inhibition (Fig. 2A). Furthermore, damage-induced RPA2 foci (marker of end resection) or RAD51 foci (marker for HR repair) were reduced in BRCA1-depleted cells but were re-established in BRCA1/Artemis-codepleted cells (Fig. 2B,C), indicating that Artemis acts to prevent end resection and suppress RAD51-dependent HR repair. In addition, after PARPi treatment, the number of radial chromosomes increased in BRCA1-deficient cells, while this phenotype was partially rescued in Artemis-codepleted cells (Fig. 2D).
Figure 2.
The nuclease Artemis prevents DNA end resection and HR repair. (A) Loss of Artemis renders BRCA1-deficient cells resistant to PARP inhibition. HeLa cells were transfected with the indicated siRNAs and treated with 1 μM PARPis. Colony formation was quantified relative to colonies formed in untreated cells from the same setting. Data are represented as the mean ± SEM (n = 3). Knockdown efficiency of Artemis or BRCA1 was confirmed by Western blotting with the indicated antibodies. (B) Artemis suppresses RAD51-dependent HR repair. HeLa cells were transfected with the indicated siRNAs. At 3 h post-irradiation (10 Gy), cells were processed for RAD51 immunofluorescence. RAD51 foci were quantified (at least 400 cells were counted for each cell). Data are represented as the mean ± SEM (n = 3). (C) Artemis acts to prevent end resection. HeLa cells were transfected with the indicated siRNAs. At 3 h post-irradiation (10 Gy), cells were processed for RPA immunofluorescence. Shown is the quantitation of RPA foci-positive cells (at least 400 cells were counted for each cell). Data are represented as the mean ± SEM (n = 3). (D) Artemis knockdown reduces radial chromosomes in BRCA1-deficient cells. HeLa cells were transfected with the indicated siRNAs and treated with 1 μM PARPis. Metaphases were analyzed for radial chromosomes (n = 50 metaphases analyzed in each case). Data are represented as the mean ± SEM (n = 3). (E) The nuclease activity of Artemis is required for the prevention of end resection and HR repair. HeLa cells were reconstituted with empty vector, siRNA-resistant Artemis wild-type, or Artemis nuclease-inactive mutant (H35AD37N). Next, HeLa cells as well as these reconstituted cells were transfected with the indicated siRNAs and treated with 1 μM PARPis. Colony formation was quantified relative to colonies formed in untreated cells from the same setting. (Left panel) Data are represented as the mean ± SEM (n = 3). (Top panel) Knockdown efficiency of Artemis or BRCA1 was confirmed by Western blotting with the indicated antibodies. SFB empty vector (Mock), SFB-tagged wild-type Artemis, and H35AD37N mutant Artemis were transfected into 293T cells, and Artemis complexes were first pulled down by streptavidin beads, then eluted by biotin buffer, and pulled down again by S protein beads. (Bottom panel) In vitro nuclease assays of wild-type and H35AD37N mutant Artemis were performed as described in the Materials and Methods.
The nuclease activity of Artemis is required for opening the hairpins that form on DNA ends during V(D)J recombination (Ma et al. 2002). To assess whether the nuclease activity of Artemis is critical for this function in counteracting HR repair, we generated HeLa cells that stably express siRNA-resistant wild-type Artemis or the nuclease-inactive mutant (H35AD37N) (Fig. 2E, right panel) of Artemis so that we can express exogenous Artemis when the endogenous Artemis is depleted by siRNA. As shown in Figure 2E (left panel), reconstitution of wild-type Artemis resensitized cells to PARPis, while the nuclease-inactive mutant (H35AD37N) of Artemis failed to do so. Additionally, we observed that reconstitution of the nuclease-inactive mutant of Artemis also increased RPA foci formation compared with those that reconstitute with wild-type Artemis (Fig. 2B). We reproducibly confirmed the above data by using Artemis knockout cells reconstituted with wild-type and nuclease-inactive mutant (H35AD37N) Artemis (Supplemental Fig. S2A), suggesting that Artemis and its nuclease activity are required for the prevention of end resection and HR repair.
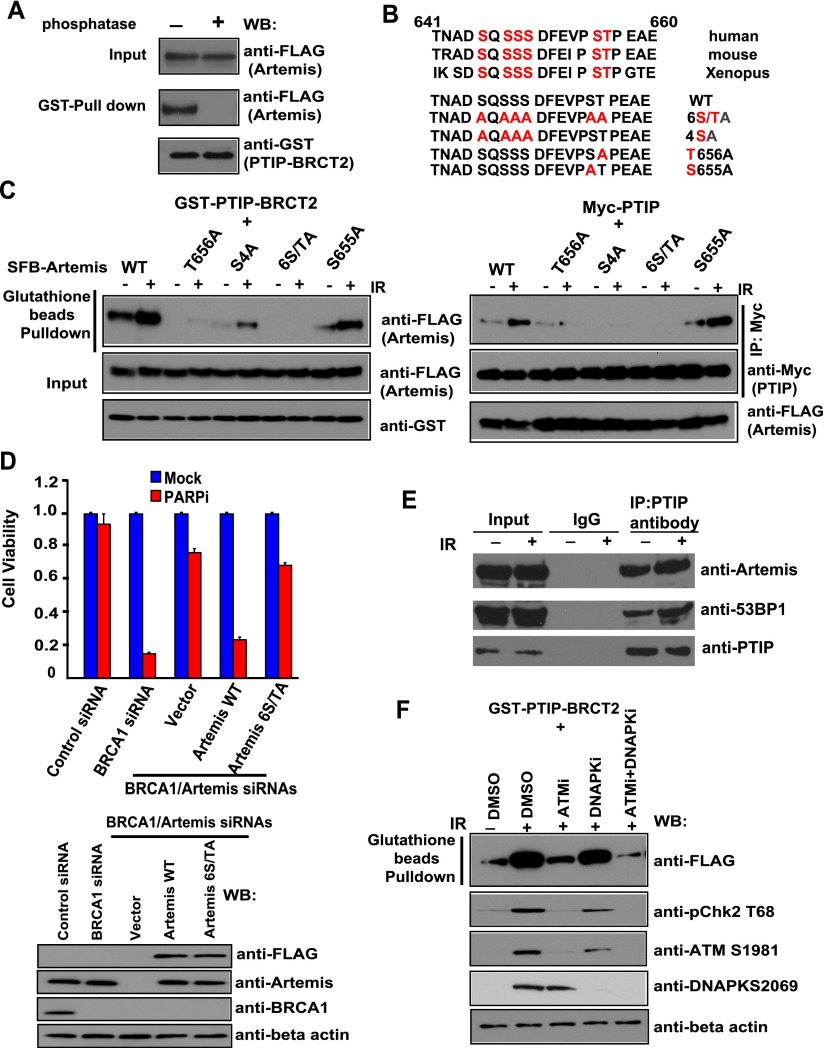
The BRCT domain is a phosphoprotein-binding domain (Manke et al. 2003; Yu et al. 2003). The interaction between PTIP and Artemis was abolished following treatment with λ protein phosphatase, suggesting that the interaction between PTIP and Artemis is phosphorylation-dependent (Fig. 3A). Using a series of Artemis deletion mutants, we mapped the PTIP-binding region to the very C terminus of Artemis (residues 641–660) (Fig. 3B; data not shown). To identify potential phosphorylation sites of Artemis, we isolated Artemis protein from cells with or without DNA damage and performed MS analysis to identify all potential phosphorylation sites on Artemis. A total of 13 phosphorylation sites were identified, with six sites showing up only in damaged cells (Supplemental Fig. S3). We mutated each of these six residues. We found that mutation of all six serine/threonine sites abolished the PTIP/Artemis interaction, while mutation of residue Thr656 greatly diminished the PTIP/Artemis interaction (Fig. 3C). Mutation of the other four serine residues also affected the PTIP/Artemis interaction (Fig. 3C), suggesting that while the Thr656 site is a major phosphorylation site involved in the PTIP/Artemis interaction, the other four residues also contribute to this interaction. We generated HeLa cells stably expressing siRNA-resistant wild-type Artemis or the substitution mutant of Artemis in which these phosphorylation sites (serine or threonine) are replaced by Alanine. As shown in Figure 3D, the expression of siRNA-resistant wild-type Artemis fully restored PARPi sensitivity in Artemis/BRCA1-codepleted cells, whereas the expression of the Artemis 6S/TA mutant failed to do so, indicating that the PTIP/Artemis interaction is critical for antagonizing HR repair. We further investigated how the phosphorylation of Artemis and its interaction with PTIP are regulated. In this regard, we found that the interaction of Artemis with PTIP was induced by DNA damage (Fig. 3E,F), which is mainly dependent on ATM kinase (Fig. 3F).
Figure 3.
The interaction between PTIP and Artemis is phosphorylation-dependent, and phosphorylation of Artemis is ATM-dependent. (A) PTIP/Artemis interaction is phosphorylation-dependent. Beads coated with bacterially expressed GST-PTIP BRCT2 fusion protein were incubated with cell lysates containing exogenously expressed, SFB-tagged Artemis that was mock-treated or treated with λ protein phosphatase. Immunoblotting experiments were carried out with the indicated antibodies. (B) Alignment of potential phospho-motif on Artemis among species. (C) Mapping the phosphorylation site on Artemis that is important for its interaction with PTIP. Beads coated with bacterially expressed GST-PTIP BRCT2 fusion protein or SFB-tagged Artemis were incubated with cell lysates containing exogenously expressed, SFB-tagged wild-type or mutant Artemis with or without IR (20 Gy). Immunoblotting experiments were carried out using the indicated antibodies. (D) The PTIP/Artemis interaction is critical for antagonizing HR repair. HeLa cells were reconstituted with empty vector, siRNA-resistant Artemis wild type, or Artemis 6S/TA mutant. Next, HeLa cells as well as these reconstituted cells were transfected with the indicated siRNAs and treated with 1 μM PARPis. Colony formation was quantified relative to colonies formed in untreated cells from the same setting. (Top panel) Data are represented as the mean ± SEM (n = 3). (Bottom panel) The knockdown efficiency of Artemis or BRCA1 was confirmed by Western blotting with the indicated antibodies. (E) Endogenous PTIP interacts with Artemis in a DNA damage-induced manner. HeLa cells were treated with or without IR (20 Gy). HeLa cell lysates were prepared and immunoprecipitated with control or PTIP antibody. Immunoprecipitates were blotted using antibodies as indicated. (F) The interaction of Artemis with PTIP was induced by DNA damage, which is mainly dependent on ATM kinase. Beads coated with bacterially expressed GST-PTIP BRCT2 fusion protein or SFB-tagged Artemis were incubated with cell lysates containing exogenously expressed, SFB-tagged Artemis mock-treated or treated with ATM inhibitors (ATMi; KU60019), DNAPK inhibitors (DNAPKi; NU7026), and a combination of ATM inhibitors and DNAPK inhibitors (ATMi+DNAPKi) with or without IR (20 Gy). Immunoblotting experiments were carried out using the indicated antibodies.
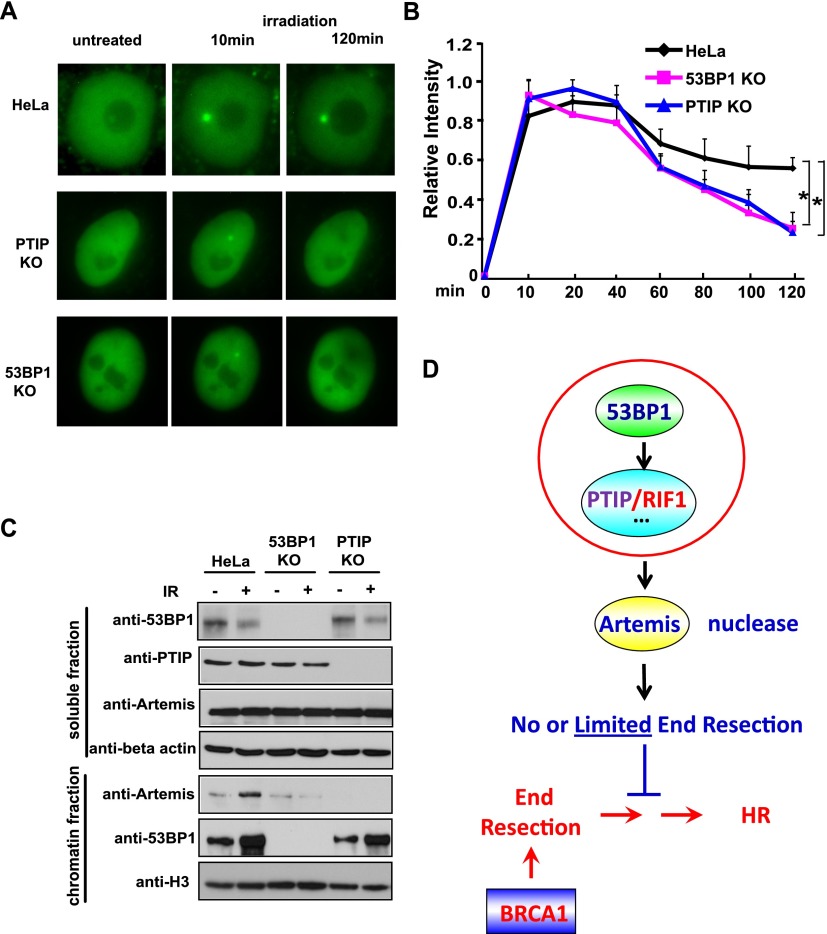
Since PTIP localizes to DNA damage sites (Munoz et al. 2007; Gong et al. 2009), we wanted to determine whether Artemis could also localize to DNA damage sites. We did not have an anti-Artemis antibody that works for immunostaining. Instead, we showed that GFP-tagged Artemis was recruited to DNA damage sites after laser-induced microirradiation (Fig. 4A). Next, we determined whether this damage-induced localization of Artemis required 53BP1 or PTIP. To do that, we used the CRISPR/Cas9-mediated gene-editing approach (Cong et al. 2013; Mali et al. 2013) to generate HeLa cell lines that are specifically defective in 53BP1 or PTIP. We further established expression of GFP-tagged Artemis in control or HeLa knockout cell lines. As shown in Figure 4A, we observed rapid accumulation of GFP-Artemis at laser-induced DNA lesions in wild-type, 53BP1 knockout, and PTIP knockout HeLa cell lines. Furthermore, continued time-lapse imaging for a period of 2 h after microirradiation followed by quantification of GFP-Artemis fluorescent intensity revealed that the fluorescent intensity of the accumulation area in HeLa cells decreased to ∼60% of the maximum level in a 2-h duration (Fig. 4B). In contrast, the Artemis fluorescent intensity was reduced to 20%–30% of the maximum level in 53BP1 or PTIP knockout HeLa cells during the same period. In addition, compared with wild-type HeLa cells, 53BP1 knockout or PTIP knockout HeLa cells displayed a substantial decrease of Artemis chromatin loading following ionizing radiation (IR) treatment (Fig. 4C). These results demonstrate that 53BP1 and PTIP are required for the retention of Artemis at DNA damage sites.
Figure 4.
Artemis is a downstream effector of the 53BP1/PTIP-dependent pathway. (A) Accumulation of Artemis at laser-induced DNA DSBs is reduced in 53BP1 knockout (KO) and PTIP knockout cells. GFP-Artemis is stably expressed in HeLa, 53BP1 knockout, and PTIP knockout cells. DSBs were generated by microirradiation, and GFP-Artemis accumulation was monitored before and after irradiation. A CRISPR/Cas9-mediated gene-editing approach was used to generate a panel of HeLa cell lines specifically defective in 53BP1 and PTIP, which was verified by Western blotting and DNA sequence (data not shown). (B) Quantitative analysis of GFP-Artemis accumulation at DSBs in HeLa, 53BP1 knockout, and PTIP knockout cell lines. Signal intensities were quantified, and a mean value for each time point was calculated from five independent measurements. (C) Both 53BP1 and PTIP are required for Artemis chromatin loading following IR treatment. HeLa, 53BP1 knockout, and PTIP knockout cell lines were treated with or without IR (20 Gy). Cells were harvested. The soluble and chromatin fractions were prepared and immunoblotted with antibodies as indicated. (D) A revised model of Artemis functions downstream from 53BP1/PTIP to counteract HR repair.
In this study, we showed that Artemis is a major nuclease that acts downstream from the 53BP1 pathway and participates in the prevention of DNA end resection, thereby antagonizing HR repair (Fig. 4D). Mechanistically, we demonstrated that Artemis binds to PTIP and accumulates at sites of DNA damage (Figs. 1B,C, 4A). The interaction between PTIP and Artemis is phosphorylation-dependent and is induced by DNA damage (Fig. 3A,C). Excitingly, just like 53BP1, loss of Artemis expression in BRCA1-deficient cells leads to PARPi resistance (Fig. 2A). Furthermore, loss of Artemis increased RAD51 foci and RPA foci formation in BRCA1-depleted cells (Fig. 2B,C), indicating that Artemis normally acts to limit end resection and inhibit RAD51-dependent HR repair. We propose that this function of Artemis is similar to its role in V(D)J recombination. The nuclease activities of Artemis are important for its function in antagonizing HR repair (Fig. 2E). Our working hypothesis is that the 53BP1/PTIP pathway facilitates the accumulation of Artemis at the sites of DNA damage. Artemis acts as an endonuclease and trims DNA ends to promote NHEJ and therefore at the same time prevents end resection and HR repair.
It remains uncertain whether Artemis is the only nuclease that acts in the 53BP1 pathway. It will be interesting to explore whether, like 53BP1, Artemis loss would rescue embryonic lethality in BRCA1-deficient mice. In addition, future studies should be devoted to taking advantage of this knowledge and developing strategies to overcome PARPi resistance in BRCA1 patients.
Materials and methods
Additional methods are described in the Supplemental Material.
Generation of DNA damage with laser microirradiation
Live-cell imaging combined with laser microirradiation was carried out as described previously (Uematsu et al. 2007). Briefly, HeLa cells and 53BP1 knockout, PTIP knockout, and Artemis knockout HeLa cells stably expressing EGFP-Artemis were grown on glass-bottomed 35-mm dishes for 48 h. A 365-nm pulsed nitrogen laser at 10 Hz (Spectra-Physics), coupled to the epifluorescent path of a Zeiss microscope, was used to introduce DNA damage in a defined area within the nuclei of EGFP-Artemis-expressing cells. To generate the same amount of DNA damage in each experiment, we used optimal irradiation conditions (80% laser output at 10 Hz for 400 msec). Time-lapse images of living cells were taken by an Axiovert 200M microscope with a Plan Apochromat 63×/NA 1.40 oil immersion objective, equipped with an AxioCam HRm camera (Carl Zeiss MicroImaging).
CRISPR–Cas9 gene-editing approach to generate 53BP1 knockout, PTIP knockout, and Artemis knockout HeLa cells
Vectors encoding guide RNA (gRNA) were digested with AfIII, and a pair of annealed oligonucleotides for each targeting site was cloned into the gRNA vector according to the protocol (Cong et al. 2013). Designs were chosen to target the gene in one of the first few exons and were tested for obvious potential off-targets by bioinformatics analysis. HeLa cells were transfected with vectors expressing hCas9 gRNAs along with EGFP expression vector. Cells were then sorted and seeded as single colonies in 96-well plates. After 2–3 wk, clones were selected based on Western blotting with the appropriate antibodies. In addition, genomic DNA was extracted from cell lines arising from single clones. PCR reactions to amplify targeted loci were performed, and agarose gel electrophoresis was used to confirm the correct size of PCR products. PCR products were then cloned into the pCR2.1-TOPO vector and transformed into DH5α competent cells. Plasmid DNA was isolated from multiple colonies of each transformation and sequenced to ensure frameshift mutations in the targeted region.
Chromatin extraction preparation
HeLa, 53BP1 knockout, and PTIP knockout cell lines were harvested after treatment with or without IR (20 Gy) and subsequently lysed with 10 vol of NTEN buffer with low salt (20 mM Tris-HCl at pH 8.0, 10 mM NaCl, 1.5 mM MgCl2, 1 mM EDTA, 0.5% nonidet P-40, 20 mM NaF, 1mM Na3VO4, 1 µg/mL aprotinin, 1 µg/mL pepstatin). The chromatin-enriched pellet was washed three times with PBS. The insoluble chromatin fractions were resuspended in 0.2 M HCl for 30 min on ice. The resultant soluble extraction was neutralized with 1 M Tris-HCl (pH 8.5) for further analysis.
In vitro nuclease activity assay
The in vitro nuclease assay was performed as previously reported (Goodarzi et al. 2006). Briefly, to prepare the nuclease substrate, complementary oligonucleotides (5′-AAGCTTGCATGCCTGCAGGTCGACTTTTTTTTTTTTTTT-3′ and 5′-GGTCGACCTGCAGGCATGCAAGCTT-3′) were annealed and labeled with 32P-α-dATP using polynucleotide kinase. Radiolabeled oligonucleotides were purified and stored at 4°C in sterile water. 293T cells transfected with SFB-tagged wild-type Artemis and mutants of Artemis were lysed in lysis buffer (NETN + protease inhibitor). The supernatant was incubated with streptavidin Sepharose beads on a shaker for 1 h in a cold room after centrifugation. Beads were washed three times with NETN buffer. Bound proteins were eluted with 2 mg/mL biotin. Next, eluates were incubated with S beads on a shaker for 1 h in a cold room. The wild-type Artemis and mutant of Artemis proteins were incubated with buffer containing 25 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 1 mM DTT, 50 ng/μL BSA, ∼0.15–0.25 pmol of 32P-labeled DNA, 0.25 mM ATP, and 75 mM KCl. Reactions were carried out for 10 min at 37°C before being stopped with 5 μL of formamide loading buffer (96% [v/v] formamide, 10 mM EDTA at pH 8.0, 0.2% Orange-G dye), incubated for 1 min at 100°C, and cooled rapidly on ice. DNA fragments were resolved at 300 V on 0.5-mm thick, 15% PAGE minigels containing 1× TBE buffer and 7 M urea. Resolved gels were fixed in 10% (v/v) methanol and 10% (v/v) acetic acid before being dried and exposed to film.
Supplementary Material
Acknowledgments
We thank the Cytometry and Cellular Imaging Core Facility of the M.D. Anderson Cancer Center for their technical assistance. We thank all members of the Chen laboratory for their advice and technical assistance. This work was supported in part by grants from the National Institutes of Health to J.C. (CA089239, CA092312, and CA100109). J.C. is also a recipient of the Basser Team Science Award and an Era of Hope Scholar award from the Department of Defense (W81XWH-05-1-0470) and is a member of the M.D. Anderson Cancer Center (CA016672).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.252478.114.
References
- Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N, et al. 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 376: 245–251. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. [DOI] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. 2013. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153: 1266–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattell E, Sengerova B, McHugh PJ. 2010. The SNM1/Pso2 family of ICL repair nucleases: from yeast to man. Environ Mol Mutagen 51: 635–645. [DOI] [PubMed] [Google Scholar]
- Chan SL, Mok T. 2010. PARP inhibition in BRCA-mutated breast and ovarian cancers. Lancet 376: 211–213. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49: 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, et al. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339: 711–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkáč J, Cook MA, Rosebrock AP, Munro M, Canny MD, et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1–RIF1 and BRCA1–CtIP controls DNA repair pathway choice. Mol Cell 49: 872–883. [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J. 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J Biol Chem 288: 11135–11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Cho YW, Kim JE, Ge K, Chen J. 2009. Accumulation of Pax2 transactivation domain interaction protein (PTIP) at sites of DNA breaks via RNF8-dependent pathway is required for cell survival after DNA damage. J Biol Chem 284: 7284–7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Kim JE, Leung CC, Glover JN, Chen J. 2010. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell 37: 438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, et al. 2006. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J 25: 3880–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, Drost R, Wientjens E, Ji J, Aly A, et al. (2013). Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov 3: 68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639. [DOI] [PubMed] [Google Scholar]
- Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, et al. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105: 177–186. [DOI] [PubMed] [Google Scholar]
- Munoz IM, Jowsey PA, Toth R, Rouse J. 2007. Phospho-epitope binding by the BRCT domains of hPTIP controls multiple aspects of the cellular response to DNA damage. Nucleic Acids Res 35: 5312–5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR. 2007. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 13: 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. 2010. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376: 235–244. [DOI] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177: 219–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong Z, Chen J. 2011. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J Cell Biol 193: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Chen J, Gong Z. 2013. TopBP1 controls BLM protein level to maintain genome stability. Mol Cell 52: 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642. [DOI] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339: 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.